Abstract
Culturing newly identified human lung pathogens from clinical sample isolates can represent a daunting task, with problems ranging from low levels of pathogens to the presence of growth suppressive factors in the specimens, compounded by the lack of a suitable tissue culture system. However, it is critical to develop suitable in vitro platforms to isolate and characterize the replication kinetics and pathogenesis of recently identified human pathogens. HCoV-HKU1, a human coronavirus identified in a clinical sample from a patient with severe pneumonia, has been a major challenge for successful propagation on all immortalized cells tested to date. To determine if HCoV-HKU1 could replicate in in vitro models of human ciliated airway epithelial cell cultures (HAE) that recapitulate the morphology, biochemistry, and physiology of the human airway epithelium, the apical surfaces of HAE were inoculated with a clinical sample of HCoV-HKU1 (Cean1 strain). High virus yields were found for several days postinoculation and electron micrograph, Northern blot, and immunofluorescence data confirmed that HCoV-HKU1 replicated efficiently within ciliated cells, demonstrating that this cell type is infected by all human coronaviruses identified to date. Antiserum directed against human leukocyte antigen C (HLA-C) failed to attenuate HCoV-HKU1 infection and replication in HAE, suggesting that HLA-C is not required for HCoV-HKU1 infection of the human ciliated airway epithelium. We propose that the HAE model provides a ready platform for molecular studies and characterization of HCoV-HKU1 and in general serves as a robust technology for the recovery, amplification, adaptation, and characterization of novel coronaviruses and other respiratory viruses from clinical material.
About 335 new or emerging infectious diseases have been identified since 1940 (23), and while many threaten human health, the global economy, and national security, respiratory pathogens are of particular public health concern. Using modern methods, several previously unknown viruses have been identified, including respiratory pathogens (1, 18, 27, 54, 57), yet research remains restricted to prevalence and disease association studies since a virus culture system is oftentimes lacking. Immortalized tissue culture cells are adapted to growth in laboratory conditions and, as such, display altered gene expression patterns, which may not be optimal for the replication of fastidious viruses. Primary cell-differentiated culture models provide alternative in vitro model systems closer in nature to the in vivo host tissue environment for infection studies and amplification of pathogens for further characterization. Here, we use an in vitro model of human ciliated airway epithelial cell cultures (HAE) that mimic the properties of the cartilaginous airway epithelium (17) to culture the previously unculturable human coronavirus HKU1 (HCoV-HKU1).
Coronaviruses are important pathogens of humans and animals, causing a range of symptoms depending on the host. Following the severe acute respiratory syndrome (SARS)-CoV epidemic, several new strains of human coronaviruses were identified by molecular techniques, including HCoV-NL63, identified in the Netherlands from an infant with bronchiolitis (54), and HCoV-HKU1, identified in an adult patient with severe pneumonia in Hong Kong (57). HCoV-NL63 has been demonstrated to infect and replicate in both conventional immortalized cells and human ciliated airway cell cultures, producing sufficient amounts of virus for characterization studies of viral replication and pathogenesis and the successful development of an infectious clone (3, 13, 22, 41). In contrast, little is known about HCoV-HKU1, as no in vitro replication model has been identified to date, limiting further investigations of the virus.
Clinical isolates of previously isolated human coronaviruses have been adapted to replicate in standard transformed cell culture; for example, SARS-CoV and HCoV-NL63 replicate efficiently in epithelial monkey kidney cells (VeroE6 and LLC-MK2), HCoV-OC43 in BHK21 cells, and HCoV-229E in MRC5 cells (14, 24, 35, 47, 54, 59). Despite the successful amplification of these human coronaviruses in cell lines, all attempts to date to culture a clinical isolate of HCoV-HKU1 have failed. No HCoV-HKU1 genomic replication was observed after inoculation of standard cell lines previously utilized for virus propagation, including RD (human rhabdomyosarcoma cells), HRT-18 (colorectal adenocarcinoma cells), HEp-2 (human epithelial carcinoma cells), MRC-5 (human lung fibroblast cells), A549 (human lung epithelial adenocarcinoma cells), Caco2 (human colorectal adenocarcinoma cells), Huh-7 (human hepatoma cells), B95a (marmoset B-lymphoblastoid cells), mixed neuron-glia culture, LLC-MK2 (rhesus monkey kidney cells), FRhK-4 (rhesus monkey kidney cells), BSC-1 (African green monkey kidney cells), Vero E6 (African green monkey kidney cells), MDCK (Madin-Darby canine kidney cells), I13.35 (murine macrophage cells), and L929 (murine fibroblast cells) (57).
Here, we use human ciliated airway epithelial cell cultures to successfully propagate HCoV-HKU1 for the first time in vitro. In this culture model, HCoV-HKU1 genome copy numbers increased by several logs over the initial three-day incubation period and electron micrograph, Northern blot, and immunofluorescence data confirmed HKU1 replication in HAE and that ciliated cells were the preferential target for virus infection, the same cell type infected by all human coronaviruses tested so far in these model systems.
MATERIALS AND METHODS
Human tracheobronchial epithelial cultures and clinical virus isolate.
Human tracheobronchial epithelial cells were obtained from airway specimens resected from patients undergoing surgery under University of North Carolina Institutional Review Board-approved protocols by the Cystic Fibrosis Center Tissue Culture Core. Primary cells were expanded on plastic to generate passage 1 cells and plated at a density of 2.5 × 105 cells per well on permeable Transwell-COL (12-mm-diameter) supports. Human airway epithelium cultures were generated by provision of an air-liquid interface for 4 to 6 weeks to form well-differentiated, polarized cultures that resemble in vivo pseudostratified mucociliary epithelium (17). A sample containing HCoV-HKU1 virus (nasal aspirate) was obtained as a clinical specimen derived from an individual suffering from an upper respiratory tract infection. The sample was collected in March 2005 at the pediatric department of the University Hospital of Caen, France, and is designated here as HCoV-HKU1 strain Caen1.
