Abstract
Patients with diarrhea, defined as loose or watery stool, and two or more Clostridium difficile tcdB PCR tests within 14 days of each other were investigated. Repeat PCR for 293 patients with a prior negative result yielded negative results in 396 (97.5%) of 406 tests. Ten new positives were detected, including one false positive. Repeat PCR within 7 days appears rarely useful, except for patients with evidence of a new infection.
Currently, the most sensitive method for diagnosis of Clostridium difficile infection is anaerobic toxigenic stool culture; however, in order to provide simpler and faster testing, the majority of hospitals in the United States rely on enzyme immunoassay (EIA) (3, 8). More recently, real-time PCR assays targeting the tcdB toxin B gene of C. difficile have been developed and made commercially available as an alternative to EIA or cell culture cytotoxicity testing of stool (2, 7, 9, 13, 17, 18, 19). Due to the historically low sensitivity of EIAs, a common practice has been to order multiple EIAs to increase the test's sensitivity, despite recent studies suggesting that this may not be helpful and could result in increased false positives (5, 11, 12). Similarly, given the higher sensitivity of PCR testing and the role of toxin B as the essential virulence factor of C. difficile, multiple PCR tests may not be necessary (10, 15).
Although no studies have examined the utility of repeat tcdB PCR, one study evaluated repeat PCR testing for the tcdC toxin regulatory gene and found that only 1.7% of patients with an initial negative result gave a positive result on subsequent testing over 7 days (1). After implementation of tcdB PCR at our institution, many physicians continued to request duplicate or triplicate testing, similar to ordering patterns for the cell culture cytotoxicity assay or EIA. The purpose of the current study was to examine the clinical utility of repeating tcdB PCR within 14 days of a negative test result and to provide evidence on when repeat testing may be warranted.
A retrospective cohort study, approved by the institutional review board, was performed at the Stanford Hospital and Clinics clinical microbiology laboratory on patients with real-time PCR results for C. difficile in stool from July 2009 to December 2009. Stool samples were routinely accepted for PCR in patients who had diarrhea and were over 1 year of age, although exceptions were made upon medical director approval. Diarrhea was defined as loose or watery stool which took the shape of the collection container. All patients with two or more tcdB PCR tests within 14 days of each other were included in the study. Testing intervals for each patient were calculated as the number of days between two consecutive tests. Patient samples which showed conversions from negative to positive results upon repeat PCR testing were tested with a commercial tcdB PCR assay (Cepheid Xpert C. difficile) (9, 13). Discrepancies between the in-house-PCR (discussed below) and commercial-PCR results were resolved with anaerobic toxigenic culture (discussed below). A retrospective chart review was done to identify if patients were hospitalized, had been on any antibiotics when the test was ordered, and had any history of immunosuppression, organ transplantation, malignancy, or gastrointestinal manipulation within a week of PCR testing. Descriptive statistics and chi-square testing were done using Stata analysis software (College Station, TX).
The in-house real-time PCR primers were selected using tcdB sequences from the NCBI database and included forward (5′TTGGAAGTACAAAAAGAAGAACTTGA) and reverse (5′TTTTAATGTTGTTATTAAAGCATCAGC) primers. For each stool specimen, 0.1 ml was diluted 20-fold in 50% Vertrel XF (Miller Stephenson Chemical Company, Danbury, CT) and subjected to the NucliSens easyMAG platform for DNA extraction (bioMérieux, Durham, NC) by the on-board lysis protocol according to the instructions of the manufacturer. For real-time PCR, each 10-μl reaction mixture on the Rotor-Gene 6000 (Corbett Life Science, Sydney, Australia) contained 5 μl of 2× FastStart SYBR green master mix (Roche Diagnostics, Indianapolis, IN), 3 μl of extracted DNA, and 2 μl of the 10 μM tcdB primer mix. A second reaction mixture, containing 5 μl of 2× FastStart SYBR green master mix, 3 μl of extracted DNA, 2 μl of ∼300 copies of an internal control plasmid, and 10 μM primer mix specific for the plasmid, was used to check for inhibition of PCR. The PCR conditions included initial denaturation at 95°C for 5 min and 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. The final step involved a 60°C-to-95°C temperature ramp at a rate of 0.1°C/second to generate the melting curve. Amplified products in the melting curve plot were analyzed for their intensity and specificity. A peak with a signal intensity of ≥0.5 units and a melting temperature (Tm) of 76°C ± 2°C was considered indicative of positivity for tcdB. If a specimen demonstrated PCR inhibition without positive tcdB amplification, the DNA extract was diluted 1:10 and both reactions were repeated. Specimens with unresolved inhibition were called indeterminate.
