Abstract
Detection of methicillin-resistant Staphylococcus aureus (MRSA) by single-locus PCR assays that target the extremity of the staphylococcal cassette chromosome-mec (SCCmec) and part of the adjacent S. aureus-specific open reading frame gene (orfX) is a significant diagnostic advancement, since it provides real-time detection directly from screening specimens. However, isolates harboring mecA deletions within SCCmec may result in false-positive identification of MRSA in these assays. We characterized 24 methicillin-susceptible S. aureus (MSSA) isolates that tested positive in one such assay to investigate this phenomenon. Seven isolates resembled USA100 and carried SCCmec II elements with mecA deletions that spanned 20 to 46 kbp. The mecA excisions in USA100-resembling isolates appeared to be linked with IS431 transposable elements present in SCCmec II. For 17 isolates that resembled USA400 and/or MSSA476, the identity and possible excision of SCC elements could not be confirmed. The downstream common sequence (dcs) shared by SCCmec I, II, and IV elements was detected in these isolates. Sequence analysis of the chromosomal regions flanking the missing SCC element revealed an intact SCC integration site, a duplicate dcs, and the enterotoxin gene cluster downstream of orfX. An annealing sequence for one of the SCCmec-specific primers (mecii574) in the single-locus PCR assay was identified in the duplicate dcs. In the absence of SCC, a 176-bp amplicon can be generated from this mecii574 annealing sequence to yield a false-positive result. In conclusion, partial SCCmec II excisions via IS431 elements in strains that resembled USA100 and the presence of a duplicate mecii574 annealing sequence in strains that resembled USA400/MSSA476 were identified as causes for false-positive results in a single-locus PCR assay that targets the SCCmec/orfX junction.
Methicillin-resistant Staphylococcus aureus (MRSA) is a multidrug-resistant pathogen associated with significant morbidity, mortality, and hospitalization costs (3, 8, 9, 19, 41). Since 2003, more than 60% of S. aureus infections were caused by MRSA in intensive care units of hospitals belonging to the U.S. National Nosocomial Infection Surveillance system (20). In the last decade, MRSA has also emerged in community settings, causing skin and soft tissue infections and other deep-seated infections in individuals without conventional health care-associated risk factors for MRSA (10). The resistance to β-lactam antibiotics in MRSA is mediated by an altered penicillin-binding protein (PBP2a) encoded by the mecA gene (13). This resistance determinant resides on a mobile genetic element termed the staphylococcal cassette chromosome-mec (SCCmec) that integrates downstream of a S. aureus-specific open reading frame (orfX) (11, 14). Eight major SCCmec types ranging from 22 to 64 kbp in size have been described to date (1, 16-18, 26, 34, 44).
A single-locus PCR assay, utilizing a S. aureus-specific orfX gene primer (Xsau325) and a combination of SCCmec-specific primers that anneal at the extremity of SCCmec to amplify the SCCmec/orfX junction, was first proposed by Huletsky et al. for MRSA detection (15). Since mecA resides on SCCmec, detection of the SCCmec/orfX junction is considered a surrogate for the detection of MRSA. There are now a number of commercially available assays that identify MRSA based on the site-specific integration of SCCmec at orfX (36, 38, 43). Shortly after the introduction of these tests, there were reports of assay-positive specimens that only contained methicillin-susceptible strains of S. aureus (2, 4, 12, 32, 35-38, 40, 43). Although investigators mentioned SCC homologues and partial SCCmec deletions as plausible reasons for discrepant results, few studies have determined why these “false-positive” test results occurred. In an effort to determine possible causes, we characterized 24 methicillin-susceptible S. aureus (MSSA) isolates, collected from 10 healthcare institutions in the United States and Canada, which produced “false-positive” results in one such single-locus PCR assay.
(This study was presented in part at the American Society for Microbiology 109th General Meeting, Philadelphia, PA, 17 to 21 May 2009.)
MATERIALS AND METHODS
Bacterial isolates.
Twenty-four Staphylococcus aureus that were identified as MRSA by the BD GeneOhm MRSA assay (version 3; BD Diagnostics, Quebec, Quebec, Canada) originating from hospitals in six states (Illinois, Maryland, Massachusetts, Ohio, Indiana, and North Carolina) and four Canadian hospitals in two provinces (Ontario and Quebec) were examined. The S. aureus isolates were grown on Columbia agar supplemented with 5% sheep blood (Oxoid, Nepean, Ontario, Canada) for 16 to 18 h at 35°C. Suspension cultures were prepared by inoculating a single bacterial colony into 5 ml of BD BBL brain heart infusion broth (Becton Dickinson, Sparks, MD) and grown for 16 to 18 h at 35°C with agitation. The S. aureus control strains for molecular analyses included MRSA strain N315 (SCCmec II) (17), USA100 MRSA and USA400 MRSA (27), and MSSA476 (an MSSA strain that resembles the USA400 SmaI pulsed-field type) (7).
