Abstract
Salmonella enterica serovar Typhi is highly homogeneous. Single nucleotide polymorphisms (SNPs) have been shown to be valuable markers for molecular typing of S. enterica serovar Typhi. Here, we used a hairpin primer real-time PCR assay for SNP typing of S. enterica serovar Typhi isolates. Forty-two SNPs were selected from a comparison of 19 published S. enterica serovar Typhi genomes and sequences from other studies. The SNPs were used to type 71 global S. enterica serovar Typhi isolates and differentiated these S. enterica serovar Typhi isolates and the 19 genome sequenced strains into 25 SNP profiles. Phylogenetic analysis revealed that these SNP profiles were grouped into six major clusters. These clusters can be identified by using five SNPs, while the full differentiation of the 25 SNP profiles requires a minimum of 24 SNPs. This real-time PCR-based SNP typing method will be useful for global epidemiological analysis.
Salmonella enterica serovar Typhi is highly homogeneous (10, 17, 18). The lack of genetic diversity is a major challenge to the development of suitable typing methods to differentiate S. enterica serovar Typhi isolates for both phylogenetic and epidemiological purposes. Single nucleotide polymorphisms (SNPs) are considered the most valuable markers, particularly for studying the evolutionary relationships of isolates of homogeneous pathogenic clones, such as Bacillus anthracis (16), Mycobacterium tuberculosis (4), and Yersinia pestis (1).
SNPs have been used as markers for molecular typing of S. enterica serovar Typhi in a large study by Roumagnac et al. (17). A total of 88 SNPs, found from analysis of 200 gene fragments from 105 diverse S. enterica serovar Typhi isolates, could differentiate 481 global S. enterica serovar Typhi isolates into 85 haplotypes (SNP profiles) and five major clusters (17). However, despite the large number of SNPs used, each of the five clusters was supported by only a single SNP and there was little resolution of the relationships of the haplotypes within a cluster. Eighty of the SNPs have also been used to differentiate 140 Indonesian S. enterica serovar Typhi isolates into nine haplotypes (2).
We have also shown that genome-wide SNPs are useful for molecular typing and determining the relationships of global S. enterica serovar Typhi isolates (14). Thirty-seven SNPs selected from a comparison of the genomes of S. enterica serovar Typhi strains CT18 (15) and Ty2 (3) were typed using restriction enzyme digestion to differentiate 73 global S. enterica serovar Typhi isolates into 23 SNP profiles and four distinct genetic groups. As the SNPs were selected by comparison of only two S. enterica serovar Typhi genomes, this resulted in a phylogenetic bias which revealed the full path of the last common ancestors connecting strains CT18 and Ty2 but only the node locations for the other SNP profiles (14).
Advances in technology, such as high-throughput sequencing, allow SNPs to be discovered to obtain a full resolution of the phylogenetic relationships of isolates. A recent study by Holt et al. (8) utilized 454 and/or Solexa technologies to sequence 19 S. enterica serovar Typhi isolates selected from the five major clusters found by Roumagnac et al. (17). There were more than 1,700 SNPs found, and these gave a fully resolved phylogenetic tree of these isolates. These SNPs are invaluable resources for investigation of the evolutionary history of global S. enterica serovar Typhi isolates. This study aimed to select a better set of SNPs on the basis of the genome tree and the previous SNP studies by Roumagnac et al. (17) to differentiate and establish the phylogenetic relationships of global S. enterica serovar Typhi isolates, using real-time (R-T) PCR assays based on hairpin (HP) primers (6).
MATERIALS AND METHODS
Bacterial isolates.
The 71 global S. enterica serovar Typhi isolates used were the same set of isolates from a previous study of Octavia and Lan (14). Genome sequenced strains CT18 and Ty2 were also used as controls. The SNP data for the remaining 17 genome sequenced strains were obtained from GenBank and were included in the analysis.
SNP selection and primer design.
A total of 42 SNPs were selected from a comparison of 19 S. enterica serovar Typhi genomes (11) and are listed in Table S1 in the supplemental material. Locus tags were used as SNP names, as there was only one SNP per gene selected. The SNP location in the gene is shown in Table S1 in the supplemental material. The primers for HP real-time PCR assay were designed to produce small amplicons (about 100 bp) and to optimally anneal at 60°C (see Table S1 in the supplemental material). Hairpin primers were designed on the basis of the principles described by Hazbon and Alland (6). A secondary mismatch was introduced in some of the primers (noted in Table S1 in the supplemental material) to further decrease the affinity of the mismatched primers, therefore permitting a larger threshold cycle (CT) value difference between the matched and mismatched HP primers.
