Abstract
Because the human skin microbiota may play roles in the causation or modification of skin diseases, we sought to provide initial quantitative analysis from different cutaneous locations. We developed quantitative PCRs to enumerate the total bacterial and fungal populations, as well as the most common bacterial and fungal genera present in six locales, in eight healthy subjects. We used a set of primers and TaqMan MGB probes based on the bacterial 16S rRNA and fungal internally transcribed spacer region, as well as bacterial genus-specific probes for Propionibacterium, Corynebacterium, Streptococcus, and Staphylococcus and a fungal genus-specific probe for Malassezia. The extent of human DNA contamination of the specimen was determined by quantitating the human housekeeping GAPDH gene. The highest level of 16S rRNA copies of bacteria was present in the axilla (4.44 ± 0.18 log10 copies/μl [mean ± standard error of the mean]), with normalization based on GAPDH levels, but the other five locations were similar to one another (range, 2.48 to 2.89 log10 copies/μl). There was strong symmetry between the left and right sides. The four bacterial genera accounted for 31% to 59% of total bacteria, with the highest percent composition in the axilla and the lowest in the forearm. Streptococcus was the most common genus present on the forehead and behind the ear. Corynebacterium spp. were predominant in the axilla. Fungal levels were 1 to 2 log10 lower than for bacteria, with Malassezia spp. accounting for the majority of fungal gene copies. These results provide the first quantitation of the site and host specificities of major bacterial and fungal populations in human skin and present simple methods for their assessment in studies of disease.
Human skin harbors a diverse group of microorganisms that form complex communities and occupy specific niches and microenvironments (5, 9, 12, 14, 15). While most organisms colonizing the human body may be beneficial for health (4, 9, 18), some relationships with the host can change from commensal to pathogenic for reasons that are poorly understood (1). The global composition and presence of specific organisms also is relevant to the biological effects of the skin microbiota. The human cutaneous surface includes regions with diverse pH, temperature, moisture, and sebum content (14, 17), and skin structures such as hair follicles, sebaceous, eccrine, and apocrine glands comprise subhabitats that may be associated with their own unique microbiota (20).
Until recently, our knowledge of the bacterial biota in human skin has been based mostly on cultivation studies, which are insufficient because many organisms cannot be cultured (6). The direct PCR amplification and sequencing of bacterial genes encoding the small subunit rRNA (16S rRNA) or variable region fragments thereof has been a powerful method to analyze the enormous variation in the human microbiome (8, 24, 30), showing important differences among a number of cutaneous sites (5, 9, 12, 15). However, analysis of ribosomal genes (16S rRNA or intergenomic 18S and 23S rRNA) by using clone libraries or high-throughput sequencing may not allow for an accurate enumeration due to a variety of artifacts (19, 31). However, quantitative real-time PCR (qPCR) may be used as an alternative methodology (7, 23).
Based on our prior studies, which characterized the human forearm cutaneous microbiota (12, 13, 25, 26), we sought to quantify the human cutaneous microbiota from several body locations. The aim of the present study was to develop qPCRs to enumerate total bacterial and fungal populations, as well as to determine the most common bacterial and fungal genera in different locations on human skin.
MATERIALS AND METHODS
Subjects.
Specimens from skin were obtained from eight healthy adult subjects (four males and four females) from 11 body locations, including the forehead, left and right axillae, left and right inner elbows, left and right forearms, left and right forelegs, and behind the left and right ears. To maximize the continuity of our investigations, two of the eight subjects (subjects 1 and 3) had been examined in a prior study (12), but new samples were obtained for this study. From two of the subjects (subjects 1 and 2), a second set of samples was collected 1 month later at the same time of day (afternoon). The subjects were not provided with any specific instructions before the samples were taken. The mean age of the subjects was 38 years (range, 25 to 58); all were in good health and had not received antibiotics for at least 4 months. The study was approved by the NYU Institutional Review Board, and all subjects provided written informed consent.
Specimen processing.
Methods for specimen processing have been described elsewhere (12). In brief, a 2- by 2-cm area of the cutaneous surface at each of the 11 locations was sampled by swabbing the skin with a cotton pledget that had been soaked in sterile 0.15 M NaCl with 0.1% Tween 20 (Fisher Scientific, Fair Lawn, NJ). DNA was extracted from the swab suspensions in a PCR-free clean room by using the DNeasy blood and tissue kit (Qiagen, Chatsworth, CA); glass beads (0.5 to 1 mm) were added to the specimens and vortex mixed at maximum speed for 40 s, followed by DNA extraction, using the manufacturer's protocol for genomic DNA isolation from Gram-positive bacteria, and samples were eluted in 100 μl AE buffer (DNeasy Blood and Tissue kit; Qiagen). To eliminate bacterial or DNA contamination, lysozyme (Sigma-Aldrich, St. Louis, MO) was passed through a microcentrifuge filter (molecular mass threshold, 30,000 Da; Amicon, Bedford, MA) at 18,514 × g in a centrifuge (Eppendorf, Germany) for 20 min before adding to the enzymatic lysis buffer.
Quantitative PCR.