Determination of methicillin susceptibility.
Methicillin susceptibility was determined by oxacillin broth microdilution testing in accordance with Clinical and Laboratory Standards Institute guidelines (5, 6). Methicillin susceptibility was further confirmed by PCR detection of the mecA determinant (25) (see below).
PCR template preparation.
DNA templates were prepared by suspending S. aureus colonies in 0.5 ml of neutralizing buffer (30 mM Tris [pH 8.4], 2 mM EDTA [pH 9.0]) that contains 50 μl of 0.1-mm glass beads (Scientific Industries, Bohemia, NY) and heating at 100°C for 2 min. The lysates were vortexed for 2 min and clarified by centrifugation at 20,800 × g for 5 s. Templates for long-range PCR were prepared with a High-Pure PCR template purification kit (Roche Diagnostics, Laval, Quebec, Canada).
Primer synthesis and PCR amplification.
Custom oligonucleotide primers were purchased from Invitrogen (Burlington, Ontario, Canada) (Table 1). PCR amplifications were performed using the GeneAmp PCR System 9700 (Applied Biosystems, Inc., Foster City, CA). Unless otherwise specified, monoplex and multiplex PCR assays for amplicons <1.5 kbp were performed in 25-μl reactions containing 1.25 U of AmpliTaq DNA polymerase, 0.1 mM deoxynucleoside triphosphates, 1.5 mM MgCl2 (Roche Diagnostics), and 0.5 μM concentrations (each) of forward and reverse primers with the following thermocycling parameters: 94°C for 2 min, 30 cycles of 94°C for 1 s, 55°C for 15 s, and 72°C for 7 min. Long-range PCRs were performed in 50-μl reactions using an Expand Long Template PCR system (Roche Diagnostics) with the following parameters: 92°C for 2 min; 10 cycles of 92°C for 10 s, 50°C for 15 s, and 68°C for 10 min; followed by 20 cycles of 92°C for 10 s, 50°C for 15 s, and 68°C for 10 min (with an additional 20-s extension time incorporated per subsequent cycle), and then 68°C for 7 min. PCR amplification products were resolved on 0.5× Tris-borate EDTA agarose gels containing 0.5 μg of ethidium bromide/ml for digital photography under UV illumination.
TABLE 1.
Primers used in this study
| Test and primer | Sequence (5′-3′) | Description | Source or reference |
|---|---|---|---|
| MRSA/PVL detection | |||
| 16S-F | AGA GTT TGA TCA TGG CTC AG | 16S rRNA | 25 |
| 16S-R | GGA CTA CCA GGG TAT CTA AT | ||
| mecA-F | AAA ATC GAT GGT AAA GGT TGG C | mecA | 25 |
| mecA-R | AGT TCT GCA GTA CCG GAT TTG C | ||
| nuc-F | GCG ATT GAT GGT GAT ACG GTT | nuc | 25 |
| nuc-R | AGC CAA GCC TTG ACG AAC TAA AGC | ||
| luk-F | ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A | lukS/F (PVL) | 23 |
| luk-R | GCA TCA AST GTA TTG GAT AGC AAA AGC | ||
| SCC typing | |||
| KDP F1 | AAT CAT CTG CCA TTG GTG ATG C | Locus B (SCCmec II) | 33 |
| KDP R1 | CGA ATG AAG TGA AAG AAA GTG G | ||
| DCS F2 | CAT CCT ATG ATA GCT TGG TC | Locus D (SCCmec II) | 33 |
| DCS R1 | CTA AAT CAT AGC CAT GAC CG | ||
| IS431 P4 | CAG GTC TCT TCA GAT CTA CG | Locus G (SCCmec II) | 33 |
| pUB110 R1 | GAG CCA TAA ACA CCA ATA GCC | ||
| ccr typing | |||
| α1 | AAC CTA TAT CAT CAA TCA GTA CGT | ccrA1 (SCCmec I) | 22 |
| α2 | TAA AGG CAT CAA TGC ACA AAC ACT | ccrA2 (SCCmec II) | 22 |
| βc | ATT GCC TTG ATA ATA GCC ITC T | ccrB1 (SCCmec I) and ccrB2 (SCCmec II) | 22 |
| spa typing | |||
| 932-F | GCC AAA GCG CTA ACC TTT TA | Protein A (spa) | B. Kreiswirth, unpublished |
| 1740-R | TCC AGC TAA TAA CGC TGC AC | data | |
| J1 PCR | |||
| LIR2-F | CGA ATT AAA CAC GCT TGC TC | J1 (SCCmec II) | This study |
| LIR2-R | CAG AAG TCT ACA GGT CTT GC | ||
| Long-range PCR | |||
| 3.0-R | CTC AGA CAG CAA TTT CCC G | Downstream of SCCmec | This study |