HP R-T PCR assay.
Each PCR mixture contained 10 ng of chromosomal DNA, 3 μl of SYBR green (Quantace), 0.5 μl each of 10 μM forward and reverse primers (see Table S1 in the supplemental material), and Milli-Q water to a final volume of 10 μl. All reactions were performed in a Rotor-gene 6000 sequence detector system (Corbett Life Science, Australia). Thermal cycling conditions were as follows: stage 1, 95°C for 10 min and then hold; stage 2, 10 cycles of 72°C for 30 s, 95°C for 15 s, and 69°C for 30 s, with the temperature being lowered 1°C in the last step for every cycle; and stage 3, 72°C for 30 s, 95°C for 20 s, and 60°C for 30 s, repeated 40 times. Data for analysis were collected at the last step of stage 3.
Bioinformatic analyses.
The PAUP program (19) was used to construct a maximum-parsimony tree from the SNP data and calculate the homoplasy index. SNPT software (4) was used to determine the minimum number of SNPs required for typing.
RESULTS AND DISCUSSION
Selection of SNPs.
A total of 42 SNPs were selected for typing on the basis of the data from genome sequencing of 19 S. enterica serovar Typhi isolates (8) and the SNP typing results of Octavia and Lan (14) and Roumagnac et al. (17). We previously mapped the SNPs from these three studies onto the genome tree of the 19 strains whose genomes were sequenced (11), and SNP selection was based on this mapping. SNPs showing reverse/parallel changes were avoided. We first selected SNPs that have been used in previous studies to divide S. enterica serovar Typhi isolates into clusters, including four SNPs (STY0778, STY1397, STY1583, and STY4105) by us (14) and four (STY2513, STY2629, STY3196, and STY3622) by Roumagnac et al. (17). We then selected SNPs to cover branches with at least one SNP for branches with 40 or more SNPs. The SNPs typed by Roumagnac et al. (17) were selected as the first preference. In total, 15 and 22 SNPs were selected from the internal nodes and terminal branches of the genome tree, respectively (see Fig. S1 in the supplemental material). Finally, we used the haplotype frequency data from Roumagnac et al. (17) to select SNPs so that common haplotypes can be separated. There were 16 most common haplotypes, with each haplotype having from 3 to 156 isolates. SNPs separating 11 haplotypes (haplotype 1 [H1], H8, H42, H45, H46, H50, H52, H55, H58, H59, and H85) have already been selected on the basis of the criteria described above. Five additional SNPs (STY1503, STY1919, STY2389, STY3876, and STY4562) were selected to separate the remaining five common haplotypes (H39, H81, H15, H56, and H84), respectively (17). The 42 SNPs selected resolved the 19 genomes into 13 groups, and the terminal branches between strains E03-4983 and 404ty collapsed into a single branch. As these two branches represent the distinctive isolates with the z66 flagellar antigen, the collapse of divisions between these two lineages has no impact on their identification.
Out of the 42 SNPs, 25 were synonymous SNPs (sSNPs), 14 were nonsynonymous SNPs (nsSNPs), and 3 were nonsense mutations. The SNPs selected were mostly from genes of known functions. Only 4 of the 42 SNPs (STY0321, STY0336, STY1583, and STY1720) are in genes that might be related to virulence, and these SNPs can potentially be under selection pressure, giving rise to reverse or parallel changes in different lineages. These SNPs were selected, as they were from previous studies and are key SNPs required for resolving the lineages involved (11, 17). However, the SNP data (see below) showed no reverse or parallel changes in these SNPs, and such changes overall are rare, suggesting that reverse or parallel changes are not an issue in inferring relationships on the basis of the SNPs selected.
Reliability of HP R-T PCR for SNP typing.