Inoculation of HAE and RNA extraction.
Prior to infection, the apical surfaces of HAE were washed three times with phosphate-buffered saline (PBS) and then inoculated via the apical surface with 200 μl of a 1:2-diluted nasal aspirate or a 1:10-diluted viral stock (generated from apical washes from cultures infected with the clinical sample isolate), which was obtained from HAE harvest at 96 h postinfection. Following a 2-h incubation at 32°C, the unbound virus was removed by washing with 500 μl for 10 min at 32°C, and the HAE were maintained at an air-liquid interface for the remainder of the experiment at 32°C. HCoV-HKU1 replication kinetics were determined at specific time points postinoculation (4, 24, 48, 72, and 96 h), 120 μl of culture medium was applied to the apical surface of HAE, and after 10 min of incubation at 32°C the apical sample was harvested. RNA was isolated from the samples by the silica affinity-based Boom extraction method (6) or by Trizol extraction (Invitrogen). The RNA was analyzed by real-time reverse transcriptase (RT) PCR to determine whether viral RNA was present. RNA was also extracted from HAE by Trizol extraction to determine the presence of viral replication-specific subgenomic (sg) mRNA species from total cell lysates.
Real-time reverse transcriptase PCR quantification.
Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase (200 U per reaction; Invitrogen) and 25 ng of random hexamers (Amersham Biosciences) in 10 mM Tris (pH 8.3), 50 mM KCl, 0.1% Triton X-100, 5 mM MgCl2, and 20 μM each deoxynucleoside triphosphate at 37°C for 90 min in a total volume of 40 μl.
Virus yield was determined by real-time PCR, using the Platinum quantitative PCR SuperMix-uracil-DNA glycosylase (UDG; Invitrogen). Five microliters of cDNA was amplified in 50 μl 1× Platinum quantitative PCR SuperMix-uracil-DNA glycosylase (Invitrogen) with 125 nM MgCl2, 10 μM specific probe labeled with FAM (6-carboxyfluorescein) and TAMRA (6-carboxytetramethylrhodamine), and 45 μM each primer. The following primers targeting the N gene were used for HCoV-HKU1 quantification: sense, HKUqPCR5 (5′-CTGGTACGATTTTGCCTCAA-3′); antisense, HKUqPCR3 (5′-CAATCACGTGGACCCAATAAT-3′); and probe, HKUqPCRP (5′-FAM-TTGAAGGCTCAGGAAGGTCTGCTTCTAA-TAMRA-3′). Following UDG treatment for 2 min at 50°C and a denaturation step of 10 min at 95°C, 45 cycles of amplification were performed for 15 s at 95°C and 60 s at 60°C on a Prism 7000 real-time PCR machine (ABI).
Northern blotting.
Total RNA was extracted from HCoV-HKU1-infected HAE and purified using Oligotex mRNA spin column reagents according to the manufacturer's directions (Qiagen, Valencia, CA). RNA was separated on an agarose gel using Northern-Max-Gly (Ambion/Applied Biosystems), transferred to a BrightStar-Plus membrane (Ambion/Applied Biosystems) for 4 h, and cross-linked to the membrane with UV light. The blot was prehybridized and probed with an HCoV-HKU1 nucleocapsid (N)-specific oligodeoxynucleotide probe (CCTGAACGATTTCCAGAGGAGCTbTbCTbACTb), where biotinylated nucleotides are designated with a superscript b. Blots were hybridized overnight and washed with low- and high-stringency buffers as recommended by the manufacturer. Filters were incubated with streptavidin-alkaline phosphatase (AP), washed, and then developed using the chemiluminescent substrate CDP-STAR.
HCoV-HKU1-active TRS elements and sg mRNA generated during viral replication.
The subgenomic mRNA leader (L)-body transcription regulatory sequence (TRS) junctions were identified by sequencing RT-PCR-amplified cDNA from total RNA obtained from an HCoV-HKU1-infected HAE (Trizol, Invitrogen). Briefly, we performed 35-cycle PCR with the 5′ L primer (HKL1_5) and gene-specific 3′ primers (S gene, SL3′ [ACT ACG GTG ATT ACC AAC ATC AAT ATA]; ORF3-4L3′ [CAA GCA ACA CGA CCT CTA GCA GTA AG]; E gene, EL3′ [TAT TTG CAT ATA ATC TTG GTA AGC]; M gene, ML3′ [GAC CCA GTC CAC ATT AAA ATT GAC A]; N gene, 3-163-F15 [ATT ACC TAG GTA CTG GAC CT]), which were designed from full-length sequence and predicted annotations of downstream open reading frames (ORFs) and putative leader-body junctions as previously described in the literature (36, 60). After amplification, the sample was analyzed by electrophoresis on a 0.8% agarose gel and products of discrete sizes were used for sequencing by using the BigDye Terminator kit (ABI) and a model 3100 genetic analyzer (Applied Biosystems). Raw data were processed and analyzed with CodonCode Aligner version 1.52 software (CodonCode Corporation).
Antibody blockade assay.
To determine whether HCoV-HKU1 infection of HAE required human leukocyte antigen C (HLA-C) (8), the apical surfaces of HAE were rinsed and 300 μl of a 1:10 dilution of monoclonal anti-HLA-C (Santa Cruz), a mixture of monoclonal anti-HLA-B/C (Santa Cruz), or a control antibody (anti-angiotensin-converting enzyme 2 [ACE2]) (45) was applied to the apical surfaces of HAE. After 2 h incubation, excess antibody was removed and HCoV-HKU1 (200 μl stock virus diluted 1:1,000) was inoculated onto the apical surface and incubated for an additional 2 h at 32°C. Viral inocula were then removed, cultures were washed, and 30 μl of the original diluted antiserum was returned to the apical surface of HAE. Apical washes were collected at 6, 20, 30, 44, 54, and 72 h postinoculation, RNA was extracted, and titers were assessed by real-time PCR.