We used sets of primers and TaqMan MGB probes (Table 1), based on the bacterial 16S rRNA sequences (12), the fungal internally transcribed spacer (ITS) sequences (25; http://pmb.berkeley.edu/∼bruns/tour/primers.html), and the human glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) (28). In addition, we developed genus-specific approaches for Propionibacterium, Staphylococcus, Streptococcus, Corynebacterium, and Malesezzia, because these have been the most common genera identified in studies using universal primers (5, 9, 12, 15, 25, 26). PCRs were performed using 3.5 mM MgCl2, 0.4 ng/μl bovine serum albumin, 0.2 mM each deoxynucleoside triphosphate, 10 pmol of each primer, 5 pmol of each probe, 0.625 U Taq DNA polymerase (Qiagen), and 2 μl extracted DNA in a final 25-μl volume. PCR conditions were 5 min at 94°C and 45 cycles of 10 s at 94°C, 45 s at 54°C (for primers 8F/Eub361R) or 56°C (for primers Eub519R/U785R), and 60 s at 72°C. The assays were performed using a Rotor-Gene 6000 system (Corbett Life Science, Sydney, Australia), and a standard curve was constructed by using serial dilutions of cloned PCR products corresponding to each tested species. Each sample was tested at least twice, and the results were analyzed using the Rotor-Gene 3000 v.6.1.81 software.
TABLE 1.
Primers and probes used in this study
| Designation | Primer or probe | Target | Sequence (5′→3′) |
|---|---|---|---|
| 8F | Primer | Bacteria | AGAGTTTGATYMTGGCTCAG |
| Eub361R | Primer | Bacteria | CGYCCATTGBGBAADATTCC |
| Eub519F | Primer | Bacteria | CAGCAGCCGCGGTRATA |
| U785R | Primer | Bacteria | GGACTACCVGGGTATCTAAKCC |
| ITS1F | Primer | Fungi | CTYGGTCATTTAGAGGAAGTAA |
| ITS2 | Primer | Fungi | RCTGCGTTCTTCATCGWTG |
| Mal1F | Primer | Genus Malassezia | TCTTTGAACGCACCTTGC |
| Mal1R | Primer | Genus Malassezia | AHAGCAAATGACGTATCATG |
| GAPF | Primer | Human GAPDH | GGGCTCTCCAGAACATCATCC |
| GAPR | Primer | Human GAPDH | GTCCACCACTGACACGTTGG |
| Bacteria_P | Probe | Bacteria | TACGGGAGGCAGCAGT |
| Strep_P | Probe | Genus Streptococcus | AGATGGACCTGCGTTGT |
| Staph_P | Probe | Genus Staphylococcus | CTGTAACTGACGCTGATGTG |
| Prop_P | Probe | Genus Propionibacterium | CTTTCGATACGGGTTGACTT |
| Coryne_P | Probe | Genus Corynebacterium | ACAGYACTCHAGTHATGCCCGT |
| Fungal_P | Probe | Fungi | TCYGTAGGTGAACCTGCRG |
| Mala_P | Probe | Genus Malassezia | ATGCCTGTTTGWGTGC |
| GAP_P | Probe | Human GAPDH | CCTCTACTGGCGCTGCCAAGGCT |
Statistical analysis.
Statistical analysis was performed with the Paleontological Statistics Software Package for Education and Data Analysis (http://palaeo-electronica.org/2001_1/past/issue1_01.htm). All qPCR results are presented as the means of the log10 value ± the standard errors of the means (SEM). Statistically significant differences were determined by t tests and one-way analysis of variance (ANOVA) tests for independent samples. ANOVA tests also were conducted using the SAS 9.2 statistical package for Windows (SAS Institute Inc., Cary, NC) with the PROC GLM procedure. Total variations accounted for by the model and by site, subject, side, and time were based on estimates for the omega-square and semipartial omega-square values. Statistical significance was defined as a P value of <0.05. The linear regression method was used to assess the correlation between the proportions of bacteria and fungi from samples obtained from the subjects' left and right sides.
RESULTS
Validation of TaqMan MGB probes.
To examine the specificity of the probes, all probes first were validated through the Probe Match algorithm of the Ribosomal Database Project (RDP; http://www.rdp.cme.msu.edu/probematch). Subsequently, 16S rRNA gene PCR products from the most common skin bacteria genera (12) were used to evaluate the specificity of the probes. The products studied included members of the genus Propionibacterium as well as Streptococcus, Staphylococcus, Corynebacterium, Rothia, Gemella, Micrococcus, and Kocuria species (12). In each case, as expected, the genus-specific probes recognized the cloned DNA from the species within the same genus but not the other common skin genera (data not shown). Assessment of sensitivity was performed using 10-fold dilutions of the DNA templates used for the standard curves, corresponding to 3 × 101 to 3 × 107 16S rRNA gene copies per reaction mixture. The techniques could detect up to 107 competing organisms. The limit of detection for the genus-specific probes ranged from 101 to 102 16S rRNA gene copies per reaction mixture. All probes performed well, with correlation (R) values of >0.99 for the standard curve and PCR efficiencies of 80%.
Contamination by human DNA.