We used two sets of SNPs to test the reliability of HP R-T PCR SNP typing. We first typed the four cluster-dividing SNPs (STY0778, STY1397, STY1583, and STY4105) from Octavia and Lan (14) in the 73 isolates, including the 71 global S. enterica serovar Typhi isolates and 2 genome sequenced strains, CT18 and Ty2, which have previously been typed using restriction enzyme digestion. The typing results for these SNPs were consistent with previous results. The average differences in the CT values were 5.36, 12.53, 8.22, and 5.41 for STY0778, STY1397, STY1583, and STY4105, respectively.
We then tested the four cluster-dividing SNPs (STY2513, STY2629, STY3196, and STY3622) from Roumagnac et al. (17). Twenty-nine of the 73 isolates have previously been independently typed for these four SNPs by Roumagnac et al. (17). All except three nucleotides were consistent with previous data. The three discrepancies observed were in the assignment of nucleotides for two of the four SNPs. The isolate of ST60 was typed as H50 by Roumagnac et al. (17), SNP STY2629 should be a G nucleotide for this isolate but was typed as nucleotide A in the present study. Similarly, isolates R1962 and T189 were both typed as H42 by Roumagnac et al. (17) due to allele T in SNP STY3196, but both had nucleotide C in the present study. To determine whether our HP R-T PCR results were correct, we sequenced the SNPs from these isolates, which confirmed our HP R-T PCR results.
For the remaining 34 SNPs selected, we had only strains CT18 and Ty2 as controls. The HP R-T PCR typing results for these two strains matched the genome data, which further confirmed the reliability of the HP R-T PCR for SNP typing. We typed the remaining SNPs for the 71 isolates using the HP R-T PCR assay. The average CT value difference for the SNPs typed was 7.92 (see Table S1 in the supplemental material), and the CT values between the two sets of primers were distinctive, which correlates with the matched and mismatched primer pairs for each nucleotide.
Polymorphisms among the S. enterica serovar Typhi isolates.
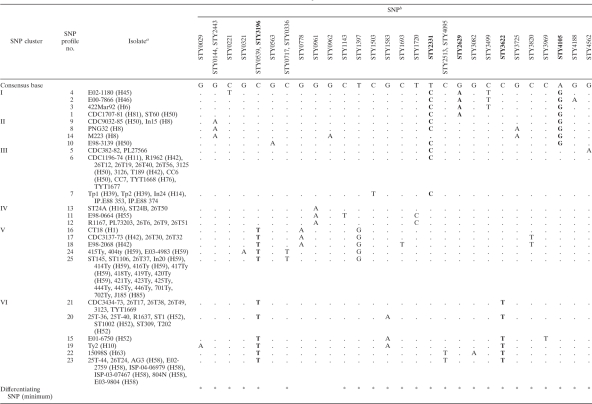
The SNP data for the 71 isolates, 2 fully sequenced genome strains (CT18 and Ty2), and 17 partially sequenced genome strains (8) were combined for comparison (Table 1). Twenty-three of the 42 SNPs were informative. Four pairs of SNPs, STY0539 and STY3196, STY2513 and STY4095, STY336 and STY717, and STY144 and STY2443, gave the same patterns of polymorphisms among the SNP profiles. The 90 isolates were distinguished into 25 SNP profiles. The same level of differentiation can also be achieved by typing 24 SNPs (indicated by asterisks in Table 1). Twelve profiles were unique to a single isolate, while 13 others were shared by more than one isolate. SNP profiles 6 and 25 contained more than 10 isolates, and SNP profile 25 was the largest SNP profile, containing 19 isolates.
TABLE 1.
SNP profiles
a The haplotypes according to Roumagnac et al. (17) are in parentheses.
b The SNPs typed are designated with the locus name, according to the S. enterica serovar Typhi CT18 genome. See Table S1 in the supplemental material for the locations of the SNPs. The SNP profiles were arranged according to the parsimony tree in Fig. 1. A minimum of five SNPs (highlighted in boldface), STY2629 for cluster I, STY4105 for cluster II, STY2331 for cluster III, STY3196 for cluster IV, and STY3622 for cluster V, can be used to divide the SNP profiles into the respective clusters. Consensus base data represent the predominant nucleotide for each column. Dots indicate nucleotides identical to the consensus base nucleotides. *, the minimum SNPs required for full differentiation into individual SNP profiles.