Prior to its implementation, the performance of PCR was compared to that of an anaerobic toxigenic culture and a cell culture cytotoxicity assay. For the anaerobic culture, stool was diluted 1:1 in 95% ethanol and incubated at an ambient temperature for 45 min. The sample was then sedimented briefly, one drop of the sediment was added to anaerobic chopped-meat carbohydrate broth (CMB) (Anaerobe Systems, Morgan Hill, CA), and a second drop was streaked onto prereduced cycloserine-cefoxitin-fructose agar medium (CCFA-HT formulation; Anaerobe Systems). The plates were incubated at 37°C for 48 h in the anaerobic chamber. The plates were screened for flat, grayish, and shiny colonies with spreading edges. Colonies giving positive results with the use of the RapID ANA system (Remel, Lenexa, KS), a typical horse manure smell, and fluorescence under UV light were considered to be C. difficile. If CCFA-HT plates did not yield C. difficile colonies, CMB cultures were subcultured on CCFA-HT and rescreened as described above. All C. difficile isolates were inoculated into a new CMB and incubated for 4 days, and the supernatants were tested for cytotoxin. Only isolates showing cytotoxicity were considered positive for production of toxin. The cell culture cytotoxicity assay was performed directly on stool samples and also on CMB. Sterilized filtrates were added to MRC-5 cells (Diagnostic Hybrids, Athens, OH) with and without C. difficile toxin B antitoxin (Techlab, Blacksburg, VA). Samples demonstrating cytopathic effect only in the absence of antitoxin were considered to be positive for C. difficile toxin B.
When compared to anaerobic toxigenic culture and cell culture cytotoxicity assay, PCR was shown to have a sensitivity of 87.2% and specificity of 98.6% on 47 positive and 70 negative liquid stool samples (Table 1). The sensitivity was found to be similar to the range of sensitivities for other real-time PCR assays when toxigenic culture was used as the “gold standard.” In those studies, the sensitivities of real-time PCR were 86% and 93% with two in-house assays (14, 16) and between 77% and 94% with commercially available FDA-approved assays (2, 7, 13, 17, l8). Thus, the performance of the real-time PCR assay in the current study is comparable to that of other reported assays, and the results obtained from this study can be applied to institutions using either in-house or commercial assays.
TABLE 1.
Performance of tests for diagnosis of Clostridium difficile infectiona
| Test | % (no. of positive results/total no. of results) |
|
|---|---|---|
| Sensitivity | Specificity | |
| Anaerobic culture | 95.7 (45/47) | 100 (70/70) |
| Direct cytotoxicity | 83.0 (39/47) | 98.6 (69/70) |
| Real-time PCR | 87.2 (41/47) | 98.6 (69/70) |
The reference standard was based on anaerobic culture or ≥2 positive test results.
