Abstract
Distinguishing nontypeable Haemophilus influenzae and Haemophilus haemolyticus isolates by outer membrane protein (OMP) P6 gene sequencing is complicated by sequence variants in isolates. Further testing using RapID NH and multilocus sequence analysis may not help identify some isolates. Translated OMP P6 gene sequences are not conserved among all isolates presumed to be H. influenzae.
Recently, Murphy et al. (9) identified Haemophilus haemolyticus as a nasopharyngeal (NP) colonizer of humans and cautioned that this commensal might be confused with Haemophilus influenzae using routine microbiological methods. In that study, nontypeable H. influenzae (NTHi) was differentiated from H. haemolyticus using molecular techniques relying on the conserved sequence of P6 at 4 specific key residues. Residues 33, 42, 59, and 61 of outer membrane protein (OMP) P6 are alanine, alanine, aspartate, and threonine, respectively, in NTHi, while the corresponding residues in H. haemolyticus are glycine, serine, asparagine, and glutamate. In particular, the conformation of the monoclonal antibody (MAb) 7F3 epitope has been shown to depend on amino acids 59 and 61. A conclusion from that work was that the two Haemophilus species should be correctly identified in studies evaluating immune responses following NTHi colonization or infection (7, 9).
Our group is conducting a multiyear, prospective study to evaluate the immune response of children to NP colonization and acute otitis media (AOM) caused by NTHi from children aged 6 months to 30 months (1, 11). Following the recommendation of Murphy et al. (9), we undertook to distinguish NTHi and H. haemolyticus during NP and oropharyngeal (OP) colonization and AOM among our pediatric samples. Presumptive identification of NTHi involved standard microbiologic protocols, but these proved inadequate to segregate NTHi and H. haemolyticus. The addition of Haemophilus QuadID plates that differentiate species of Haemophilus by their use of heme factors and hemolytic capabilities was also not sufficient (3, 5). Therefore, we elected to distinguish NTHi strains from H. haemolyticus strains based on the sequence of P6 that differs between NTHi and H. haemolyticus. A total of 163 viable isolates were tested, with 62 from OP samples, 50 from NP samples, and 51 from middle ear fluid (MEF) samples from children with AOM. All isolates from the NP and MEF samples were shown to have the same translated amino acids at the 4 key residues as NTHi. Of the 63 from the OP, 51 isolates were NTHi, 12 were H. haemolyticus, and 6 could not be determined by OMP P6 gene sequencing.
We also found that 8 of the 163 NTHi isolates had variations in their OMP P6 translated amino acid sequence at sites outside the 4 key residues. For these 8 isolates, the amino acid substitutions could affect protein structure because the substitutions are nonconservative. OP samples were the source of isolation of 5 of these 8 isolates; others were isolated from NP (n = 2) or MEF (n = 1) samples.
Initial tests using the RapID NH system identified all 6 isolates that could not be determined by OMP P6 gene sequencing as being NTHi. However, the accuracy of the RapID NH system was not sufficient in identification (2). To better identify these isolates, we amplified the 7 housekeeping genes in the multilocus sequence typing (MLST) scheme for H. influenzae (6). Upon sequencing, we were able to amplify all 7 genes for 3 of the isolates (HH13, HH22, and HH39). We were able to amplify all but the adk gene (HH75 and HH77) in two other isolates and all but the fucK gene (HH86) in the remaining isolate. In addition, all 7 genes were amplified and sequenced from the two isolates (HH2 and HH5) from NP samples that had amino acid variations outside the 4 key residues.
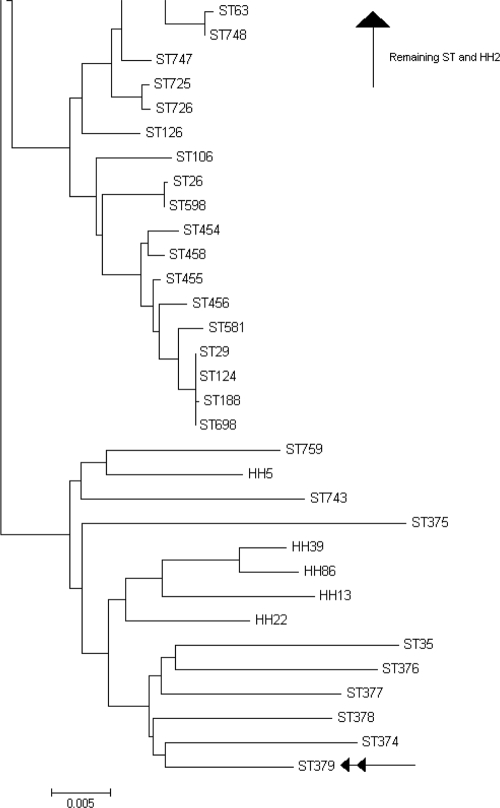
The sequences of 6 of the housekeeping genes (all except fucK) of the 4 isolates (HH13, HH22, HH39, and HH86) that could not be determined by OMP P6 gene sequencing and the two isolates (HH2 and HH5) with amino acid variations outside the 4 key residues were concatenated (2,712 nucleotides). In addition, 760 sequence types from the MLST website, including some H. haemolyticus strains, as well as cryptic genospecies of H. influenzae, were concatenated. A neighbor-joining dendrogram was generated from the concatenated sequences using MEGA version 4.0 (12). The results presented in Fig. 1 show HH5, HH13, HH22, HH39, and HH86 clustering with a known H. haemolyticus strain and cryptic genospecies of NTHi (8, 9). HH2 is a new strain, but it clusters with other H. influenzae sequence types.
