Abstract
Five Mycobacterium tuberculosis isolates were obtained from three body sites from a Dutch patient. The isolates displayed a single genotype by 24-locus MIRU-VNTR typing (except for a single locus not amplified from one isolate) but were differentiated by small variations in IS6110 fingerprints, spoligotypes, 6 hypervariable MIRU-VNTR loci, and/or DiversiLab profiles, revealing patterns of microevolution in a clonal infection.
CASE REPORT
A 56-year-old woman born in the Netherlands, a low-tuberculosis (TB)-incidence country, presented with diffuse nodular abnormalities on her chest X-ray. She had no history of TB. The tuberculin skin test result was 0 mm. Bronchoscopy was performed, and sarcoidosis was diagnosed. Treatment of sarcoidosis was started and was given for a year and a half. Five years later, she returned with an inflammatory finger. Under suspicion of relapse of her sarcoidosis, treatment with prednisone was started. At the start of treatment with prednisone, a urine sample (23 May 1986) was collected. Due to deterioration of the patient's condition, 2 months later, 2 more urine samples were collected on the same day (30 July 1986). The following day, samples from sputum (31 July 1986) and pus from her finger (31 July 1986) were also collected. The isolates were cultured on Lowenstein-Jensen medium after routine decontamination with 4% Na-OH. Cultures of the sputum, urine, and pus were all positive for Mycobacterium tuberculosis. At that time, antituberculosis treatment with isoniazide (INH), rifampin, and pyrazinamide was started, after a 4-month period of prednisone treatment. All five isolates were sensitive to INH (MIC, 0.2 μg/ml), ethambutol (MIC, 4 μg/ml), and rifampin (MIC, 0.5 μg/ml); the result for susceptibility to pyrazinamide was not available. Treatment was ended successfully after 12 months.
For IS6110 restriction fragment length polymorphism (RFLP) and DiversiLab analysis, DNA was extracted and purified, respectively, from M. tuberculosis cultures on Middlebrook 7H10 slants, using proteinase K and NaCl-cetyltrimethylammonium bromide (24). IS6110-RFLP was performed according to the standardized protocol recommended by van Embden et al. (24). DiversiLab analysis, a fingerprinting method based on repetitive sequence-based PCR (rep-PCR), was used as described by Cangelosis et al. (4) and following the manufacturer's instructions. Analysis of the isolate patterns was performed with the Web-based DiversiLab system software (version 2.1.66), using a virtual gel image and corresponding dendrogram.
For spoligotyping and mycobacterial interspersed repetitive unit-variable number of tandem repeats (MIRU-VNTR) typing, DNA was extracted by suspending approximately 10 mg of wet bacterial cells in 100 μl of sterile water and subsequently heating at 100°C for 30 min to inactivate and lyse the cells. PCR-based spoligotyping targets polymorphism of various spacer sequences present in the direct repeat (DR) region by a reverse dot blot procedure. It was performed as described by van der Zanden et al. (23). MIRU-VNTR typing relies on PCR amplification of multiple loci containing tandemly repeated sequences using primers specific for the flanking regions (6, 9, 17, 20, 21). Standard 24-locus-based MIRU-VNTR typing and 6-hypervariable-locus-based typing were performed as described previously (17, 19) using a quality-controlled commercial service (Genoscreen, Lille, France).
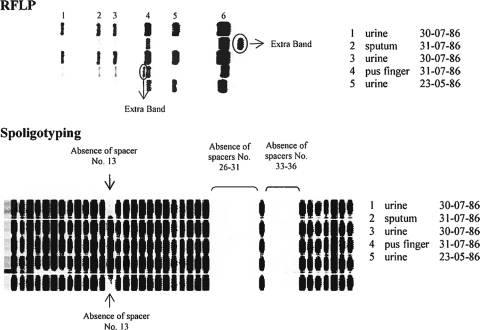
All five isolates showed identical patterns by standard MIRU-VNTR typing, parsimoniously accounting the missing data in locus 154 (MIRU 02) of isolate 1 as noninformative (Table 1). Submission of this 24-locus-based genotype to the MIRU-VNTRPlus database according to the strategy described by Allix-Beguec et al. (1) predicted that the corresponding genotype belongs to the Haarlem family, a member of the Euro-American superlineage. This was fully supported by the typical signatures of the two variant spoligotypes detected among the five isolates (3) (Fig. 1).