A total of 1,949 PCR tests were done on stool samples from 1,287 unique patients during the study period. The patient ages ranged from 1 month to 108 years, with 84.7% being over 18 years of age. The number of PCR tests per patient ranged from one to six, with a mean of 1.5 tests per patient. One hundred seventy-one tests (8.8%) were positive, 1,740 (89.3%) were negative, and 38 (1.9%) were indeterminate due to the presence of PCR inhibitors. Of all the tests, 461 (23.7%) were repeat tests for patients who already had at least one PCR result within the last 14 days. Of these 461 repeat tests, 406 were on 293 patients with a prior negative result, 37 were on 33 patients with a prior positive result, and 18 were on 18 patients with a prior indeterminate result. The results for 25 (67.6%) tests for patients with a prior positive result converted from positive to negative on repeat testing, the results for 11 (29.7%) remained positive, and the result for 1 (2.7%) was indeterminate. The results for all patients with initial indeterminate test results were negative on the second PCR except for one patient, for whom the result remained indeterminate.
For the 293 patients with a prior negative result, the highest frequency of repeats (22%) occurred within the first 24 h, followed by a gradual decline during the following 13 days (Fig. 1). The age range of these patients was from 9 months to 92 years. In total, the results for 10 of 406 repeat tests (2.5%) turned positive, while the results for 396 of 406 repeat tests (97.5%) stayed negative. As shown in Fig. 1, the positives were detected on repeat interval days 2, 5, 6, 7 (n = 2), 9, 11, 12 (n = 2), and 14. Figure 1 also demonstrates that while the majority of repeat tests were ordered during the first 6 days after the initial negative result, the majority of positives occurred after repeat interval day 6. Only 1.1% (n = 3) of 267 repeat tests done less than 7 days after the initial negative result gave positive results. New positives were found for both pediatric (n = 2) and adult (n = 8) patients, with the ages of these 10 patients ranging from 19 months to 81 years. Only 2 of the 10 patients whose results converted from negative to positive had more than two tests. The patient whose positive result was detected on day 6 had another negative result on day 6. The patient whose result turned positive on day 9 had another negative test result 13 days earlier. Additional testing with a commercial PCR assay was done on 8 of the 10 samples which had turned positive, with the remaining samples unavailable for testing. Seven were confirmed as true positives, while the one from repeat interval day 5 was a false positive, as it was negative by the commercial PCR assay and also by toxigenic culture.
FIG. 1.
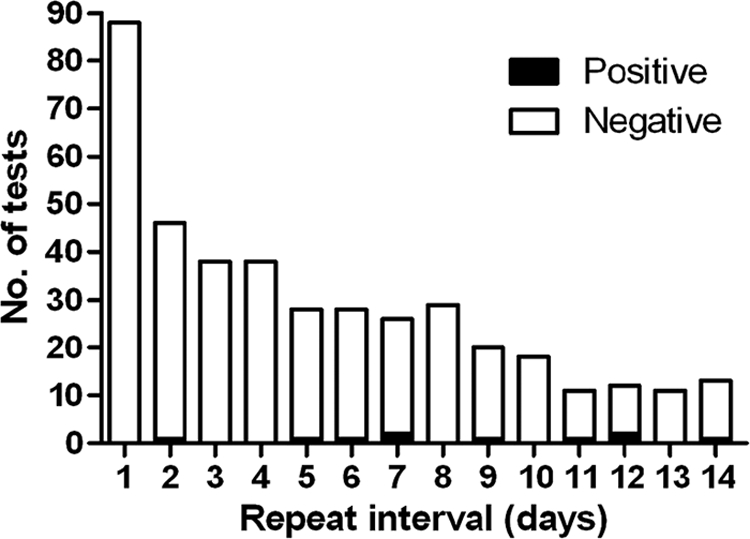
Results for repeat PCR tests following a negative result. The PCR results per day for all patients who underwent repeat testing 1 to 14 days following a prior negative result are shown.