The 29 isolates also typed by Roumagnac et al. (17) were previously differentiated into 14 haplotypes using 88 biallelic polymorphisms. In the present study, the 29 isolates were distinguished into 13 SNP profiles. Some haplotypes previously defined by Roumagnac et al. (17) were further differentiated using our set of 42 SNPs. H8, H50, H52, and H59 were each further divided into two or three profiles. However, some other haplotypes collapsed into the same SNP profile in the present study; for example, SNP profile 6 contained four different haplotypes (H50, H11, H42, and H76), with the last three being less frequent haplotypes (17). SNP profile 6 also contained multiple divergent types, determined by multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) (13), suggesting that there is more diversity within this SNP profile. A genome from this lineage should be sequenced for better phylogenetic coverage of its diversity.
Two of our isolates, isolates 25T-44 and 26T24, were grouped together with six genome sequenced strains of H58. According to Roumagnac et al. (17), H58 was mostly isolated from Southeast Asia (17), and recent H58 isolates, particularly those from Vietnam, are associated with resistance to nalidixic acid (12). Both of our isolates were from Canada. There was no information on whether these two isolates were imported cases, but multidrug-resistant isolates from travelers to Southeast Asia have been reported in Canada (5).
Eighteen isolates with the z66 flagellar antigen were differentiated into two SNP profiles, SNP profiles 24 and 25. SNP profile 24 contained three isolates, including 2 of the genome sequenced strains, 404ty and E03-4983, while SNP profile 25 contained 19 isolates, including 1 genome sequenced strain, J185. Two strains from SNP profile 25, including J185, were z66 negative (2, 8, 17) and ST1106.
Evolutionary relationships of the SNP profiles.
A maximum-parsimony phylogenetic tree was constructed to determine the relationships of the 25 SNP profiles. There was only one most parsimonious tree found, and the homoplasy index was very low (0.003) due to a T nucleotide rather than a C nucleotide for SNP STY2331 in SNP profile 14 (Table 1). As the profile was represented by the genome strain M223 only, the change was potentially a sequencing error and remained to be confirmed. The observation of a low homoplasy index was concordant with observations from previous two SNP studies by Roumagnac et al. (17) and Holt et al. (8).
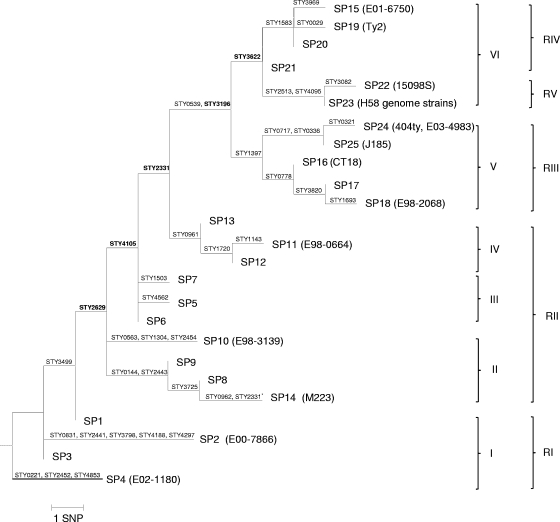
The SNP profiles could be divided into six clusters (clusters I to VI), with at least one SNP supporting each cluster (Fig. 1). Cluster I consisted of four SNP profiles (profiles 1 to 4) supported by STY2629; cluster II contained four SNP profiles (profiles 8, 9, 10, and 14) supported by STY4105; cluster III contained SNP profiles 5 to 7 supported by STY2331; cluster IV had three SNP profiles (profiles 11 to 13) supported by STY0539, STY3196, and STY0961; and cluster V had five SNP profiles (profiles 16 to 18, 24, and 25) supported by STY3622 and STY1397, which also separate cluster V from cluster VI, which contained six SNP profiles (profiles 15, 19, and 20 to 23). A minimum of five SNPs (STY2629, STY4105, STY2331, STY3196, and STY3622) is required to differentiate these six clusters. Note that three of the SNPs (STY2629, STY3196, and STY3622) were also cluster-dividing SNPs from Roumagnac et al. (17). The five SNPs act like dichotomous keys sequentially differentiating the clusters.
FIG. 1.