| 4.2-R | CCC TAA AAA TAC CTG TAG GGG | ||
| Detection of MGE targets | |||
| MW756-F | TGG TTA GCT ATG AAT GTA GTT GC | Hypothetical protein (νSa3), | 45 |
| MW756-R | GTC CAT CCT CTG TAA TTT TGC | unique to MW2 | |
| sel2-F | ATC ATA CAG TCT TAT CTA ACG GCG | sel2 (νSa3) | This study |
| sel2-R | TCT GGA AGA CCG TAT CCT GTG | ||
| SAS0898-F | GAA GTA GCA GAT ATA TTG GGC G | Intergenic region of SAS0898/0899 | This study |
| SAS0899-R | ATC ACT GGC AAG TCC TAC C | (φSa4) | |
| SAS0922-F | GAG GGA TAA ACT AAT GAA AGA ACG | Hypothetical phage protein | This study |
| SAS0922-R | TCA TAG AAC CTA AAC GTC CCG | (φSa4), unique to MSSA476 | |
| SCCmec/orfX junction detection | |||
| Xsau325 | GGA TCA AAC GGC CTG CAC A | orfX | 15 |
| mecii574 | GTC AAA AAT CAT GAA CCT CAT TAC TTA TG | SCCmec I, II, IV | 15 |
| SCCmec II gene elements detection and sequencing | |||
| kdpB2 | TAA ACT GTG TCA CAC GAT CCA T | kdpB | 22 |
| II-48600R | TCT TAG ATA GTA GCC AAC GGC | J3 of SCCmec II | This study |
MRSA PVL multiplex PCR.
Detection of 16S rRNA, mecA, lukS/F, and nuc genes was performed as a multiplex PCR assay with the following concentrations of primer pairs: 0.5 μM luk, 0.4 μM mecA, 0.3 μM 16S rRNA (internal control), and 0.2 μM nuc (23, 25).
SCCmec type determination.
SCCmec types were determined by using a combination of SCCmec typing assays as described previously (22, 33). The J1 region of SCCmec II was detected by monoplex PCR using the LIR2 primers.
Detection of mobile genetic elements.
The S. aureus pathogenicity island νSa3 present in MW2 (GenBank accession no. BA000033) and the bacteriophage φSa4 associated with MSSA476 (GenBank accession no. BX571857) were detected by PCR (24). The gene targets MW756 (nucleotides [nt] 824637 to 825009) and sel2 (nt 828744 to 829329) were surrogates for νSa3. The intergenic region of SAS0898/SAS0899 (nt 985159 to 985508) and SAS0922 (nt 994863 to 995288) were surrogates for φSa4.
DNA sequence determination.
Long-range PCR products were purified by using a High-Pure PCR product purification kit (Roche Diagnostics) for sequence determination at the TCAG sequencing facility (Hospital for Sick Children, Toronto, Ontario, Canada).
spa typing.
Strain typing based on the polymorphic X-region of the protein A gene (spa) was performed essentially as described previously (39), except that the primers 932-F and 1740-R were used for amplification and sequencing of the spa amplicon. The Kreiswirth spa nomenclature obtained by the spa typing tool (http://fortinbras.us/cgi-bin/spatyper/spaTyper.pl) was adopted for the present study.
PFGE.
The Canadian standardized protocol for pulsed-field gel electrophoresis (PFGE) typing of MRSA was adopted for MSSA typing in the present study (28). SmaI-restricted DNA profiles were digitized and analyzed with BioNumerics v6.0 (Applied Maths, Austin, TX). XbaI (Roche Diagnostics)-digested Salmonella serotype Braenderup H9812 DNA was used as the reference standard for band size determination in BioNumerics. Alternatively, the Lambda Ladder PFG Marker (New England Biolabs, Pickering, Ontario, Canada) was used as the reference standard in Southern analyses.
Southern hybridization.
Southern blot hybridization and detection were performed by using an ECL direct nucleic acid labeling and detection system (GE Healthcare, Piscataway, NJ). Briefly, SmaI-digested chromosomal DNA was resolved by PFGE and transferred onto Hybond-N+ nylon membrane (GE Healthcare). DNA probe corresponding to the 342-bp locus D (33) was used for hybridization, and Kodak BioMax Light film (Rochester, NY) was used for signal detection.