Microscopy studies.
For detection of HCoV-HKU1 antigens in HAE, cultures inoculated with HCoV-HKU1 (∼105 genome copies/ml) were fixed in 4% paraformaldehyde (PFA) for 24 h, transferred to 70% ethanol, and prepared as paraffin-embedded histological sections by the UNC Cystic Fibrosis Center Morphology and Morphometrics Core. After deparaffinization, histological sections were incubated for 1 h in phosphate-buffered saline (PBS) containing 3% bovine serum albumin (BSA). Primary antibodies (rabbit polyclonal serum directed against whole mouse hepatitis virus virions, kindly provided by Mark Denison at Vanderbilt University, and mouse monoclonal serum directed against β-tubulin IV [Sigma]) were applied at a 1:50 dilution in PBS with 1% BSA for 2 h and detected with anti-rabbit antibody conjugated to Alexa 488 (Invitrogen) or anti-mouse antibody conjugated to Alexa 594 (Invitrogen). Rabbit preimmune serum was used as a control for the immune serum. Immunofluorescence was visualized with a Leica Leitz DM IRB inverted fluorescence microscope equipped with a cooled color charge-coupled digital camera (MicroPublisher; QImaging).
For visualization of virus by transmission scanning electron microscopy, HAE were apically inoculated as described above, and cultures were fixed in 2% PFA-0.2% glutaraldehyde at 72 h postinoculation exactly as previously described (45).
Full genome sequencing.
HCoV-HKU1 strain Caen1 RNA was reverse transcribed as described above and amplified in a 35-cycle PCR with sets of primers covering the complete genome (primer sequences are available upon request). Amplicons ranging in size from 96 to 660 bp were analyzed on the 0.8% agarose gel and purified, and discrete products were subjected to sequencing. Sequence reactions were performed without column purification, according to the manufacturer's protocol for BigDye Terminator version 1.1 cycle sequencing. Electrophoresis and data collection were performed employing a model 3100 genetic analyzer (Applied Biosystems). Raw data were processed and analyzed with the CodonCode Aligner version 1.52 software (CodonCode Corporation).
Phylogenetic comparison of HKU1 genomes.
Selected genomes from different HKU1 strains were downloaded from GenBank and compared by multiple sequence alignment using ClustalX version 2.011 (26).
Maximum likelihood phylogenetic trees were generated using the PhyML program (20) as implemented in the Geneious package. All trees were generated using the MtREV substitution model with estimated transition/transversion ratios for DNA models and the proportion of invariable sites fixed at zero. Bootstrapping was conducted and generated 100 bootstrapped data sets, and a consensus tree was generated using Consensus from the Phylip package (16). Trees were visualized in the Geneious tree viewer, and the Seaview (19) tool was used for editing and rearranging branches. Sequences selected for comparison included AY597011.2, HKU1_A; AY884001.1, HKU1_B; DQ415896.1, HKU1.N19; DQ415897.1, HKU1.N20; DQ415898.1, HKU1.N21; DQ415899.1, HKU1.N22; DQ415900.1, HKU1.N23; DQ415902.1, HKU1.N25; DQ415903.1, HKU1.N3; DQ415904.1, HKU1.N6; DQ415905.1, HKU1.N7; DQ415906.1, HKU1.N9; DQ415907.1, HKU1.N10; DQ415908.1, HKU1.N11; DQ415909.1, HKU1.N13; DQ415910.1, HKU1.N14; DQ415911.1, HKU1.N15; DQ415912.1, HKU1.N16; DQ415913.1, HKU1.N17; and DQ415914.1, HKU1.N18, NC_006577.2, HKU1.
Nucleotide sequence accession number.
The complete genome sequence of HCoV-HKU1 strain Caen1 was deposited in GenBank (accession number HM034837).
RESULTS
HCoV-HKU1 replication kinetics in HAE.
HCoV-HKU1 was identified in clinical isolates as a human coronavirus in 2005. RNA isolated from clinical samples provided the basic gene order of the HCoV-HKU1 genome and the number of putative viral subgenomic RNA species (Fig. 1). However, all efforts to culture this virus in traditional human and nonhuman cell lines have proven unsuccessful. Previous studies in our laboratory and those of others have demonstrated that in vitro models of well-differentiated airway epithelial cells, including those for human lung cells (HAE), provide a robust model system for studying replication and pathogenesis of human coronaviruses, including SARS-CoV, HCoV-NL63, HCoV-229E, and HCoV-OC43 (3, 13, 45, 56). We and others have shown that such cell models make an excellent primary culturing resource for propagating respiratory virus isolates (3, 11, 13, 45).
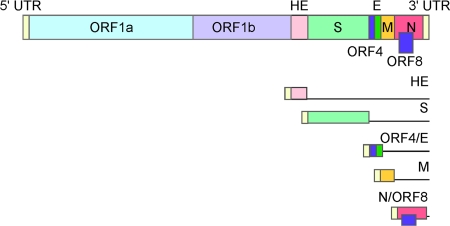
FIG. 1.
HCoV-HKU1 genome and subgenomic RNA species schematic. All open reading frames (ORFs) in the HCoV-HKU1 genome are shown as rectangles in the top schematic, and the proteins expressed from each are indicated. Viral leader sequences are shown as small yellow rectangles at the 5′ end of each line representation of the viral subgenomic RNA species at the bottom of the figure. The 5′-most ORF is translated from each subgenomic RNA, and this ORF is indicated as a colored rectangle for each RNA. UTR, untranslated region; HE, hemagglutinin; S, spike; AP, accessory open reading frame/protein; E, envelope; M, membrane; N, nucleocapsid.