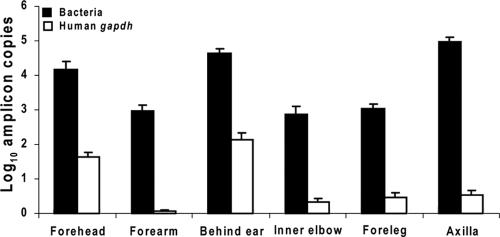
To assess the level of contamination of the skin swab material by human DNA, we performed qPCR for a human housekeeping gene (GAPDH) which is present in all nucleated cells of human origin (10). The GAPDH qPCR was performed in parallel with the bacterial qPCRs. In total, for each site, the log10 mean GAPDH amplicons per μl of sample ranged from 0.07 ± 0.03 (forearm) to 2.14 ± 0.20 (behind ear) (Fig. 1 and Table 2). In general, the standard deviations were low across the 8 to 16 specimens sampled per site, indicating that the variation was site specific, rather than individual specific or technique related. The highest GAPDH values were obtained in specimens from sebaceous sites on the head (behind the ear and on the forehead) and were significantly greater than from the other dry sites (elbow, forearm, and foreleg) or moist sites (axilla) (P < 0.001).
FIG. 1.
Representation of bacterial and human DNA in skin swab specimens. For each of six assayed locales, skin swab specimens from 8 (forehead) to 16 (all other locales) were obtained, and qPCR was performed to detect human GAPDH or 16S rRNA using universal bacterial primers. From each site, bacterial amplicons outnumbered human amplicons by >2 log10/μl.
TABLE 2.
Composition of human skin microbiota from six cutaneous locations
| Data treatment and body location | No. of samples | Mean log10 copies/μl ± SEM |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| All bacteria | Corynebacterium | Streptococcus | Staphylococcus | Propionibacterium | All fungi | Malassezia | Human GAPDH | ||
| Nonnormalized data | |||||||||
| Forehead | 8 | 4.16 ± 0.25 | 0.77 ± 0.50 | 3.32 ± 0.22 | 2.62 ± 0.23 | 2.65 ± 0.19 | 2.79 ± 0.22 | 2.71 ± 0.31 | 1.62 ± 0.16 |
| Forearm | 16 | 2.97 ± 0.17 | 0.06 ± 0.06 | 2.00 ± 0.13 | 1.84 ± 0.26 | 1.85 ± 0.22 | 1.70 ± 0.14 | 1.32 ± 0.22 | 0.07 ± 0.03 |
| Behind ear | 16 | 4.62 ± 0.14 | 0.60 ± 0.32 | 3.76 ± 0.17 | 3.57 ± 0.14 | 3.56 ± 0.14 | 2.78 ± 0.14 | 2.74 ± 0.14 | 2.14 ± 0.20 |
| Inner elbow | 16 | 2.88 ± 0.22 | 0.08 ± 0.06 | 1.92 ± 0.20 | 1.87 ± 0.31 | 1.79 ± 0.29 | 1.59 ± 0.13 | 1.21 ± 0.22 | 0.33 ± 0.10 |
| Foreleg | 16 | 3.02 ± 0.14 | 0.35 ± 0.13 | 1.97 ± 0.11 | 2.08 ± 0.23 | 2.00 ± 0.21 | 1.48 ± 0.07 | 1.06 ± 0.14 | 0.46 ± 0.13 |
| Axillaa | 10 | 4.98 ± 0.13 | 2.98 ± 0.68 | 4.16 ± 0.17 | 4.13 ± 0.14 | 3.95 ± 0.14 | 2.22 ± 0.24 | 2.11 ± 0.31 | 0.55 ± 0.13 |
| Normalized with respect to human gapdh | |||||||||
| Forehead | 8 | 2.54 ± 0.21 | −0.85 ± 0.53 | 1.70 ± 0.19 | 1.00 ± 0.20 | 1.02 ± 0.22 | 1.17 ± 0.20 | 1.09 ± 0.27 | |
| Forearm | 16 | 2.89 ± 0.17 | −0.02 ± 0.05 | 1.92 ± 0.13 | 1.77 ± 0.26 | 1.77 ± 0.22 | 1.62 ± 0.14 | 1.25 ± 0.22 | |
| Behind ear | 16 | 2.48 ± 0.19 | −1.54 ± 0.28 | 1.62 ± 0.19 | 1.43 ± 0.20 | 1.42 ± 0.18 | 0.64 ± 0.20 | 0.60 ± 0.22 | |
| Inner elbow | 16 | 2.56 ± 0.19 | −0.25 ± 0.08 | 1.59 ± 0.18 | 1.54 ± 0.26 | 1.46 ± 0.23 | 1.26 ± 0.10 | 0.88 ± 0.19 | |
| Foreleg | 16 | 2.56 ± 0.21 | −0.11 ± 0.20 | 1.51 ± 0.19 | 1.63 ± 0.31 | 1.54 ± 0.29 | 1.02 ± 0.13 | 0.60 ± 0.17 | |
| Axillaa | 10 | 4.44 ± 0.18 | 2.43 ± 0.75 | 3.62 ± 0.21 | 3.58 ± 0.20 | 3.40 ± 0.19 | 1.68 ± 0.32 | 1.57 ± 0.39 | |
Data were not available from three subjects.
b Data were normalized by dividing by the number of GAPDH amplicon copies.
Quantitative PCR detection of all bacteria and fungi in human skin samples.