A maximum-parsimony tree to illustrate the evolutionary relationships of the SNP profiles (SP). For SNP profiles containing genome strains, the strain names are in parentheses after the SNP profile numbers. H58 genome strains include AG3, E02-2759, ISP-04-06979, ISP-03-07467, 804N, and E03-9804. The roman numerals correspond to the cluster number. Clusters defined by Roumagnac et al. (17) were marked RI to RV. SNPs are labeled on the branch. Five cluster-dividing SNPs are highlighted in boldface.
Comparison of evolutionary relationships established using different molecular markers.
SNPs have also been used as molecular markers for S. enterica serovar Typhi in a similar study by Roumagnac et al. (17). A total of 88 SNPs (referred to as biallelic polymorphisms), found upon analysis of 200 gene fragments from 105 diverse S. enterica serovar Typhi isolates using denaturing high-performance liquid chromatography (dHPLC), could differentiate 481 global S. enterica serovar Typhi isolates into 85 SNP haplotypes (17). Four SNPs (STY2513, STY2629, STY3196, and STY3622) divided their haplotypes into five major clusters (17). We have also included these four SNPs for typing in the present study. We can allocate our SNP profiles according to the Roumagnac et al. (17) clustering scheme, and we designated them RI to RV (Fig. 1). The clustering was different to some extent between the two studies. Clusters I and V correspond to RI and RIII, respectively. Clusters II, III, and VI were grouped together as RII, suggesting that our selected SNPs have a better resolution to further differentiate cluster RII. Cluster VI was divided into RIV and RV. We can divide cluster VI into two subclusters corresponding to RIV and RV using either STY1583 or STY2513/ST4095. Therefore, despite the difference in cluster division, the evolutionary relationships of the SNP profiles were consistent, and there was no conflict at the cluster level.
These 73 S. enterica serovar Typhi isolates have also been typed using MLVA with nine VNTRs (13). For SNP profiles containing multiple isolates, all were divided further into multiple MLVA profiles (see Fig. S2 in the supplemental material). In addition, MLVA profiles with the same SNP profiles were not always grouped together, as was also found in our previous study (13). The number of VNTR differences was smaller between the isolates within SNP profiles 7 and 25, and their corresponding MLVA profiles belonged to the same MLVA cluster. Moreover, the SNP clusters defined in the present study were not correlated with the clusters defined using VNTR markers. The SNP clusters were no longer visible and were distributed among different MLVA clusters. Highly polymorphic VNTRs appeared to evolve too fast to have retained sufficient phylogenetic information. Evolutionary relationships based on VNTRs would best be viewed within SNP profiles (see Fig. S2 in the supplemental material). SNPs are necessary to resolve the genetic relationships of S. enterica serovar Typhi isolates, and VNTRs should be used only to achieve a further resolution of closely related isolates. Therefore, SNP typing will be more appropriate for global surveillance of S. enterica serovar Typhi, and a combination of SNP typing and VNTR typing can be used for local epidemiological analysis.
Concluding comments.
Previous studies by Roumagnac et al. (17) and Octavia and Lan (14) have shown that SNPs are very useful for typing and provide better insights into the evolutionary relationships of S. enterica serovar Typhi isolates. SNP typing in these two studies was achieved using restriction enzyme digestion (14) or dHPLC (17). We have shown in the present study that the HP R-T PCR assay is an alternative method for SNP typing and was applied to type 42 SNPs. The HP R-T PCR assay is not gel based, unlike the two aforementioned methods, and the results could be obtained directly after completion of the PCRs. Therefore, the use of this method greatly reduced the time for SNP typing. Two recent studies by Kariuki et al. (9) and Holt et al. (7) employed the GoldenGate bead array high-throughput platform (Illumina) to type 1,500 SNPs in 94 Kenyan and 62 Nepalese S. enterica serovar Typhi isolates, respectively. However, the method is far less cost efficient than the HP R-T PCR assay used in the present study and is thus less suitable for routine use in laboratories, particularly in developing countries where typhoid fever is endemic. In addition, the 1,500 SNPs used by Kariuki et al. (9) and Holt et al. (7) were highly redundant, as only eight and six haplotypes were found among the 94 and 62 isolates studied, respectively.