TABLE 1.
MIRU typing for five isolates using 24 locia
| Isolate | MIRU-VNTR allele in locus: |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 154 | 424 | 577 | 580 | 802 | 960 | 1644 | 1955 | 2059 | 2163b | 2165 | 2347 | 2401 | 2461 | 2531 | 2687 | 2996 | 3007 | 3171 | 3192 | 3690 | 4052 | 4156 | 4348 | |
| 1 | NA | 2 | 3 | 2 | 2 | 5 | 2 | 3 | 2 | 5 | 3 | 4 | 4 | 2 | 5 | 1 | 5 | 3 | 2 | 3 | 3 | 5 | 3 | 2 |
| 2 | 2 | 2 | 3 | 2 | 2 | 5 | 2 | 3 | 2 | 5 | 3 | 4 | 4 | 2 | 5 | 1 | 5 | 3 | 2 | 3 | 3 | 5 | 3 | 2 |
| 3 | 2 | 2 | 3 | 2 | 2 | 5 | 2 | 3 | 2 | 5 | 3 | 4 | 4 | 2 | 5 | 1 | 5 | 3 | 2 | 3 | 3 | 5 | 3 | 2 |
| 4 | 2 | 2 | 3 | 2 | 2 | 5 | 2 | 3 | 2 | 5 | 3 | 4 | 4 | 2 | 5 | 1 | 5 | 3 | 2 | 3 | 3 | 5 | 3 | 2 |
| 5 | 2 | 2 | 3 | 2 | 2 | 5 | 2 | 3 | 2 | 5 | 3 | 4 | 4 | 2 | 5 | 1 | 5 | 3 | 2 | 3 | 3 | 5 | 3 | 2 |
Correspondences with body sites and dates of isolation are given in Fig. 1. MIRU-VNTR markers are named according to a standardized designation (i.e., position on H37Rv chromosome). NA, nonamplified marker after one multiplex and two simplex PCR rounds.
FIG. 1.
RFLP and spoligotyping patterns. Correspondences with isolate identification numbers, body sites, and dates of isolation are indicated.
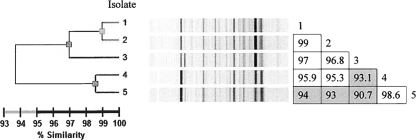
In contrast, small variations were detected among isolates with each of the four other genotyping methods. Three different patterns were obtained by using 6 hypervariable MIRU-VNTR loci, mutually differing by single-locus variations (SLVs) (Table 2). Urine isolates 1 and 3 showed different alleles at loci 3232 and 3336, respectively, compared to the genotype shared by the other three isolates. Three different IS6110 RFLP patterns differing by one-band changes were detected. Sputum (isolate 2) and pus (isolate 4) isolates, respectively, showed an additional band above or at an intermediate position compared to the 6 bands shared with the three urine isolates. Two spoligotypes were observed for the five isolates, differing in the presence of spacer 13 and distinguishing sputum (isolate 2), pus (isolate 4), and the first urine isolate (isolate 3) from urine isolates 1 and 5. Finally, three different patterns sharing more than 93% similarity were obtained by using the DiversiLab typing system (Fig. 2). Isolate 1 (urine) and isolate 2 (sputum) were indistinguishable, since they were above the 99% similarity threshold. Likewise, isolate 4 (pus) and isolate 5 (urine) displayed other patterns that were considered the same, since they were above 98% similarity. Isolate 3 (urine) displayed a unique pattern, albeit closely related to those of the two others.
TABLE 2.
MIRU-VNTR typing for five isolates using six hypervariable locia
| Isolate | MIRU-VNTR allele in locus: |
|||||
|---|---|---|---|---|---|---|
| 1982 | 2163a | 3232 | 3336 | 3820 | 4120 | |
| 1 | 4 | 7 | 9 | 7 | 3 | 5 |
| 2 | 4 | 7 | 7 | 7 | 3 | 5 |
| 3 | 4 | 7 | 7 | 1 | 3 | 5 |
| 4 | 4 | 7 | 7 | 7 | 3 | 5 |
| 5 | 4 | 7 | 7 | 7 | 3 | 5 |
Correspondences with body sites and dates of isolation are given in Fig. 1. MIRU-VNTR markers are named according to a standardized designation (i.e., position on M. tuberculosis H37Rv chromosome).