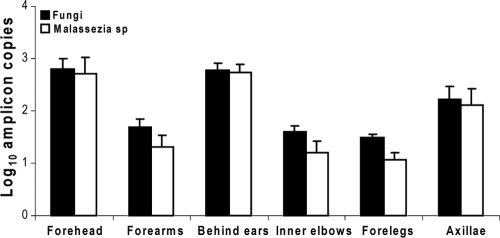
Real-time PCR analyses were performed to quantify all bacteria and fungi in the human skin samples obtained from 11 body sites (5 bilateral and 1 in the midline). All 82 samples studied from human skin showed 103 to 107 amplicons of bacterial 16S rRNA genes per extraction sample. The enumeration of the total bacteria, as well as four bacterial genera in the six cutaneous locales, is shown in Table 2 without or with normalization to GAPDH levels. Before normalization, the highest bacterial log10 levels were found in the axilla (4.98 ± 0.13 log10/μl), which were substantially greater than the levels observed in the sebaceous sites (4.16 to 4.62 log10/μl) (P = 0.03). Levels at dry sites were even lower (∼3 log10/μl) (P < 0.001). Using normalization, based on the number of copies of human GAPDH, the highest level of 16S bacterial rRNA again was found in the axilla (4.44 ± 0.18 log10/μl), and the level was >2 log10 higher than the levels for the other five body locations tested. The levels among the other five locales were similar to one another (range, 2.48 to 2.89 log10/μl). Before normalization, the range for fungal DNA was narrow (means, 1.48 to 2.79 log10/μl), with the highest levels found from the sebaceous sites on the head and with lower levels on the forelegs, forearms, and inner elbows; values for the forehead and behind the ears were virtually identical. After normalization to GAPDH, the highest level of fungi also was found in axillae (1.68 ± 0.32 log10/μl), with the lowest level found behind the ears (0.64 ± 0.20 log10/μl). In total, these results indicate substantial quantitative differences in overall bacterial and fungal populations that are site specific.
Representation of major genera in human cutaneous samples.
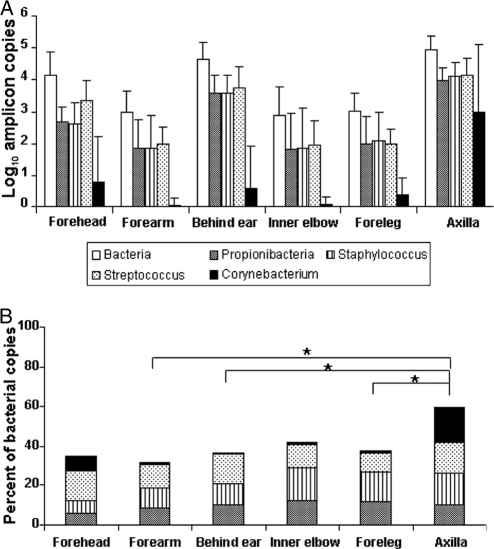
Results from our prior studies, based on analysis of ribosomal genes from clone libraries, provided evidence that four bacterial genera (Corynebacterium, Streptococcus, Staphylococcus, and Propionibacterium) and one fungal genus (Malassezia) were the most common genera on the human forearm (12, 25). Our present results show that the four bacterial genera represent 31.3 ± 9.3% (mean ± standard deviation [SD]) of the bacterial copies in the forearms (Fig. 2 A and B), confirming their dominance among the hundreds of genera identified (5, 12, 15). Corynebacterium spp. were prominently found on the axilla and forehead (Fig. 2B), but there was substantial variation between individuals (range, ≪1.0 to 5.08 log10/μl in the axilla and ≪1.0 to 3.86 log10/μl on the forehead). More consistently low numbers were detected in other body locales. The specimens from the axilla and from the head (behind the ear and the forehead) harbored more Streptococcus spp. than did other areas. In total, Streptococcus, Propionibacterium, and Staphylococcus populations were variable in the different locations but to a lesser extent than was Corynebacterium (Fig. 2B). The proportions of the four genera in the axilla in relation to the total counts were significantly higher than those on the forearm, behind the ear, or the foreleg (P < 0.01). Malassezia spp. was calculated to account for 53% to 80% of all fungi in the different body locations; the highest numbers (2.74 ± 0.14 log10/μl) and highest proportions (80%) were found behind the ear (Table 2 and Fig. 3).
FIG. 2.
Enumeration of major bacterial genera in human cutaneous samples at six locales. (A) Log10 amplification of total bacteria and four prominent cutaneous genera: Propionibacterium, Staphylococcus, Streptococcus, and Corynebacterium. The data were not normalized with respect to the human GAPDH gene. (B) Four bacterial genera as a proportion of total bacterial amplicons in six human cutaneous locales. An asterisk indicates that proportions are significantly (P < 0.01) different from other sites. For Corynebacterium, there was substantial interhost variation (from <1% to 45%).
FIG. 3.
Enumeration of fungal species in six cutaneous locales. From the eight tested subjects, a single specimen was obtained from the forehead and bilateral specimens were obtained from forearm, behind ear, inner elbow, and foreleg; samples from axillae were obtained for five subjects only. qPCR was performed using primers for universal fungal or Malassezia species. Bars indicate mean ± SD of the values across the subjects. The data were not normalized with respect to the human GAPDH gene.