Excluding the false-positive case, all nine patients for whom the PCR results converted from negative to positive had multiple risk factors for C. difficile infection (3, 4), including current hospitalization (100%), ongoing antibiotic treatment (66.7%), history of severe underlying disease such as organ transplantation, immunosuppression, and/or malignancy (88.9%), and recent gastrointestinal tube placement (11.1%). The results for two patients (22.2%) converted from negative to positive less than 7 days after the initial test, although the patient with a positive result detected on day 2 did not have a remaining sample for confirmation. The other seven (77.8%) patients who converted their PCR result did so 7 or more days after their initial negative result. Six patients had archived stool samples, and the results for all six patients were confirmed as true positives. Five of these seven patients were tested due to new-onset diarrhea after their first test. Another one of these seven patients was already being treated for a recent outside diagnosis of C. difficile infection at the time of the initial negative test, which may have led to the negative result.
The findings from this study indicate that repeating C. difficile tcdB PCR within 14 days of a negative result yields little relevant clinical data, other than confirming the negative result of the initial test, in an overwhelming majority (97.5%) of tests. Additionally, repeat testing can lead to false positives. With the false positive seen on day 5 disregarded, repeat testing less than a week after the initial negative result provided new information in only 2 (0.8%) out of 266 tests, or 2 (1.0%) out of 197 patients. However, the current study does show that repeat testing, particularly between interval days 7 and 14, can be useful in a small subset of patients with high clinical suspicion for infection. Of all the cases whose results converted to positive on repeat testing, more than half involved patients with a recurrence of diarrhea after resolution of their initial episode of loose stool, suggesting a different disease process, which would explain the change in PCR results. Additionally, all these patients had multiple risk factors for C. difficile infection, including ongoing antibiotic usage, prolonged hospitalization, severe underlying illness, immunosuppression, and/or recent gastrointestinal procedures (3, 4). Since a limitation of the current study is the lack of confirmatory toxigenic culture results and clinical data on all patients with an initial negative test result, further prospective studies with parallel PCR, toxigenic cultures, and close clinical follow-up could help determine which patients may benefit from repeat PCR testing over short intervals.
The usefulness of repeat PCR as a “test of cure” for C. difficile infection in patients with initial positive results is also unclear. Although the results for 67.6% of repeat tests for patients with prior positive results converted to negative upon repeat testing within a 14-day period, the limited sample size and lack of clinical and laboratory follow-up do not allow us to determine if these results are from clearance of the infection in response to treatment or are false negatives. Again, a prospective study with testing over short intervals would also help determine when one may expect a negative PCR result after initiation of effective treatment.
Since the majority of positive conversions occurred after 7 or more days, our findings suggest that repeat PCR testing should not be done routinely for patients with a prior negative result for at least 7 days. As the assays continue to improve, utilization of assays with higher sensitivities would only result in fewer false negatives and consequently make repeat testing even less necessary, providing even more support for our findings. Eliminating unnecessary repeat tests at our institution would have resulted in a >20% reduction in testing during this study period. Furthermore, patients suspected to have C. difficile infection are routinely placed in private rooms with contact isolation until the infection can be ruled out, and this practice on average costs $100 more per day than the use of a regular hospital bed (6). Without repeat testing, patients with negative results can be removed from isolation earlier, leading to significant savings in testing and isolation costs for the duration of study period even if we conservatively estimate that the 396 repeat negative results each resulted in only 1 more day of isolation. As real-time PCR becomes more commonplace for diagnosis of C. difficile infection, clinical ordering practices should adapt to reflect the increased sensitivity of PCR. Together, these changes will result in improved clinical care and lower health care costs.