RFLP.
Restriction fragment length polymorphism (RFLP) of long-range PCR amplicons was used to assess the similarity between the amplified products. Restriction enzymes BclI, EcoRI, HincII, HindIII, and XbaI (Roche Diagnostics) were used according to the manufacturer's instructions.
RESULTS
Molecular and phenotypic characterization.
The 24 isolates were determined to be Staphylococcus aureus by the presence of the S. aureus-specific nuc gene. Susceptibility to methicillin was determined by broth microdilution and confirmed by the absence of mecA with PCR. These methicillin-susceptible S. aureus (MSSA) did not carry the genes for PVL. The downstream common sequence (locus D, dcs) shared by SCCmec I, II, and IV was detected in all 24 MSSA isolates (Table 2). Locus B from the kdp gene cluster of SCCmec II was detected in isolates IDI2406, IDI2407, IDI2445, IDI2595, IDI2515, and IDI2643. In addition, isolates IDI2515 and IDI2683 were also positive for locus G (linearized pUB110 plasmid) that is typically present in SCCmec II. Apart from IDI2683, all SCCmec II isolates were positive for the SCCmec II-specific J1 target. The PCR target for the ccr gene complex of SCCmec II was detected in isolates IDI2406, IDI2407, IDI2445, and IDI2595.
TABLE 2.
Characteristics of S. aureus isolates
| Isolate | Source | PCR targets downstream of mecA |
PCR targets upstream of mecA |
Molecular typing |
νSa3 (MW756/sel2) | φSa4 (SAS0898-0899/ SAS0922) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus D | Locus G | ccr | Locus B | J1 regiona | PFGE | spa | ||||
| IDI2406 | Hospital A (Canada) | + | − | ccrAB2 | + | SCCmec II | USA100 | TJMBMDMGMK | NAb | NA |
| IDI2407 | Hospital A | + | − | ccrAB2 | + | SCCmec II | USA100 | TJMBMDMGMK | NA | NA |
| IDI2445 | Hospital B (United States) | + | − | ccrAB2 | + | SCCmec II | USA100 | TJMBMDMGMK | NA | NA |
| IDI2515 | Hospital C (Canada) | + | + | - | + | SCCmec II | USA100 | TJMBMDMGMK | NA | NA |
| IDI2595 | Hospital D (Canada) | + | − | ccrAB2 | + | SCCmec II | USA100 | TJMBMDMGMK | NA | NA |
| IDI2643 | Hospital E (United States) | + | − | - | + | SCCmec II | USA100 | TJMBMDMGMK | NA | NA |
| IDI2683 | Unknown | + | + | - | − | - | USA100 | TMDMGMK | NA | NA |
| IDI2235 | Hospital F (United States) | + | − | - | − | - | USA400 | TJFKBPE | -/- | -/- |
| IDI2239 | Hospital F | + | − | - | − | - | USA400 | UJFKPE | -/- | -/- |
| IDI2247 | Hospital F | + | − | - | − | - | USA400 | UJFKBPE | -/- | -/- |
| IDI2250 | Hospital F | + | − | - | − | - | USA400 | UJFKBPE | -/- | -/- |
| IDI2253 | Hospital F | + | − | - | − | - | USA400 | UJFKBPE | -/- | -/- |
| IDI2256 | Hospital F | + | − | - | − | - | USA400 | TJFKBPE | -/- | -/- |
| IDI2491 | Hospital G (United States) | + | − | - | − | - | USA400 | UJFKPE | -/- | -/- |
| IDI2492 | Hospital H (United States) | + | − | - | − | - | USA400 | ULE | -/- | -/- |
| IDI2500 | Hospital I (United States) | + | − | - | − | - | USA400 | UJFKBPE | -/- | -/- |
| IDI2545 | Hospital A | + | − | - | − | - | USA400 | UJFKBPE | -/- | -/- |
| OTT004 | Hospital J (Canada) | + | − | - | − | - | USA400 | U4FKBPE | -/- | -/- |
| OTT009 | Hospital J | + | − | - | − | - | USA400 | UJFKAFK | -/- | +/+ |
| OTT3498 | Hospital J | + | − | - | − | - | USA400 | UJFFLE | -/- | +/+ |
| OTT3573 | Hospital J | + | − | - | − | - | USA400 | UJFFKBPE | -/- | -/- |
| OTT3574 | Hospital J | + | − | - | − | - | USA400 | U4FKBPE | -/- | -/- |
| OTT3596 | Hospital J | + | − | - | − | - | USA400 | UJGBGGJAGJ | -/- | -/- |
| OTT3598 | Hospital J | + | − | - | − | - | USA400 | UJFKBPE | -/- | -/- |
| USA400 | Sunnybrook Health Sciences Centre | + | − | ccrAB2 | − | SCCmec IVa | USA400 | UJJFKBPE | +/+ | -/- |
| MSSA476 | PHRI | + | − | ccrAB1 | − | - | USA400 | UJJFKBPE | -/- | +/+ |
J1 region, upstream of the ccr genes.