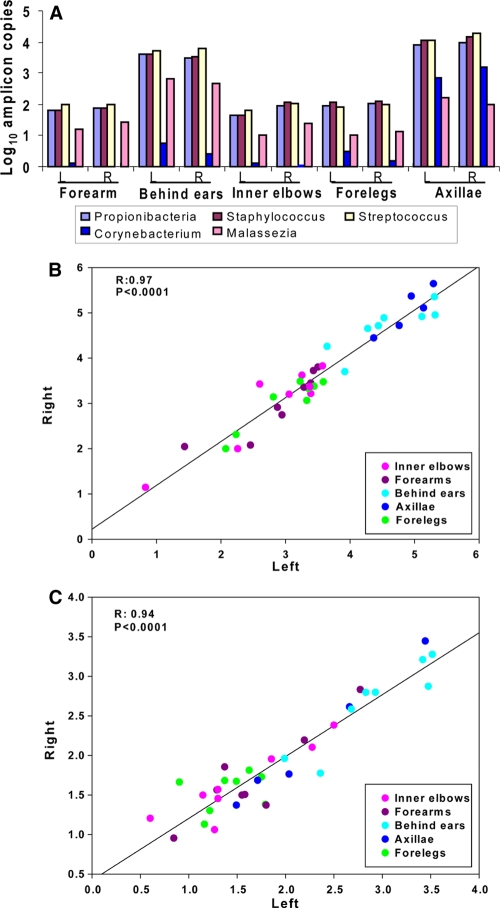
Comparison of lateral symmetries of the cutaneous human microbiota.
We studied the symmetry of the left and right sides by comparing the patterns of five genera in the five body locales for which bilateral sampling was done. There was pronounced left/right symmetry at all locations (r = 0.97 and 0.94 for bacteria and fungi, respectively) (Fig. 4).
FIG. 4.
Lateral symmetry of major microbial populations at five cutaneous locales in eight subjects, based on qPCR determination. (A) Genus-level qPCR results for five cutaneous locales. Values shown are mean log10/μl for eight subjects (five subjects for axilla samples). The genera include Propionibacterium, Staphylococcus, Streptococcus, Corynebacterium, and Malassezia. (B) Linear regression analysis of lateral symmetry of bacterial enumeration at five cutaneous locales, by individual study site. (C) An analysis similar to that in panel B, except universal fungal primers for qPCR were used.
Comparison of intra- and interpersonal patterns of four major cutaneous bacterial genera.
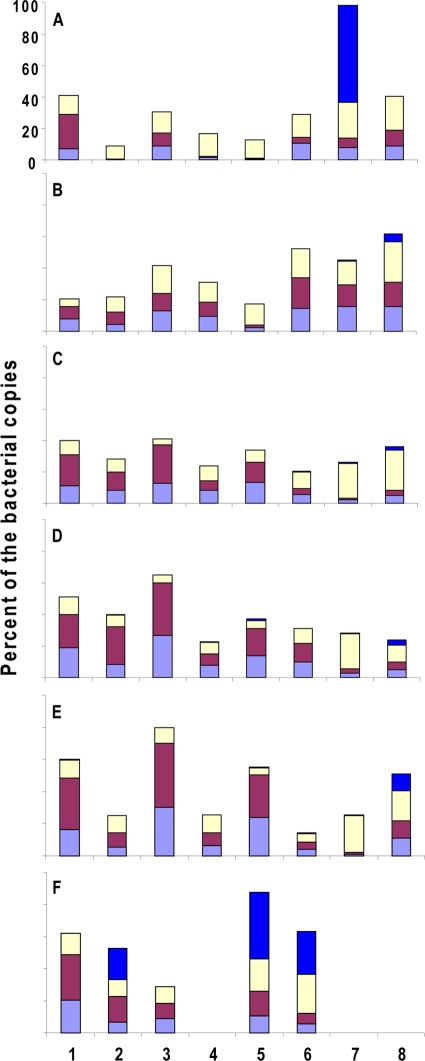
Figure 5 shows the proportions of four major bacterial genera in eight different subjects. Overall, the patterns from the six locales showed extensive intrahost variation (P < 0.001, one-way ANOVA test). The patterns in the same locales but from different individuals also differed substantially. Significant interindividual differences were observed in the samples from forearms (P = 0.004), inner elbows (P < 0.001), axilla (P = 0.003), and forelegs (P = 0.002). The composition patterns of the four bacterial genera were similar among different individuals (P > 0.05) in the samples from the head (behind the ear and the forehead). Analysis of variance also was performed across all of the specimens to estimate the effects of subject, site, and side on the overall variation (Table 3). For all four major bacterial genera, differences in site accounted for the greatest amount of variation, which was significant (P < 0.01) in each case. Although estimates were lower, the amount of variance attributable to between-subject differences also was significant for all genera except for Corynebacterium. Exploratory analyses also were conducted to assess pair-wise interactions between subjects, site, and time. The data indicated significant interactions between subjects and site for all four major bacterial populations (P < 0.01). This finding provides evidence that the relationship between subjects and the level of major bacterial populations varies by site.
FIG. 5.
Representation of four bacterial genera at six cutaneous sites in eight healthy subjects. Numbers 1 through 8 designate the enrolled subjects. (A) Forehead; (B) behind left and right ears; (C) left and right forearms; (D) left and right forelegs; (E) left and right inner elbows; (F) left and right axillae. For three subjects, no axillary specimens were included. The bacterial genera are color coded as for Fig. 4A.
TABLE 3.