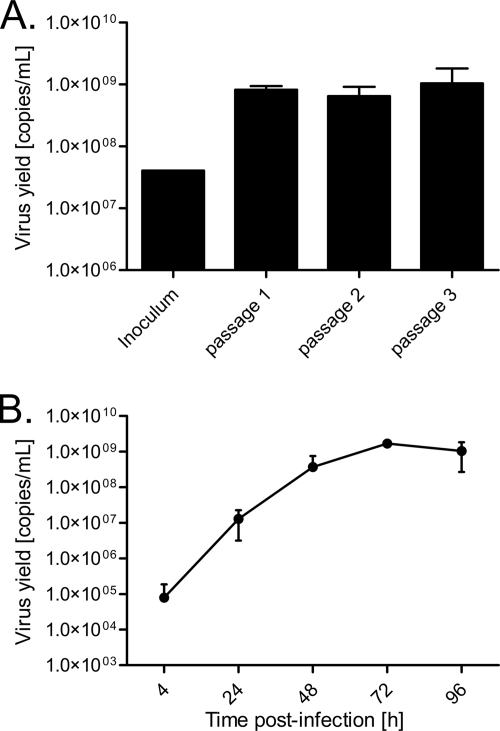
To determine if HAE could be utilized to propagate the HCoV-HKU1, the apical surface of HAE was inoculated and incubated for 2 h at 32°C with a clinical sample containing HCoV-HKU1 from France in 2005 (strain Caen1). For the passage 1 sample, RNA was extracted from both apical washes and basolateral medium at 96 h postinoculation. Real-time reverse transcriptase PCR analysis of apical medium demonstrated an increase in genome copy numbers 96 h postinoculation (Fig. 2 A), the first indication that HCoV-HKU1 in clinical isolates was infectious and could replicate in an in vitro model. Subsequent passages were performed by inoculating virgin cultures with the 96-h apical wash from the previous passage. No virus was detected in the basolateral compartments of HAE (data not shown), suggesting that virus shed only from the apical surface, a finding consistent with previous reports of other human coronaviruses in this culture model (3, 45, 56). In addition, these experiments demonstrated that the genome copies detected in the apical wash were infectious and did not represent defective particles, confirming active HCoV-HKU1 replication in HAE (Fig. 2A).
FIG. 2.
(A) Replication of an HCoV-HKU1 clinical isolate in HAE. HAE were inoculated with diluted nasal aspirates for the first passage (108 copies/ml) and with apical washes from 96 h postinoculation for the subsequent passage. The bars represent real-time PCR analysis of apical media harvested from HCoV-HKU1-infected HAE at 96 h postinoculation or from the inoculating clinical sample. (B) Replication kinetics of HCoV-HKU1 in HAE. Cultures were inoculated with passage 3 virus (108 copies/ml). Data points represent real-time PCR of apical washes from HCoV-HKU1-infected HAE, harvested at the indicated times postinoculation. Data are presented as HCoV-HKU1 RNA copies/ml and are representative of results from experiments performed in duplicate.
Having demonstrated increased genome copy numbers for HCoV-HKU1 following inoculation of the apical surface of HAE, we next determined the viral replication kinetics. HCoV-HKU1 viral stocks were used to inoculate the apical surfaces of HAE, and at 4, 24, 48, 72, and 96 h postinoculation, apical surfaces were washed, harvested directly for RNA, and assayed by real-time reverse transcriptase PCR for the presence of HCoV-HKU1 RNA. During the 7 days of incubation, we observed no major morphological changes in HAE nor any obvious cytotoxicity or leakage of basolateral media into the apical compartment, suggesting that the epithelial cell layer integrity was maintained. Multilog increases in viral genome copies were detected over the course of infection, providing the first evidence for an in vitro replication model for HCoV-HKU1 (Fig. 2B). Viral genome copy numbers increased rapidly from 4 h postinoculation, reaching peaks of >109 copies/ml within 72 h and maintaining those copy numbers at 96 h postinfection.
Analysis of HCoV-HKU1 subgenomic mRNA synthesis in HAE.
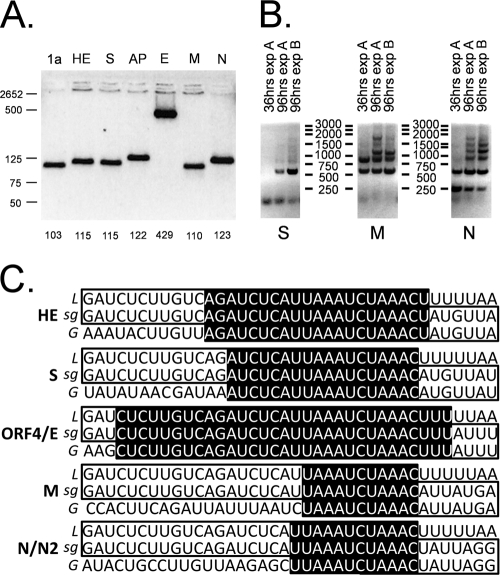
One distinction of the Coronaviridae is the synthesis of a nested set of subgenomic mRNAs (sg mRNAs) via a mechanism called discontinuous transcription (Fig. 1) (4, 39, 40). These subgenomic RNAs serve as the templates for the translation of structural and accessory proteins. We employed Northern blot analysis with a probe specific to the nucleocapsid sequence to assess the number and size of the HCoV-HKU1 subgenomic mRNA species. Five distinct RNA species were identified, with the sizes indicating the presence of RNA molecules corresponding to the hemagglutinin (HE; ∼8,226 nucleotides [nt]), spike (S; ∼7,056 nt), membrane (M; ∼2,368 nt), and nucleocapsid (N; ∼1,685 nt) structural sg mRNA as well as one species for the accessory open reading frame (AP; ∼2,954 nt) between spike and membrane, confirming all previously predicted HCoV-HKU1 subgenomic mRNAs (Fig. 3) (57).
FIG. 3.