NA, not applicable.
Strain typing and genetic deletion determination.
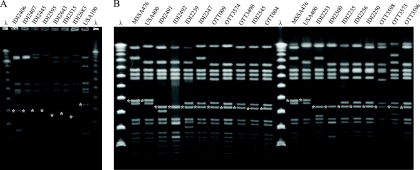
The PFGE and spa typing results are summarized in Table 2. The seven isolates with remnants of SCCmec II resembled USA100 MRSA in their pulsed-field profile and spa type; the remaining 17 isolates resembled USA400 MRSA and/or MSSA476 (Fig. 1). Southern blot hybridization with the locus D probe detected genetic deletions of approximately 18 to 45 kbp in the SCCmec II isolates that resembled USA100, and approximately 20 kbp in the USA400/MSSA476-resembling isolates (Fig. 1).
FIG. 1.
PFGE of MSSA isolates that resembled USA100 (A) and USA400/MSSA476 (B) were resolved with the Lambda Ladder PFG Marker (λ) for Southern blotting. SmaI fragments that hybridized with the locus D probe are indicated with an asterisk.
Mapping of deletion junction.
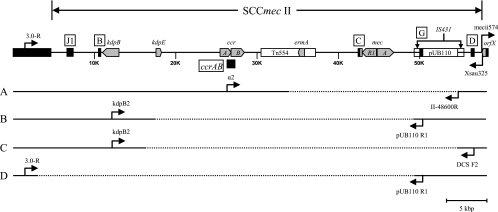
In order to map the deletion junction, long-range PCR using upstream and downstream primers based on the positive PCR targets that flanked the missing mecA determinant was performed. Four SCCmec II deletion patterns were observed among the seven isolates that resembled USA100 (Fig. 2). Long-range PCR with primers α2 (ccrAB2) and II-48600R (pUB110) for isolates IDI2406, IDI2407, IDI2445, and IDI2595 yielded an ∼8-kbp product (pattern A). RFLP analyses using EcoRI, HindIII, HincII, or XbaI indicated the PCR product from IDI2595 is different from the other three isolates (data not shown). Sequence determination revealed nt 33837 to 54641 (20.8 kbp) of SCCmec II (GenBank accession no. D86934.2) were missing in IDI2406, IDI2407, and IDI2445, and nt 34927 to 54641 (19.7 kbp) were missing in IDI2595. For IDI2515 (pattern B) the primers kdpB2 (kdpB) and pUB110 R1 (locus G) were used to successfully amplify a 9-kbp product, and for IDI2643 (pattern C) the primers kdpB2 (kdpB) and DCS F2 (locus D) amplified an 8-kbp product. Deletions from nt 17486 to 49295 (31.8 kbp) and nt 16339 to 54641 (38.3 kbp) were identified for IDI2515 and IDI2643, respectively. For IDI2683 (pattern D), PCR results suggested all SCCmec II sequences upstream of the pUB110 plasmid were missing. As such, a chromosomal primer (3.0-R) upstream of the SCC integration site was designed for amplification with primer pUB110 R1. A 2.6-kbp product was obtained; sequence data confirmed that nt 3012 to 49295 (46.3 kbp) was absent in IDI2683.
FIG. 2.
Internal deletions of SCCmec II. The prototypical SCCmec II is illustrated with key genetic elements (gray chevron) and locations of PCR targets (black box) on the top. PCR primers (arrows) used in long-range PCRs to span the deleted region (segmented line) are as indicated for the four deletion patterns: A (IDI2406, IDI2407, IDI2445, and IDI2595), B (IDI2515), C (IDI2643), and D (IDI2683).
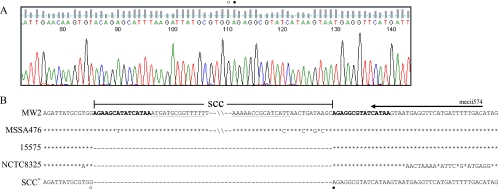
For the 17 isolates that resembled USA400/MSSA476, no SCC remnant other than locus D was detected. Therefore, a chromosomal primer (4.2-R) that anneals at nt 65312 to 65292 in the MW2 genome (equivalent to nt 64019 to 63999 in MSSA476) was designed for long-range PCR in conjunction with the Xsau325 primer that anneals at nt. 34013 to 34031 of MW2 and MSSA476. A 7-kbp PCR product with identical RFLP patterns when digested with BclI, HincII, or XbaI (data not shown) was obtained. The sequence of the amplicons was identical to MSSA476 without SCCfar (nt 34150 to 56985), MW2 without SCCmec IVa (nt 34150 to 58278), the SCC integration site of MSSA strain 15575 (GenBank accession no. EU272079), MSSA strain NCTC8325, and highly homologous to several SCCmec-excisant strains generated in vitro (16, 31) (Fig. 3).