Analysis of variance for the four major bacterial populations in the study subjects
| Source of variance |
Corynebacterium |
Propionibacterium |
Staphylococcus |
Streptococcus |
||||
|---|---|---|---|---|---|---|---|---|
| Estimate (95% CIc) | P value | Estimate (95% CIc) | P value | Estimate (95% CI) | P value | Estimate (95% CI) | P value | |
| Total variance explaineda | 45.0 (27.0-59.0) | 69.0 (57.0-77.0) | 68.0 (56.0-77.0) | 76.0 (0.7-0.8) | ||||
| Variance explained by: | ||||||||
| Subjects | 5.5 (0.0-16.7) | 0.05 | 18.8 (1.6-30.6) | <0.01 | 21.6 (3.5-33.7) | <0.01 | 7.5 (0.0-15.6) | <0.01 |
| Site | 41.4 (25.2-54.8) | <0.01 | 41.4 (23.8-53.7) | <0.01 | 36.6 (18.9-49.5) | <0.01 | 58.2 (42.9-67.6) | <0.01 |
| Side | 0.5 (0.0-6.2) | 0.57 | 0.3 (0.0-5.3) | 0.59 | 0.2 (0.0-6.0) | 0.48 | 0.0 (0.0-6.3) | 0.37 |
| Subsample with repeated measurementsb | ||||||||
| Total variance explained | 39.0 (6.0-63.0) | 78.0 (59.0-87.0) | 75.0 (52.0-85.0) | 74.0 (51.0-85.0) | ||||
| Variance explained by: | ||||||||
| Subjects | 11.8 (0.0-37.5) | 0.02 | 34.4 (7.8-55.9) | <0.01 | 14.9 (0.0-39.3) | <0.01 | 0.9 (0.0-0.13) | 0.96 |
| Site | 29.6 (4.5-53.9) | <0.01 | 25.7 (7.6-47.8) | <0.01 | 24.8 (0.5-47.3) | <0.01 | 0.0 (0.0-14.7) | 0.38 |
| Side | 2.1 (0.0-11.0) | 0.82 | 1.4 (0.0-20.9) | 0.11 | 0.6 (0.0-13.8) | 0.56 | 0.8 (0.0-10.0) | 0.76 |
| Time | 0.9 (0.0-18.6) | 0.45 | 6.0 (0.0-28.8) | 0.01 | 33.2 (7.1-55.1) | <0.01 | 76.6 (59.2-85.6) | <0.01 |
Absolute values(log10 copies/μl) for bacterial density used for this analysis.
Ratio of specific bacteria to total bacteria used for this analysis.
CI, confidence interval.
Fluctuation of the four bacterial genera in two subjects.
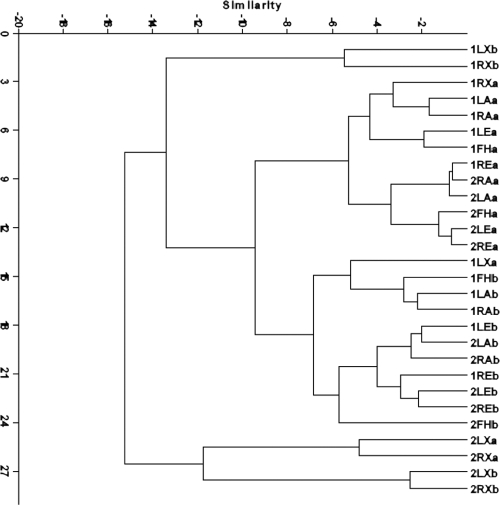
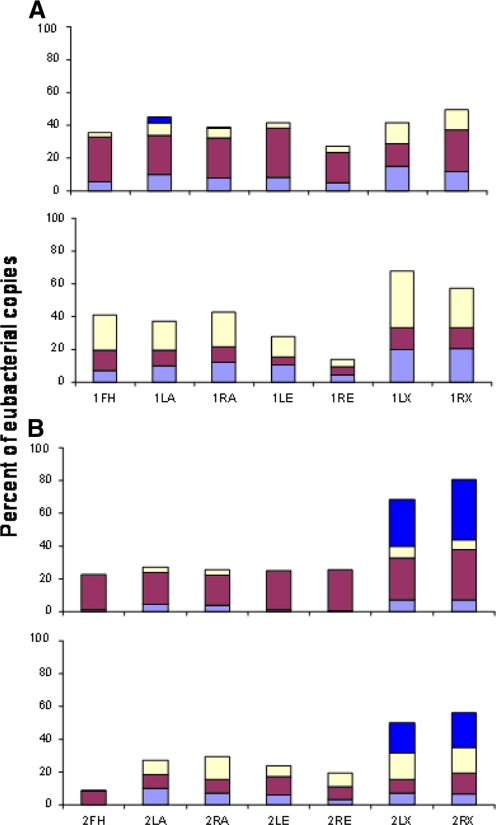
Skin samples were collected 1 month apart from two healthy subjects to examine variations in the four major bacterial genera (Fig. 6 and 7). The results of cluster analysis showed that samples from the same subject clustered closely, whereas the two time points were not clustered. Numbers of Streptococcus and Staphylococcus were significantly different for samples obtained 1 month apart, with fluctuating proportions of the two genera (P < 0.001 and P = 0.001, respectively). For Corynebacterium and Propionibacterium, time of sampling had little effect on the overall variation, whereas it was the dominant source of variation for Staphylococcus and Streptococcus. The amount of variation in the level of staphylococci and streptococci that is attributable to time (33.2% and 76.6% for staphylococci and streptococci, respectively) is greater than the variation due to subject, side, or site (all <25% and <1% for staphylococci and streptococci, respectively) (Table 3).