HCoV-HKU1 RNA species present during infection of HAE. Representative Northern blot analysis for total RNA isolated from HCoV-HKU1-infected HAE. RNA species are indicated by arrows to the right of the autoradiograph. HE, hemagglutinin; S, spike; ORF 4, accessory open reading frame; E, envelope; M, membrane; N, nucleocapsid; N2, internal ORF in N. (A) Total RNA, 0.1 μg; (B) total RNA, 1 μg.
To confirm the expression profile of sg mRNA during active virus replication, we employed a standard RT-PCR amplification technique to detect the presence of subgenomic mRNAs which were formed by the joining of 5′ genomic leader RNA sequences to the body sequences of each subgenomic mRNA (Fig. 1) (39). We used a common 5′ leader RNA primer and multiple 3′ primers targeting the first 100 nucleotides of each predicted gene. The PCR amplification products were then separated by electrophoresis on a 0.8% agarose gel, and the DNA was visualized using ethidium bromide on a Dark Reader (Clare Chemical). Bands were detected for subgenomic mRNA corresponding to the 1a, HE, S, AP, M, and N genes (Fig. 4 A). Of note, a specific subgenomic mRNA corresponding to the envelope (E) gene was not amplified, as expected from the in silico TRS analysis (57). Sequence analysis of the subgenomic mRNA PCR products revealed that all subgenomic mRNAs do contain the HCoV-HKU1 5′-end leader sequence (Fig. 4C).
FIG. 4.
HCoV-HKU1 subgenomic species. (A) Subgenomic mRNA species generated during HCoV-HKU1 replication in HAE. Each lane contains RT-PCR products from total RNA isolated from HCoV-HKU1-inoculated HAE, amplified using a 5′ HCoV-HKU1 leader primer and the indicated ORF-specific 3′ primer. Size markers are indicated on the far left of the gel, and the size of each band is shown at the base of each well. 1a, open reading frame 1a replicase proteins; HE, hemagglutinin; S, spike; AP, accessory open reading frame 4/protein; E, envelope; M, membrane; N, nucleocapsid. (B) Kinetics of subgenomic RNA species synthesis during HCoV-HKU1 replication in HAE. Each lane contains RT-PCR bands from RNA extracted from HCoV-HKU1-inoculated HAE for the first 500 nt of the structural proteins spike, membrane, and nucleocapsid. Marker sizes are indicated between the gels, times postinoculation are indicated above each well, and the gene of interest is indicated at the base of each gel. S, spike; M, membrane; N, nucleocapsid. (C) Leader-body junctions of all HCoV-HKU1 sg mRNAs. Shown on the top row is the leader (L) sequence, and the bottom row shows the specific sequences upstream of the structural genes (G). The sequence in the middle (sg) represents the mature sg mRNA generated during coronavirus replication. Sequence homology between the strands near the junction is highlighted in black. HE, hemagglutinin; S, spike; ORF4, accessory open reading frame; E, envelope; M, membrane; N, nucleocapsid; N2, internal ORF in N.
We also investigated the levels of S, M, and N subgenomic mRNA synthesis during infection employing a semiquantitative RT-PCR. Total RNA was harvested from HCoV-HKU1-infected HAE at 36 and 96 h postinoculation. RNA was analyzed by RT-PCR optimized to detect the multiple subgenomic RNA species containing the ORF of interest (Fig. 1). These data and our Northern blot analysis indicated that HCoV-HKU1 used transcription kinetics similar to that of other human coronaviruses with higher levels of the more 3′ ORFs, N and M subgenomic RNA (Fig. 3 and 4B). In contrast, we were not able to detect S subgenomic RNA at 36 h postinoculation and only the 96-h postinoculation sample was positive (Fig. 4B). These results are consistent with previous studies demonstrating that 3′-proximal transcripts are typically more abundant in coronavirus-infected cells (10, 36).
Sequencing of the HCoV-HKU1 Caen1 genome.
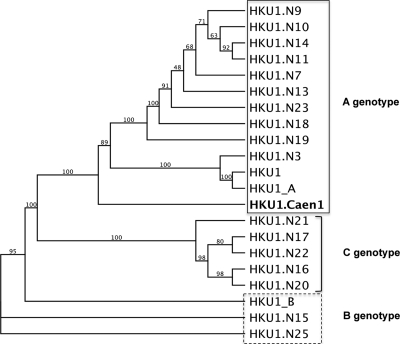
To characterize the HCoV-HKU1 isolate replicating in HAE, we sequenced the entire viral genome by using primer sets designed with sufficient overlap to verify the data for the complete genome. The genome was 29,926 nucleotides (nt) in length and comprised 8 genes (1a/b, HE, S, ORF4, E, M, N, N2). Phylogenetic analysis clearly shows that the Caen1 isolate clusters with other group A isolates (Fig. 5). Comparative genome analysis with the reference strains did not reveal any apparent deviations, suggesting that a recombination event had not occurred nor had there been extensive alteration of the sequence by insertions and/or deletions. The S gene had ∼99.6% identity with other group A HCoV-HKU1 isolates.
FIG. 5.
Maximum likelihood cladogram of HKU1 genomes. HCoV-HKU1 strain Caen1 was compared to 23 additional full-length HCoV-HKU1 virus genomes to determine its evolutionary relatedness within the HCoV-HKU1 lineage. This maximum likelihood tree generated with the PhyML package shows that HCoV-HKU1 strain Caen1 is most closely related to the A genotype. Branch points are labeled with bootstrap values, based upon 100 iterations, and the cladogram is set to be proportional.
HCoV-HKU1 cellular localization and egress during infection of HAE.