FIG. 2.
Dendrogram and similarity matrix for five M. tuberculosis isolates analyzed using the DiversiLab system. Correspondences with body sites and dates of isolation are given in Fig. 1.
Despite these variations, both the above genotyping results and the TB patient's history strongly indicate that the five isolates are not independent but represent the progeny of a single original strain and infection. The patient was born in the Netherlands, a country with one of the lowest TB incidences worldwide, the isolates had a Haarlem genotype (as seen by 24-locus based MIRU-VNTR typing and spoligotyping; see above) typically associated with this geographic region, and the patient had no known high risk of TB exposure. These features render very unlikely the scenario of mixed or superinfection by two different strains, typically seen with various frequencies in high-incidence or high-risk settings (2, 12, 14, 16, 18). Moreover, the 24 standard MIRU-VNTR loci were identical among the 5 isolates (accounting for missing data in one locus; see above). In line with the well-documented clonal stability of these markers, complete conservation of these standard MIRU-VNTR loci is the most likely situation among serial isolates from a single infection (1, 11, 13, 19). Changes in one band or one spacer and variations limited to an SLV are hallmarks of genetic drift in clonal subpopulations, as seen in (hypervariable) MIRU-VNTR loci, IS6110 RFLP, and spoligotyping, respectively, (1, 5, 10, 19). Likewise, rep-PCR patterns were highly similar if not identical in some cases. Altogether, these data thus strongly support that the observed changes result from microevolution in a clonal population of M. tuberculosis.
The relative extent of such microevolution is unprecedented. Except for the 24 standard MIRU-VNTR loci, the combination of the other markers (even without including rep-PCR results) systematically defined a unique clonal variant for each of the five isolates obtained from this single patient, regardless of body site origins. This extent may result from the probably long course between initial infection and TB diagnosis in the present case, leading to some differentiation in the clonal population. Infection with two probable clonal variants compartmentalized in two different body sites has been described (7). Here, however, we report microevolution by an unusual extent of distinct clonal variants among five isolates from three different body sites and one same site/fluid sample (urine) collected at the same day or at a 2-month interval, as monitored by using five independent genotyping methods. As suggested for cases of pulmonary TB with mixed infection (14), this might reflect some compartmentalized infection even at the suborgan level, the lesions of which could then be stochastically sampled, leading to isolation of different variants.
It is also noteworthy that the changes in the different markers were generally not associated. For instance, spoligotype changes affected two isolates that were different from those affected by RFLP changes. Likewise, the SLV in hypervariable MIRU-VNTR locus 3336 occurred in isolate 3, which was affected neither by RFLP nor by spoligotype change. This lack of association reveals stochastic evolution of independent and relatively high-turnover markers in the short term. Because of this stochastic nature, no general conclusion on the relative evolutionary rates or discriminatory power of the markers can be drawn from observations on a limited number of isolates. Nonetheless, such stochastic evolution can explain some degree of discordance between results of different genotyping methods among very closely related strains over short evolutionary distances (8). Over longer genetic distances, in populations of more distantly related strains, a higher correlation is apparent as a reflection of the clonal evolution of M. tuberculosis (15, 22).