FIG. 3.
SCC integration site in isolates that resembled USA400/MSSA476. (A) A representative chromatogram of the Xsau325/4.2-R amplicon sequenced with the Xsau325 primer. The SCC boundaries are denoted with open and closed circles. (B) Sequence alignment of the SCC borders delineated from the 17 isolates that resembled USA400/MSSA476 (SCC−) with MW2, MSSA476, and S. aureus strain 15575, and NCTC8325 (GenBank accession no. CP000253) illustrating the missing SCC element that encompassed the direct repeat sequence (in boldface) at the left border to the inverted repeat sequence (underlined) on the right border. The secondary mecii574 annealing sequence is illustrated with an arrow. The open circle corresponds to nt 34149 (MW2 and MSSA476), 608 (15575), and 34151 (NCTC8325); the closed circle corresponds to nt 58279 (MW2), 56986 (MSSA476), 609 (15575), and 34152 (NTCT8325).
Detection of mobile genetic elements.
In the absence of their SCC elements, detection of pathogenicity islands and bacteriophages that typically associate with MW2 and MSSA476 was performed to help identify the 17 USA400/MSSA476-resembling strains. The φSa4-associated genes were detected in isolates OTT009 and OTT3498. None of the gene targets present on either mobile genetic elements were detected in the remaining 15 isolates (Table 2).
DISCUSSION
Single-locus PCR assays based on the site-specific integration of SCCmec at orfX provide real-time identification of MRSA directly from specimens. There have been reports describing S. aureus isolates containing SCCmec “remnants” (without mecA), and misidentified as MRSA in single-locus PCR assays (15, 35, 36, 40). In most cases, these MSSA isolates were not fully characterized to elucidate the cause of the misidentification. In the report by Shore et al., the molecular characterization of MSSA isolates with residual SCCmec elements provided one explanation of the false-positive reactions in single-locus PCR assays, although this present study characterized only three MSSA isolates (40).
In the present study, we characterized 24 MSSA isolates, from geographically diverse regions of North America, which were detected as MRSA with a single-locus PCR assay, and identified two possible explanations for the discrepant results. Molecular typing and PCR analyses identified seven isolates that resembled USA100 (spa, TJMBMDMGMK and related) with remnants of SCCmec II. Internal deletions of SCCmec II terminated at IS431 sequences (nt 49295 to 50086 and 54642 to 55432 in SCCmec II) at the 3′ border. IS431 elements are ubiquitous in staphylococci and play a role in gene transfer (21, 42). Partial SCCmec II excisions at IS431 sequences during in vitro exposure to vancomycin have been observed, possibly as a fitness compensation mechanism (30). Therefore, it is likely the excision events in SCCmec II isolates from the present study were mediated through IS431.
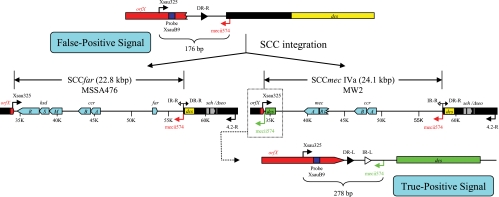
There were 17 isolates that resembled USA400/MSSA476 with undetermined SCC elements (spa, UJFKBPE and related). Characterization of these isolates led to the discovery of a second cause of false-positive results in the single-locus PCR assay. The annealing sequence for the SCCmec-specific primer (mecii574) is located in the dcs region (GenBank annotation MW0025) of SCCmec IVa in MW2. A homologue of MW0025, which also contains a mecii574 annealing sequence, exists adjacent to the SCC integration site in both MW2 and MSSA476 (GenBank annotations MW0048 and SAS0048, respectively). The mecii574 sequence from within SCCmec IVa yields the intended 278-bp amplicon for positive MRSA identification in the single-locus PCR assay. However, the absence of an intact SCCmec IVa (or SCCfar) positions the secondary mecii574 annealing sequence immediately downstream of the orfX-specific primer (Xsau325). In this configuration, a 176-bp amplicon can be generated and detected as a false-positive signal (Fig. 4).
FIG. 4.