In the present study, we selected 42 SNPs on the basis of the phylogenetic distribution of the genome-wide SNPs from 19 diverse S. enterica serovar Typhi strains (8), SNPs found by Roumagnac et al. (17), and SNPs used by Octavia and Lan (14). We have shown that this set of SNPs can distinguish many of the major SNP profiles, although they may not be able to fully resolve the less common SNP profiles found by Roumagnac et al. (17). A minimum of 5 and 24 SNPs differentiated the isolates used in the present study into six major clusters and 25 SNP profiles, respectively. Nevertheless, the full set of 42 SNPs may offer a higher level of differentiation in other S. enterica serovar Typhi populations. With the flexibility of the HP R-T PCR assay, large-scale typing of S. enterica serovar Typhi isolates can be done progressively using the five cluster-dividing SNPs first, followed by the other 19 minimum SNPs and then the remaining SNPs for full resolution. In the studies of Kariuki et al. (9) and Holt et al. (7), H58 (SNP profile 23 in the present study) was subdivided into two sublineages and several subtypes. Division into sublineages requires one SNP and can be easily added to our scheme.
We have recently shown that SNPs are well suited for global and long-term epidemiological analyses, while MLVA is suitable only for short-term epidemiological analyses (13). SNP typing has also been shown to be valuable to study the local population structure and epidemiology of S. enterica serovar Typhi in Indonesia, Kenya, and Nepal (2, 7, 9). The SNPs and strains typed in the present study form the basis of a future database for international comparison. Further studies should include testing of an expanded collection of isolates. In conclusion, the present study provided a simple and cost-efficient alternative SNP typing method and a better selected set of SNPs for epidemiological studies of S. enterica serovar Typhi.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Health and Medical Research Council of Australia.
We thank Ken Sanderson (the University of Calgary) and Gordon Dougan (Imperial College London) for generously providing us the strains. We also thank the anonymous reviewers for their constructive comments and suggestions.
Footnotes
Published ahead of print on 28 July 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, et al. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S., K. Holt, E. van de Vosse, et al. 2008. High-throughput genotyping of Salmonella enterica serovar Typhi allowing geographical assignment of haplotypes and pathotypes within an urban district of Jakarta, Indonesia. J. Clin. Microbiol. 46:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng, W., S. R. Liou, G. Plunkett III, et al. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filliol, I., A. Motiwala, M. Cavatore, et al. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harnett, N., S. McLeod, Y. AuYong, et al. 1998. Molecular characterization of multiresistant strains of Salmonella typhi from South Asia isolated in Ontario, Canada. Can. J. Microbiol. 44:356-363. [PubMed] [Google Scholar]
- 6.Hazbon, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt, K., S. Baker, S. Dongol, et al. 2010. High-throughput bacterial SNP typing identifies distinct clusters of Salmonella typhi causing typhoid in Nepalese children. BMC Infect. Dis. 10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt, K. E., J. Parkhill, C. J. Mazzoni, et al. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat. Genet. 40:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariuki, S., G. Revathi, J. Kiiru, et al. 2010. Typhoid in Kenya is associated with dominant multidrug-resistant Salmonella enterica serovar Typhi haplotype that is also widespread in Southeast Asia. J. Clin. Microbiol. 48:2171-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidgell, C., U. Reichard, J. Wain, et al. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 11.Lan, R., P. R. Reeves, and S. Octavia. 2009. Population structure, origins and evolution of major Salmonella enterica clones. Infect. Genet. Evol. 9:996-1005. [DOI] [PubMed] [Google Scholar]
- 12.Le, T. A. H., L. Fabre, P. Roumagnac, et al. 2007. Clonal expansion and microevolution of quinolone-resistant Salmonella enterica serotype Typhi in Vietnam from 1996 to 2004. J. Clin. Microbiol. 45:3485-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Octavia, S., and R. Lan. 2009. Multiple-locus variable-number tandem-repeat analysis of Salmonella enterica serovar Typhi. J. Clin. Microbiol. 47:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Octavia, S., and R. Lan. 2007. Single nucleotide polymorphism typing and genetic relationships of Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 45:3795-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill, J., G. Dougan, K. D. James, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 16.Pearson, T., J. Busch, J. Ravel, et al. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 101:13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roumagnac, P., F.-X. Weill, C. Dolecek, et al. 2006. Evolutionary history of Salmonella typhi. Science 314:1301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selander, R. K., P. Beltran, N. H. Smith, et al. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swofford, D. L. 1998. PAUP: phylogenetic analysis using parsimony, 4.0 beta ed. Sinauer Associates, Sunderland, MA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.