FIG. 6.
Clustered display of data from repeat sampling of four bacterial genera in two subjects. The distribution of four bacterial genera from two healthy subjects (subjects 1 and 2) at two time points is represented by a dendrogram based on UPGMA cluster analysis. The samples are coded as follows: 1, subject 1; 2, subject 2; a, first sampling; b, second sampling; FH, Forehead; LX, left axilla; RX, right axilla; LA, left forearm; RA, right forearm; LE, behind left ear; RE, behind right ear.
FIG. 7.
Distribution of four bacterial genera from two subjects at two time points. (A) Proportion of the total bacterial amplicons represented by the four bacterial genera from subject 1 at two time points. The bacterial genera are color coded as for Fig. 4A. The bars are coded as follows: FH, forehead; LA, left forearm; RA, right forearm; LE, left behind ear; RE, right behind ear; LX, left axilla; RX, right axilla. The data were not normalized with respect to the human GAPDH gene. (B) Results for subject 2, with color coding and abbreviations as for panel A.
DISCUSSION
To investigate the population structure of the human cutaneous microbiota, a set of primers and TaqMan MGB probes was adapted or designed, validated, and deployed to enumerate the predominant bacterial and fungal genera in several cutaneous locales from healthy human subjects. The results show that the qPCR assays are sensitive, rapid, and reproducible methods for the quantitation of the major microbial genera that populate human skin.
For accurate comparison of bacteria and fungi in different samples, the amount of total host DNA can provide a measure to facilitate standardization. Since the GAPDH gene is a conserved human housekeeping gene used to normalize gene expression data (2), a GAPDH qPCR was used to standardize the process of sampling. The relatively high numbers of 16S rRNA bacterial copies in relation to the copies of human GAPDH that we observed demonstrate that skin swabbing captures a population of DNA highly enriched for microbiota. The amount of interindividual variation at each specific locale was low, indicating the relative uniformity of the swabbing procedure. Our data suggest that standardization is site specific and differs for sebaceous locales in comparison to dry sites, which have smaller amounts of human DNA. Although further study is needed, site specificity may be presumed for now.
The results from prior studies (5, 9, 12, 13, 15, 25, 26), using analysis of ribosomal genes from clone libraries or high-throughput sequencing, provided evidence that four bacterial genera, Corynebacterium, Streptococcus, Staphylococcus, and Propionibacterium and one fungal genus, Malassezia, were the most common in the human forearm. Our observation that the total for the four bacterial genera was calculated to represent 31% to 59% of all the bacterial 16S rRNA copies in each of the different locales provides further confirmation of these four conserved major populations. That Malassezia spp. represented 53% to 80% of all the fungal RNA amplicons is consistent with our prior studies of the forearm using the clone library method (25) and provides further evidence that these organisms represent the most frequent fungal genus at the tested locales.
Many culture-based studies have reported that the composition of the normal bacterial biota varies according to body location (11, 21, 22, 29). Our results demonstrated that the axilla harbored the highest density of bacterial copies and the highest percentage of the four most common bacterial genera. Although about 1% of the total bacterial population present in all sites of all subjects was Corynebacterium spp., this genus was most highly represented in the axilla, whereas Streptococci spp. were present at higher concentrations than the other three bacterial genera on the forehead and behind the ears. Such data extend our knowledge of the site specificity of the distribution of bacterial species in human skin and point to a means for further genus-specific explorations.
To examine changes in the numbers of the four bacterial genera in the skin samples over time, we collected samples from two of the subjects at intervals 1 month apart as a pilot study. The results showed that the total levels of the four common major genera of the normal human skin biota (3, 11) did not change greatly, but that their proportions did (Fig. 6 and 7). The substantial changes in the proportions of the genera Streptococcus and Staphylococcus over time suggest that these might be highly dynamic (Fig. 6 and 7) and should be followed in larger studies. The high correlations observed across sides for each site show that the human skin microbiota has a high level of left/right symmetry, as expected (9, 12, 25); the high correlations also help validate our qPCR findings. Over time, as indicated by the resampling, the host-specific conservation persists, but without a strong relationship to the prior sample from the same site. These observations indicate that the major bacterial biota is dynamic (primarily reflecting the Streptococcus and Staphylococcus populations) over the 1-month period sampled and that the changes influenced both sides similarly.
In conclusion, we developed quantitative PCR methods to investigate the distributions of the four most common bacterial genera and one fungal genus in different human cutaneous sites. The data reported here extend previous observations (5, 9, 12, 13, 15, 25) and provide further evidence that the distributions of the major bacterial and fungal genera are both site and host specific. As the Human Microbiome Project (16, 27) progresses, quantitative PCR will enable more-detailed analyses of target taxons of the human cutaneous microbiota, complementing high-throughput sequencing.