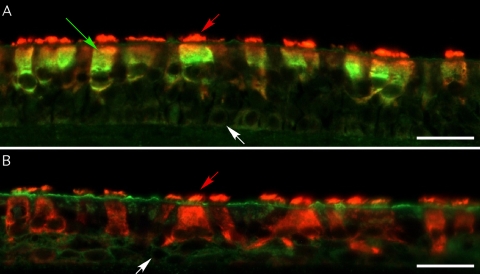
To determine the HAE cell type(s) infected by HCoV-HKU1, we inoculated HAE cultures with HCoV-HKU1 and fixed the cultures in 4% PFA 96 h postinoculation. Cultures were then processed for immunofluorescence assays as previously described (45). Sections were probed with a monoclonal antibody against a ciliated cell-specific marker, β-tubulin IV, to identify ciliated cells and rabbit antiserum generated against mouse hepatitis virus (MHV) whole virions: this antibody was used to detect HKU1 due to the significant sequence homology between the N proteins of MHV and HCoV-HKU1 N (∼70%) and because HCoV-HKU1 antibodies were not currently available. HKU1 immunoreactivity was detected only in sections of HAE inoculated with HCoV-HKU1, and colocalization of HKU1 and β-tubulin IV indicated that ciliated cells were infected by HKU1 (Fig. 6 A). No immunoreactivity for HKU1 was detected in sections of HCoV-HKU1-inoculated HAE probed with preimmune rabbit serum (Fig. 6B) or HAE not inoculated with HKU1 (not shown). Overall, significant numbers of ciliated cells were infected by HKU1, indicating robust and ciliated cell-specific infection of HAE by this virus.
FIG. 6.
HCoV-HKU1 infects ciliated cells of HAE. Representative images of HCoV-HKU1 immunoreactivity in histological sections of HAE 72 h postinoculation with HCoV-HKU1. Histological sections were probed with antisera directed against mouse hepatitis virus (MHV) whole virions (green) and β-tubulin IV (red). (A) HCoV-HKU1 immunoreactivity (green) in histological sections, demonstrating that HCoV-HKU1 infects ciliated cells. (B) HCoV-HKU1-inoculated HAE probed with preimmune rabbit serum and β-tubulin IV antibody, demonstrating no HCoV-HKU1 immunoreactivity. Original magnification, ×40. Scale bars, 100 μm; red arrows, cilial shafts of ciliated cells; green arrow, HKU1-infected cells; white arrows, basal cells present on the Transwell supports. Colocalization of HCoV-HKU1 and ciliated cell immunoreactivity (greenish-yellow) indicate ciliated cell infection by HCoV-HKU1.
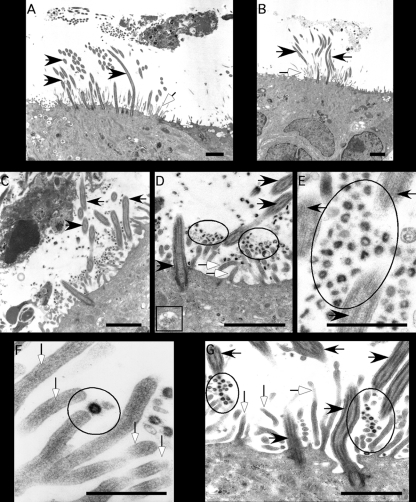
To confirm the cellular localization of HCoV-HKU1 replication and egress during infection of HAE, cultures were inoculated with HCoV-HKU1 (Fig. 7 C to G) or mock inoculated (Fig. 7A and B) for 72 h and then processed for transmission electron microscopy. These studies revealed large numbers of viral particles in the apical compartment of HAE, largely associated with the apical surface of ciliated cells (Fig. 7C to G). Viruses associated with the microvilli and cilial shafts and were present in the airway surface liquid microenvironment. The localization of HCoV-HKU1 to ciliated cell apical structures is similar to the previously reported localization of SARS-CoV after infection of HAE (Fig. 7C to G). Although fewer numbers of viral particles were observed inside cells, when present, virions were observed in vesicular structures in the cytoplasm of ciliated cells (Fig. 7D, box). These data further support our immunofluorescence studies showing that HCoV-HKU1 replicates in ciliated cells of HAE, the same cells infected by all other human coronaviruses tested to date.
FIG. 7.
Ultrastructural localization of HCoV-HKU1 in HAE. Representative transmission electron photomicrographs of HAE inoculated with HCoV-HKU1. (A and B) HAE mock inoculated with the vehicle alone, demonstrating the typical morphological features of the apical surfaces of HAE with cilia (black arrows) and microvilli (white arrows). (C to G) HAE infected with HCoV-HKU1 for 96 h showing the presence of the large numbers of virions (circled) associated with the surfaces of ciliated cells or shed into pericilial regions (black arrows, cilia; white arrows, microvilli). Intracellular virions were also noted inside vesicular structures in the cytoplasm of ciliated cells (D, box). F represents a high-power image of a virion associated with the tip of a microvillus. Scale bars are shown in the lower right of each panel and represent 2 μm in panels A to D and G and 1 μm in panels E and F.
Antibody blockade assay.
Recent studies using viral pseudoparticles containing HCoV-HKU1 spike protein have suggested that HCoV-HKU1 uses the major histocompatibility complex class I C/human leukocyte antigen C (HLA-C) as a receptor for entry into permissive cells (8). To determine whether HCoV-HKU1 utilizes HLA-C as a receptor in HAE cultures, the apical surfaces of cultures were incubated either individually or in combination with a monoclonal antibody for HLA-C or a monoclonal antibody that detects both HLA-B and HLA-C for 3 h prior to inoculation. To ensure the validity of results, the same antibodies as previously reported by Chan et al. (8) to block entry of HCoV-HKU1 spike pseudoparticles into permissive cells were used. We have previously used this approach with HAE to successfully ablate SARS-CoV infection using antibodies to block virus binding to angiotensin converting enzyme 2 (ACE2) (45). In the continued presence of HLA or ACE2 antibodies, HAE were inoculated with HCoV-HKU1 (stocks diluted 1:1,000) and apical washes were collected over time until 72 h postinoculation. RNA was isolated from the apical washes, and samples were analyzed by real-time reverse transcriptase PCR. These data demonstrated that viral genomic RNA increased over time for all treatment groups at all time points, indicating that the presence of antibody, whether control (ACE2) or directed against HLA-C or HLA-B/C, had no effect on virus replication kinetics (Fig. 8). Based on these results, we suggest that in HAE, a relevant human ciliated airway epithelium model, HLA-C is not an absolute requirement for efficient HCoV-HKU1 infection.