Absence of SCC elements in USA400 or MSSA476 strains. In the absence of an SCC, the secondary mecii574 annealing sequence (in red) is located 176 bp downstream of the Xsau325 primer to yield a false-positive signal in single-locus PCR assay (top). The acquisition of SCC in MW2 and MSSA476 prohibits amplification between Xsau325 and the secondary mecii574 sequence (middle). However, the mecii574 sequence (in green) from within SCCmec IVa in MW2 does yield the intended 278-bp amplicon for positive MRSA identification (bottom).
The identity of the USA400/MSSA476-resembling isolates remains unclear. Sequence data identified the presence of the putative transposase and enterotoxin genes (seh and Δseo) downstream of an intact SCC integration site. The enterotoxin gene cluster has been proposed to block the excision of SCCmec IVa in MW2 (29). Apart from two isolates that contained the MSSA476-associated bacteriophage (φSa4), mobile genetic elements associated with MW2 (νSa3) and MSSA476 were not detected in the USA400/MSSA476 isolates. Therefore, it is plausible that the irreversible integration of SCC upstream of the enterotoxin gene cluster had never occurred, and these USA400/MSSA476-resembling isolates may be MSSA strains where the acquisitions of mobile genetic elements such as SCC, pathogenicity islands, and bacteriophages have not taken place to derive the contemporary MW2 and MSSA476 strains. A comprehensive surveillance and characterization of MSSA isolates that test positive in single-locus PCR assays would be beneficial in determining the frequency of false MRSA identification due to the presence of dcs at the SCC integration site and in elucidating the identity of these isolates.
In summary, we determined that the single-locus PCR assay for MRSA detection performed as it was designed, to detect a SCCmec/orfX junction. However, detection of the SCCmec/orfX junction by itself may not be sufficient to confirm the presence of an intact SCCmec element. Our investigation of SCCmec II excisant strains revealed that IS431 elements may have been responsible for partial excisions in SCCmec type II that encompassed the mecA determinant. In addition, we determined that USA400/MSSA476-resembling isolates where SCC integration has not taken place would also yield a positive reaction in the single-locus PCR assay due to the presence of an additional mecii574 annealing sequence at the SCC integration site. Ongoing surveillance and molecular characterization of such isolates would be beneficial for future improvement in the performance of these MRSA screening tools.
Acknowledgments
This study was supported by BD Diagnostics.
We thank BD Diagnostics and Marc Desjardins (The Ottawa Hospital, Ontario, Canada) for contributing MSSA strains for this study, Teruyo Ito (Juntendo University, Tokyo, Japan) for strain N315, and Barry Kreiswirth (Public Health Research Institute, New Jersey) for strain MSSA476.
Footnotes
Published ahead of print on 28 July 2010.
REFERENCES
- 1.Berglund, C., T. Ito, M. Ikeda, X. X. Ma, B. Söderquist, and K. Hiramatsu. 2008. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 52:3512-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, E. J., E. A. Grabsch, S. A. Ballard, B. Mayall, S. Xie, R. Martin, and M. L. Grayson. 2006. Concurrent analysis of nose and groin swab specimens by the IDI-MRSA PCR assay is comparable to analysis by individual-specimen PCR and routine culture assays for detection of colonization by methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2904-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot, S. I., K. H. Vandewoude, E. A. Hoste, and F. A. Colardyn. 2002. Outcome and attributable mortality in critically ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch. Intern. Med. 162:2229-2235. [DOI] [PubMed] [Google Scholar]
- 4.Brenwald, N. P., N. Baker, and B. Oppenheim. 2010. Feasibility study of a real-time PCR test for methicillin-resistant Staphylococcus aureus in a point of care setting. J. Hosp. Infect. 74:245-249. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Corkill, J. E., J. J. Anson, P. Griffiths, and C. A. Hart. 2004. Detection of elements of the staphylococcal cassette chromosome (SCC) in a methicillin-susceptible (mecA gene negative) homologue of a fucidin-resistant MRSA. J. Antimicrob. Chemother. 54:229-231. [DOI] [PubMed] [Google Scholar]
- 8.Cosgrove, S. E., Y. Qi, K. S. Kaye, S. Harbarth, A. W. Karchmer, and Y. Carmeli. 2005. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 26:166-174. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 10.Deresinski, S. 2005. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin. Infect. Dis. 40:562-573. [DOI] [PubMed] [Google Scholar]
- 11.Deurenberg, R. H., C. Vink, S. Kalenic, A. W. Friedrich, C. A. Bruggeman, and E. E. Stobberingh. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13:222-235. [DOI] [PubMed] [Google Scholar]
- 12.Farley, J. E., P. D. Stamper, T. Ross, M. Cai, S. Speser, and K. C. Carroll. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J. Clin. Microbiol. 46:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., Y. Katayama, H. Yuzawa, and T. Ito. 2002. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 292:67-74. [DOI] [PubMed] [Google Scholar]
- 15.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, T., P. I. Oh, and A. E. Simor. 2001. The economic impact of methicillin-resistant Staphylococcus aureus in Canadian hospitals. Infect. Control Hosp. Epidemiol. 22:99-104. [DOI] [PubMed] [Google Scholar]
- 20.Klevens, R. M., J. R. Edwards, F. C. Tenover, L. C. McDonald, T. Horan, R. Gaynes, and the National Nosocomial Infections Surveillance System. 2006. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin. Infect. Dis. 42:389-391. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, N., M. M. Alam, and S. Urasawa. 2001. Genomic rearrangement of the mec regulator region mediated by insertion of IS431 in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45:335-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay, J. A. 2008. Staphylococcus aureus evolution: lineages and mobile genetic elements (MGEs), p. 45-69. In J. A. Lindsay (ed.), Staphylococcus molecular genetics. Caister Academic Press, Norfolk, United Kingdom.