Acknowledgments
This work was supported in part by the Michael Saperstein Medical Scholars Fund, the Diane Belfer Program in Human Microbial Ecology, and UH2-AR057506 from the National Institutes of Health.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Avila, M., D. M. Ojcius, and O. Yilmaz. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, R. D., D. W. Harmer, R. A. Coleman, and B. J. Clark. 2005. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics 21:389-395. [DOI] [PubMed] [Google Scholar]
- 3.Chiller, K., B. A. Selkin, and G. J. Murakawa. 2001. Skin microflora and bacterial infections of the skin. J. Invest. Dermatol. Symp. Proc. 6:170-174. [DOI] [PubMed] [Google Scholar]
- 4.Cogen, A. L., V. Nizet, and R. L. Gallo. 2008. Skin microbiota: a source of disease or defence? Br. J. Dermatol. 158:442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello, E., and R. Knight. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen, L., M. McFall-Ngai, and D. A. Relman. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fierer, N., M. Hamady, C. L. Lauber, and R. Knight. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994-17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foldager, C. B., S. Munir, M. Ulrik-Vinther, K. Søballe, C. Bünger, and M. Lind. 2009. Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol. Biol. 10:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredricks, D. N. 2001. Microbial ecology of human skin in health and disease. J. Invest. Dermatol. Symp. Proc. 6:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Gao, Z., C. H. Tseng, Z. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U. S. A. 104:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, Z., C. H. Tseng, B. Strober, Z. Pei, and M. J. Blaser. 2008. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One 3:e2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grice, E. A., H. H. Kong, G. Renaud, A. C. Young, G. G. Bouffard, R. W. Blakesley, T. G. Wolfsberg, M. L. Turner, and J. A. Segre. 2008. A diversity profile of the human skin microbiota. Genome Res. 18:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grice, E. A., H. H. Kong, S. Conlan, C. B. Deming, J. Davis, A. C. Young, G. G. Bouffard, R. W. Blakesley, P. R. Murray, E. D. Green, M. L. Turner, and J. A. Segre. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamady, M., and R. Knight. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland, K. T., and R. A. Bojar. 2002. Cosmetics: what is their influence on the skin microflora? Am. J. Clin. Dermatol. 3:445-449. [DOI] [PubMed] [Google Scholar]
- 18.Hornef, M. W., M. J. Wick, M. Rhen, and S. Normark. 2002. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 3:1033-1040. [DOI] [PubMed] [Google Scholar]
- 19.Hurd, P. J., and C. J. Nelson. 2009. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief. Funct. Genomic Proteomic 8:174-183. [DOI] [PubMed] [Google Scholar]
- 20.Kearney, J. N., D. Harnby, G. Gowland, and K. T. Holland. 1984. The follicular distribution and abundance of resident bacteria on human skin. J. Gen. Microbiol. 130:797-801. [DOI] [PubMed] [Google Scholar]
- 21.Leyden, J. J., K. J. McGinley, E. Holzle, J. N. Labows, and A. M. Kligman. 1981. The microbiology of the human axilla and its relationship to axillary odour. J. Invest. Dermatol. 77:413-416. [DOI] [PubMed] [Google Scholar]
- 22.Leyden, J. J., K. J. McGinley, K. M. Nordstrom, and G. F. Webster. 1987. Skin microflora. J. Invest. Dermatol. 88:65s-72s. [DOI] [PubMed] [Google Scholar]
- 23.Nasidze, I., J. Li, D. Quinque, K. Tang, and M. Stoneking. 2009. Global diversity in the human salivary microbiome. Genome Res. 19:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakley, B. B., T. L. Fiedler, J. M. Marrazzo, and D. N. Fredricks. 2008. The diversity of human vaginal bacterial communities and their association with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulino, L., C. H. Tseng, B. Strober, and M. J. Blaser. 2006. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 44:2933-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulino, L., C. H. Tseng, and M. J. Blaser. 2008. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 8:460-471. [DOI] [PubMed] [Google Scholar]
- 27.Peterson, J., S. Garges, M. Giovanni, P. McInnes, L. Wang, J. A. Schloss, V. Bonazzi, J. E. McEwen, K. A. Wetterstrand, C. Deal, C. C. Baker, V. Di Francesco, T. K. Howcroft, R. W. Karp, R. D. Lunsford, C. R. Wellington, T. Belachew, M. Wright, C. Giblin, H. David, M. Mills, R. Salomon, C. Mullins, B. Akolkar, L. Begg, C. Davis, L. Grandison, M. Humble, J. Khalsa, A. R. Little, H. Peavy, C. Pontzer, M. Portnoy, M. H. Sayre, P. Starke-Reed, S. Zakhari, J. Read, B. Watson, and M. Guyer. 2009. The NIH Human Microbiome Project. Genome Res. 19:2317-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regis, S., S. Grossi, S. Lualdi, R. Biancheri, and M. Filocamo. 2005. Diagnosis of Pelizaeus-Merzbacher disease: detection of proteolipid protein gene copy number by real-time PCR. Neurogenetics 6:73-78. [DOI] [PubMed] [Google Scholar]
- 29.Roth, R. R., and W. D. James. 1988. Microbial ecology of the skin. Annu. Rev. Microbiol. 42:441-464. [DOI] [PubMed] [Google Scholar]
- 30.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight, and J. I. Gordon. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, H., J. K. DiBaise, A. Zuccolo, D. Kudrna, M. Braidotti, Y. Yeisoo, P. Parameswaran, M. D. Crowell, R. Wing, B. E. Rittmann, and R. Krajmalnik-Brown. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]