FIG. 8.
Efficient infection of HAE by HCoV-HKU1 does not require HLA-C. Replication kinetics of HCoV-HKU1 were assessed by real-time RT-PCR of RNA present in apical washes from HCoV-HKU1-infected HAE cultures pretreated with either no antibody (yellow), HLA-C-specific antibody (blue), HLA-BC antibody (pink), a combination of the antibodies (green), or ACE2 antibody (black). Titers are represented by virus copies per milliliter. No significant differences in HCoV-HKU1 infection and replication were measured in any treatment group. n = 3 for each treatment group.
DISCUSSION
To date, five human coronavirus strains have been identified (HCoV-229E, HCoV-OC43, SARS-CoV, HCoV-NL63, and HCoV-HKU1), and all have been associated with mild to severe respiratory tract illnesses. Although more recent epidemiologic studies have demonstrated a significant association with respiratory tract infections (12, 25, 52), HCoV-229E and HCoV-OC43 were first described in the mid-1960s as human coronavirus strains that caused relatively mild common cold symptoms (21, 30, 50). The previously considered mild nature of human coronavirus-associated respiratory disease was revisited after the isolation of SARS-CoV in 2003 in the Guandong province of China (14, 24, 33). SARS-CoV was the first human coronavirus strain that caused lethal infections in otherwise healthy individuals. Soon after the SARS-CoV epidemic was controlled in early July 2003 by stringent quarantine measures, HCoV-NL63 was identified in 2004, employing a novel VIDISCA tool from patients presenting with bronchiolitis (37, 54). HCoV-NL63 is currently associated with croup and respiratory illness, causing significant illness in infants, the immunocompromised, and elderly patients (34, 53-55). Another human coronavirus strain, HCoV-HKU1, was identified in 2005 by direct analysis of clinical samples from patients with respiratory infections using degenerate primer-based PCR techniques (57). HCoV-HKU1-positive samples have been detected worldwide, and the virus has been shown to cause a wide variety of respiratory tract disease symptoms and severity of disease (7, 35, 48, 51, 57). A previous report by Chan et al. has proposed that HCoV-HKU1 utilizes HLA-C for cell attachment and entry; however, despite significant efforts, HCoV-HKU1 had proven unculturable using laboratory-based cell lines in vitro (8).
We hypothesized that the successful infection and replication of HCoV-HKU1 clinical isolates in vitro require a platform that more accurately reproduces the environment of the human airway epithelium, a likely initial site for coronavirus inoculation and spread. We have achieved this goal using a human ciliated airway epithelial cell model. This in vitro culture model is derived from freshly isolated human tracheobronchial airway epithelial cells and mirrors the morphology and physiology of the human cartilaginous airway epithelium. Following 6 to 8 weeks of cell culture growth, a fully differentiated respiratory epithelium is established, with predominant ciliated cells interspersed with goblet cells overlying a basal epithelial cell population. This model system has been shown previously to support infection and replication of a number of human respiratory viruses, including parainfluenza viruses, influenza viruses, respiratory syncytial virus (RSV), human metapneumovirus, human bocavirus, and coronaviruses (2, 3, 5, 11, 13, 30, 42, 45, 56, 61). Here, we demonstrate for the first time that this model provides a relevant and appropriate cellular environment conducive for the successful infection and growth of HCoV-HKU1. HAE cultures have been previously identified as an excellent culture system for propagation of clinical isolates (11), and here we show that HAE represents the only culture system tested so far that supports HCoV-HKU1 infection and replication. Furthermore, previous studies from our laboratory based on other coronavirus strains have demonstrated that passage of virus in HAE adapts the virus for better growth in human cell lines (44). Despite this, we have still been unable to generate HAE-derived HCoV-HKU1 stocks that are capable of replication on LLC-MK2, VeroE6, or Calu3 cell lines (data not shown).
We also demonstrate that HCoV-HKU1 does not require the HLA-C molecule for efficient infection, contradicting previously published data. As reported by Chan et al., HLA-C on the surface of A549 cells greatly increased the rate of attachment of HCoV-HKU1 spike containing pseudotyped particles but was not sufficient for entry of these particles into any cell line tested (8). In our studies, using the same blocking antibodies as Chan et al., we were unable to reduce HCoV-HKU1 infection in HAE. Consistent with these findings, we have also found that HCoV-HKU1 replication was not detected in cell lines expressing HLA-C (data not shown), suggesting that additional coreceptors may be required or that replication of HCoV-HKU1 is not supported in these cells due to particular cell line characteristics. In the present study, antibodies against HLA-C or ACE2 did not reduce HCoV-HKU1 infection of HAE. While anti-ACE2 antibody can interfere with SARS-CoV or HCoV-NL63 docking and entry into cells, the HLA-C antibodies did not prevent HKU1 infection. Further work to study HLA-C and/or identify the HCoV-HKU1 receptor(s) for docking and entry will be required to clarify the entry mechanisms and receptor complexes used by HCoV-HKU1 for entry into HAE.