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Aichinger, E., C. D. Schleck, W. S. Harmsen, L. M. Nyre, and R. Patel. 2008. Nonutility of repeat laboratory testing for detection of Clostridium difficile by use of PCR or enzyme immunoassay. J. Clin. Microbiol. 46:3795-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbut, F., M. Braun, B. Burghoffer, V. Lalande, and C. Eckert. 2009. Rapid detection of toxigenic strains of Clostridium difficile in diarrheal stools by real-time PCR. J. Clin. Microbiol. 47:1276-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. G., and D. N. Gerding. 2008. Clinical recognition and diagnosis of Clostridium difficile infection. Clin. Infect. Dis. 46:S12-S18. [DOI] [PubMed] [Google Scholar]
- 4.Bignardi, G. E. 1998. Risk factors for Clostridium difficile infection. J. Hosp. Infect. 40:1-15. [DOI] [PubMed] [Google Scholar]
- 5.Cardona, D. M., and K. H. Rand. 2008. Evaluation of repeat Clostridium difficile enzyme immunoassay testing. J. Clin. Microbiol. 46:3686-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubberke, E. R., and A. I. Wertheimer. 2009. Review of current literature on the economic burden of Clostridium difficile infection. Infect. Control Hosp. Epidemiol. 30:57-66. [DOI] [PubMed] [Google Scholar]
- 7.Eastwood, K., P. Else, A. Charlett, and M. Wilcox. 2009. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J. Clin. Microbiol. 47:3211-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerding, D. N., S. Johnson, L. R. Peterson, M. E. Mulligan, and J. Silva, Jr. 1995. Clostridium difficile-associated diarrhea and colitis. Infect. Control Hosp. Epidemiol. 16:459-477. [DOI] [PubMed] [Google Scholar]
- 9.Huang, H., A. Weintraub, H. Fang, and C. E. Nord. 2009. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of Clostridium difficile infections. J. Clin. Microbiol. 47:3729-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyras, D., J. R. O'Connor, P. M. Howarth, S. P. Sambol, G. P. Carter, T. Phumoonna, R. Poon, V. Adams, G. Vedantam, S. Johnson, D. N. Gerding, and J. I. Rood. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan, S. S., B. P. McDermott, S. Parchuri, and B. A. Cunha. 2006. Lack of value of repeat stool testing for Clostridium difficile toxin. Am. J. Med. 119:356e7-356e8. [DOI] [PubMed] [Google Scholar]
- 12.Nemat, H., R. Khan, M. S. Ashraf, M. Matta, S. Ahmed, B. T. Edwards, R. Hussain, M. Lesser, R. Pekmezaris, Y. Dlugacz, and G. Wolf-Klein. 2009. Diagnostic value of repeated enzyme immunoassays in Clostridium difficile infection. Am. J. Gastroenterol. 104:2035-2041. [DOI] [PubMed] [Google Scholar]
- 13.Novak-Weekley, S. M., E. M. Marlowe, J. M. Miller, J. Cumpio, J. H. Nomura, P. H. Vance, and A. Weissfeld. 2010. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J. Clin. Microbiol. 48:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson, L. R., R. U. Manson, S. M. Paule, D. M. Hacek, A. Robicsek, R. B. Thomson, Jr., and K. L. Kaul. 2007. Detection of toxigenic Clostridium difficile in stool samples by real-time polymerase chain reaction for the diagnosis of C. difficile-associated diarrhea. Clin. Infect. Dis. 45:1152-1160. [DOI] [PubMed] [Google Scholar]
- 15.Peterson, L. R., and A. Robicsek. 2009. Does my patient have Clostridium difficile infection? Ann. Intern. Med. 151:176-179. [DOI] [PubMed] [Google Scholar]
- 16.Sloan, L. M., B. J. Duresko, D. R. Gustafson, and J. E. Rosenblatt. 2008. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 46:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamper, P. D., R. Alcabasa, D. Aird, W. Babiker, J. Wehrlin, I. Ikpeama, and K. C. Carroll. 2009. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J. Clin. Microbiol. 47:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamper, P. D., W. Babiker, R. Alcabasa, D. Aird, J. Wehrlin, I. Ikpeama, L. Gluck, and K. C. Carroll. 2009. Evaluation of a new commercial TaqMan PCR assay for direct detection of the Clostridium difficile toxin B gene in clinical stool specimens. J. Clin. Microbiol. 47:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Berg, R. J., N. Vaessen, H. P. Endtz, T. Schülin, E. R. van der Vorm, and E. J. Kuijper. 2007. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J. Med. Microbiol. 56:36-42. [DOI] [PubMed] [Google Scholar]