FIG. 1.
Neighbor-joining dendrogram based on concatenated sequences of the housekeeping genes adk, atpG, frdB, mdh, pgi, and recA. Seven hundred sixty H. influenzae sequences were retrieved from the MLST database and are identified by the letters “ST” followed by a number. Samples that had their OMP P6 gene sequenced are identified with the letters “HH” followed by a number. ST379, identified by a double arrow, is the concatenated sequence of ATCC 33390, an H. haemolyticus strain.
As mentioned before, the fucK gene in HH13, HH22, and HH39 was successfully sequenced. It has been shown that fucK can be a marker to identify H. influenzae, because other Haemophilus spp. do not carry this gene (10). However, at this time, we cannot fully discriminate the identity of these isolates.
This is the first study to identify variations in the OMP P6 gene among NTHi respiratory isolates in humans, and the results further complicate the use of OMP P6 gene sequencing as a tool for identification. Our discovery that ∼10% of NTHi strains in a pediatric population have variations in P6 protein structure suggests that while the amino acid sequencing method proposed by Murphy et al. (9) to distinguish NTHi from H. haemolyticus is informative, it should not be relied upon alone.
Previous studies have also relied upon the reactivity between MAb 7F3 and OMP P6 as a method to distinguish NTHi from H. haemolyticus (7, 9). A previous study by McCrea et al. (5) found that 3 of 88 (3.4%) NTHi strains they studied did not react with MAb 7F3; these results may have been due to amino acid substitutions at key 7F3 antibody residues, similar to what we have seen in this study. However, McCrea et al. did not comment or pursue this finding further; one of the NTHi isolates with atypical OMP P6 in our study population did not bind to MAb 7F3. This NTHi isolate with variant OMP P6 differs from a previously described invasive urogenital NTHi isolate which was also incapable of binding to MAb 7F3 (8).
In summary, our study shows that not all strains of NTHi can be reliably distinguished from H. haemolyticus based on amino acid sequencing and MAb 7F3 reactivity. Other genes shown to be common in most NTHi strains but not in H. haemolyticus, including iga, sodC, and hap, may be better molecular traits than OMP P6 to identify NTHi bacteria (4, 5, 10). Studies are now under way by our group to specifically evaluate the effect of structural variations in OMP P6 induced by the amino acid substitutions we have identified.
Acknowledgments
This work was supported by NIH grant no. NIDCD RO1 008671.
The authors have no conflicts to declare in relationship to this article.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Casey, J. R., D. G. Adlowitz, and M. E. Pichichero. 2010. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 29:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doern, G. V., and K. C. Chapin. 1984. Laboratory identification of Haemophilus influenzae: effects of basal media on the results of the satellitism test and evaluation of the RapID NH system. J. Clin. Microbiol. 20:599-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fung, W. W. M., C. A. O'Dwyer, S. Sinha, A. L. Brauer, T. F. Murphy, J. S. Kroll, and P. R. Langford. 2006. Presence of copper- and zinc-containing superoxide dismutase in commensal Haemophilus haemolyticus isolates can be used as a marker to discriminate them from nontypeable H. influenzae isolates. J. Clin. Microbiol. 44:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilian, M., J. Mestecky, and R. E. Schrohenloher. 1979. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCrea, K. W., J. Xie, N. LaCross, M. Patel, D. Mukundan, T. F. Murphy, C. F. Marrs, and J. R. Gilsdorf. 2008. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J. Clin. Microbiol. 46:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukundan, D., Z. Ecevit, M. Patel, C. F. Marrs, and J. R. Gilsdorf. 2007. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J. Clin. Microbiol. 45:3207-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy, T. F., C. Kirkham, and D. J. Sikkema. 1992. Neonatal, urogenital isolates of biotype 4 nontypeable Haemophilus influenzae express a variant P6 outer membrane protein molecule. Infect. Immun. 60:2016-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81-89. [DOI] [PubMed] [Google Scholar]
- 10.Nørskov-Lauritsen, N., M. D. Overballe, and M. Kilian. 2009. Delineation of the species Haemophilus influenzae by phenotype, multilocus sequence phylogeny, and detection of marker genes. J. Bacteriol. 191:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabirov, A., J. R. Casey, T. F. Murphy, and M. E. Pichichero. 2009. Breast-feeding is associated with a reduced frequency of acute otitis media and high serum antibody levels against NTHi and outer membrane protein vaccine antigen candidate P6. Pediatr. Res. 66:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]