In conclusion, this report constitutes additional but puzzling evidence against the dogma of a systematically homogenous within-host population of M. tuberculosis. Besides mixed infections, previous studies have shown the existence of clonal subpopulations in some patients with a single infection by a single strain, as reflected by small genotypic heterogeneity detected in the isolate mycobacterial population (e.g., see references 5, 10, 13, 14, and 16). In almost all cases, observations were limited to two or three isolates from the same body site (i.e., pulmonary), and genotypic variations were limited to small changes seen in a single genotyping method. Our data, based on five isolates from three different body sites of a same TB patient, as monitored by using five independent genotyping methods, suggest that microevolution might be of a greater extent than previously assumed, at least in some cases. In the present case, this phenomenon was probably driven by genetic drift favored both by a long disease incubation period leading to a large infectious population size and random sampling/compartmentalization effects linked to extrapulmonary dissemination.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Allix-Beguec, C., M. Fauville-Dufaux, and P. Supply. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden, C. R., G. P. Morlock, C. L. Woodley, K. R. Johnson, A. C. Colombel, M. D. Cave, Z. Yang, S. E. Valway, I. M. Onorato, and J. T. Crawford. 2001. Simultaneous infection with multiple strains of Mycobacterium tuberculosis strains. Clin. Infect. Dis. 33:e42-e47. [DOI] [PubMed] [Google Scholar]
- 3.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cangelosi, G. A., R. J. Freeman, K. N. Lewis, D. Livingston-Rosanoff, K. S. Shah, S. J. Milan, and S. V. Goldberg. 2004. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J. Clin. Microbiol. 42:2685-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 6.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144(Pt. 5):1189-1196. [DOI] [PubMed] [Google Scholar]
- 7.Garcia de Viedma, D., M. Marin, S. Andres, G. Lorenzo, M. J. Ruiz-Serrano, and E. Bouza. 2006. Complex clonal features in an Mycobacterium tuberculosis infection in a two-year-old child. Pediatr. Infect. Dis. J. 25:457-459. [DOI] [PubMed] [Google Scholar]
- 8.Hanekom, M., G. D. van der Spuy, N. C. Gey van Pittius, C. R. McEvoy, K. G. Hoek, S. L. Ndabambi, A. M. Jordaan, T. C. Victor, P. D. van Helden, and R. M. Warren. 2008. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J. Clin. Microbiol. 46:3338-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis strains molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemann, S., E. Richter, and S. Rusch-Gerdes. 1999. Stability of IS6110 restriction fragment length polymorphism patterns of multidrug-resistant Mycobacterium tuberculosis strains. J. Clin. Microbiol. 37:3078; author reply, 3078-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oelemann, M. C., R. Diel, V. Vatin, W. Haas, S. Rusch-Gerdes, C. Locht, S. Niemann, and P. Supply. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45:691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson, M., N. M. Carroll, E. Engelke, G. D. Van Der Spuy, F. Salker, Z. Munch, R. P. Gie, R. M. Warren, N. Beyers, and P. D. Van Helden. 2002. Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J. Clin. Microbiol. 40:2750-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shamputa, I. C., L. Jugheli, N. Sadradze, E. Willery, F. Portaels, P. Supply, and L. Rigouts. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamputa, I. C., J. Lee, C. Allix-Beguec, E. J. Cho, J. I. Lee, V. Rajan, E. G. Lee, J. H. Min, M. W. Carroll, L. C. Goldfeder, J. H. Kim, H. S. Kang, S. Hwang, S. Y. Eum, S. K. Park, H. Lee, P. Supply, S. N. Cho, L. E. Via, and C. E. Barry III. 16 December 2009. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary tuberculosis hospital in South Korea. J. Clin. Microbiol. doi: 10.1128/JCM.02167-09. [DOI] [PMC free article] [PubMed]
- 16.Shamputa, I. C., L. Rigouts, L. A. Eyongeta, N. A. El Aila, A. van Deun, A. H. Salim, E. Willery, C. Locht, P. Supply, and F. Portaels. 2004. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J. Clin. Microbiol. 42:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis strains complex bacteria using novel VNTR-PCR targets. Microbiology 148(Pt. 2):519-528. [DOI] [PubMed] [Google Scholar]
- 18.Sonnenberg, P., J. Murray, J. R. Glynn, S. Shearer, B. Kambashi, and P. Godfrey-Faussett. 2001. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet 358:1687-1693. [DOI] [PubMed] [Google Scholar]
- 19.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 21.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 22.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. Van Der Spuy, L. A. Lewis, M. Tibayrenc, P. D. Van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 23.van der Zanden, A. G., K. Kremer, L. M. Schouls, K. Caimi, A. Cataldi, A. Hulleman, N. J. Nagelkerke, and D. van Soolingen. 2002. Improvement of differentiation and interpretability of spoligotyping for Mycobacterium tuberculosis complex isolates by introduction of new spacer oligonucleotides. J. Clin. Microbiol. 40:4628-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]