- 25.Louie, L., J. Goodfellow, P. Mathieu, A. Glatt, M. Louie, and A. E. Simor. 2002. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 40:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulvey, M. R., L. Chui, J. Ismail, L. Louie, C. Murphy, N. Chang, M. Alfa, and the Canadian Committee for the Standardization of Molecular Methods. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noto, M. J., and G. L. Archer. 2006. A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec) type IV is deficient in CcrAB-mediated SCCmec excision. Antimicrob. Agents Chemother. 50:2782-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noto, M. J., P. M. Fox, and G. L. Archer. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 52:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noto, M. J., B. N. Kreiswirth, A. B. Monk, and G. L. Archer. 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 190:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberdorfer, K., S. Pohl, M. Frey, K. Heeg, and C. Wendt. 2006. Evaluation of a single-locus real-time polymerase chain reaction as a screening test for specific detection of methicillin-resistant Staphylococcus aureus in ICU patients. Eur. J. Clin. Microbiol. Infect. Dis. 25:657-663. [DOI] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira, D. C., C. Milheiriço, and H. de Lencastre. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 50:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paule, S. M., D. M. Hacek, B. Kufner, K. Truchon, R. B. Thomson, Jr., K. L. Kaul, A. Robicsek, and L. R. Peterson. 2007. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus test before and during high-volume clinical use. J. Clin. Microbiol. 45:2993-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossney, A. S., C. M. Herra, G. I. Brennan, P. M. Morgan, and B. O'Connell. 2008. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J. Clin. Microbiol. 46:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossney, A. S., C. M. Herra, M. M. Fitzgibbon, P. M. Morgan, M. J. Lawrence, and B. O'Connell. 2007. Evaluation of the IDI-MRSA assay on the SmartCycler real-time PCR platform for rapid detection of MRSA from screening specimens. Eur. J. Clin. Microbiol. Infect. Dis. 26:459-466. [DOI] [PubMed] [Google Scholar]
- 38.Sherlock, O., A. Dolan, and H. Humphreys. 2010. MRSA screening: can one swab be used for both culture and rapid testing? An evaluation of chromogenic culture and subsequent Hain GenoQuick PCR amplification/detection. Clin. Microbiol. Infect. 16:955-959. [DOI] [PubMed] [Google Scholar]
- 39.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shore, A. C., A. S. Rossney, B. O'Connell, C. M. Herra, D. J. Sullivan, H. Humphreys, and D. C. Coleman. 2008. Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA. Antimicrob. Agents Chemother. 52:4407-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shorr, A. F., Y. P. Tabak, V. Gupta, R. S. Johannes, L. Z. Liu, and M. H. Kollef. 2006. Morbidity and cost burden of methicillin-resistant Staphylococcus aureus in early onset ventilator-associated pneumonia. Crit. Care 10:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada, A., Y. Katayama, K. Hiramatsu, and T. Yokota. 1991. Southern hybridization analysis of the mecA deletion from methicillin-resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 176:1319-1325. [DOI] [PubMed] [Google Scholar]
- 43.Warren, D. K., R. S. Liao, L. R. Merz, M. Eveland, and W. M. Dunne, Jr. 2004. Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. J. Clin. Microbiol. 42:5578-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, K., J. A. McClure, S. Elsayed, and J. M. Conly. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, K., J. A. McClure, S. Elsayed, J. Tan, and J. M. Conly. 2008. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J. Infect. Dis. 197:195-204. [DOI] [PubMed] [Google Scholar]