Robust HCoV-HKU1 replication provided a model to identify the subgenomic mRNA molecules synthesized during infection and revealed a characteristic coronaviral discontinuous replication process (4, 38-40, 46). HCoV-HKU1 replication results in the production of five subgenomic mRNA species, corresponding to genes HE, S, AP, M, and N, as reported previously based on in silico studies. Genome organization is similar to that of the other group 2 human coronavirus, HCoV-OC43. We were not able to detect an E sg mRNA in infected cells, which agrees with a previous report on the absence of a transcription regulatory sequence (TRS) specific for the E gene (57). The E protein forms pores and plays a role in membrane contortion during virus assembly and release. The E protein is absolutely essential for the production of group 1 coronavirus particles; however, group 2 coronaviruses, like SARS-CoV and MHV, can replicate in the absence of the E protein (9, 32). Although speculative, the E gene may be expressed by either a translation-scanning mechanism or by internal ribosome entry sites. Both strategies are employed by other coronaviruses to express specific downstream ORFs (15, 28, 29, 31, 43, 49, 58). Sequence analysis of the amplified leader-body fusion mRNA molecules revealed that the core TRS is indeed AAUCUAAAC, as predicted, and is present in the 5′ region of all identified mRNA molecules (57).
Electron micrographs and immunofluorescence data with HCoV-HKU1-infected HAE indicate that HCoV-HKU1, like all human coronaviruses studied to date, infects and replicates efficiently within ciliated cells of HAE and that virus is released only from the apical surface of the cultures. We continue to study the relevance of ciliated cell infection by these and other respiratory viruses.
In conclusion, we have established an in vitro culture system for culturing HCoV-HKU1, the most fastidious human coronavirus identified to date. Little is known about the replication or pathogenesis characteristics of this virus, since previous research was limited to analysis of clinical isolates. Future studies on HCoV-HKU1 will focus on the identification of the HCoV-HKU1 cellular receptor in the appropriate models, replication strategies of the virus, and evaluation of potential anticoronavirus drugs. Development and evaluation of a culture system such as HAE as described in this study represents a milestone in the ongoing research on this relatively new member of the Coronaviridae family.
Acknowledgments
We thank the directors and teams of the UNC Cystic Fibrosis Center Tissue Culture Core, the Morphology and Morphometry Core, and the Michael Hooker Microscopy Facility for supplying reagents and technical expertise. We gratefully acknowledge Susan Burkett for technical assistance.
This project was supported by the National Institutes of Health (NIH) grants NIH R01 HL77844 (R.J.P.) and U54-AI057157 (R.S.B. and R.J.P.). A.C.S. is supported by grants NIH R21 AI076159 and NIAID/NHLB R21 AI079521. K.P. is supported by the Foundation for Polish Science within the Homing Programme and a grant from the Ministry of Scientific Research, Poland (0095/B/P01/2009/37). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of the structural funds from the European Union (grant POIG.02.01.00-12-064/08, “Molecular biotechnology for health”). L.V.D.H. and R.D. are supported by VIDI grant 016.066.318 from the Netherlands Organization for Scientific Research (NWO) and by the sixth framework grant LSHM-CT-2005TC-037276 from the European Union.
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print on 18 August 2010.
REFERENCES
- 1.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayora-Talavera, G., H. Shelton, M. A. Scull, J. Ren, I. M. Jones, R. J. Pickles, and W. S. Barclay. 2009. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One 4:e7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banach, S., J. M. Orenstein, L. M. Fox, S. H. Randell, A. H. Rowley, and S. C. Baker. 2009. Human airway epithelial cell culture to identify new respiratory viruses: coronavirus NL63 as a model. J. Virol. Methods 156:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, E. J., M. Hennessey, M. H. Skiadopoulos, A. C. Schmidt, P. L. Collins, B. R. Murphy, and R. J. Pickles. 2008. Role of interferon in the replication of human parainfluenza virus type 1 wild type and mutant viruses in human ciliated airway epithelium. J. Virol. 82:8059-8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosis, S., S. Esposito, H. G. Niesters, E. Tremolati, S. Pas, N. Principi, and A. D. Osterhaus. 2007. Coronavirus HKU1 in an Italian pre-term infant with bronchiolitis. J. Clin. Virol. 38:251-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, C. M., S. K. Lau, P. C. Woo, H. Tse, B. J. Zheng, L. Chen, J. D. Huang, and K. Y. Yuen. 2009. Identification of major histocompatibility complex class I C molecule as an attachment factor that facilitates coronavirus HKU1 spike-mediated infection. J. Virol. 83:1026-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeDiego, M. L., E. Alvarez, F. Almazan, M. T. Rejas, E. Lamirande, A. Roberts, W. J. Shieh, S. R. Zaki, K. Subbarao, and L. Enjuanes. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 81:1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., L. van Genne, J. N. Stoop, H. Volders, and P. J. Rottier. 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77:11312-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkman, R., S. M. Koekkoek, R. Molenkamp, O. Schildgen, and L. van der Hoek. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 83:7739-7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez, S. R., C. C. Robinson, and K. V. Holmes. 2009. Detection of four human coronaviruses in respiratory infections in children: a one-year study in Colorado. J. Med. Virol. 81:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson, E. F., B. Yount, A. C. Sims, S. Burkett, R. J. Pickles, and R. S. Baric. 2008. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 82:11948-11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 15.Enjuanes, L., I. Sola, S. Alonso, D. Escors, and S. Zuniga. 2005. Coronavirus reverse genetics and development of vectors for gene expression. Curr. Top. Microbiol. Immunol. 287:161-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Fulcher, M. L., S. Gabriel, K. A. Burns, J. R. Yankaskas, and S. H. Randell. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107:183-206. [DOI] [PubMed] [Google Scholar]
- 18.Gaynor, A. M., M. D. Nissen, D. M. Whiley, I. M. Mackay, S. B. Lambert, G. Wu, D. C. Brennan, G. A. Storch, T. P. Sloots, and D. Wang. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouy, M., S. Guindon, and O. Gascuel. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221-224. [DOI] [PubMed] [Google Scholar]
- 20.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 21.Hamre, D., and J. J. Procknow. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121:190-193. [DOI] [PubMed] [Google Scholar]
- 22.Herzog, P., C. Drosten, and M. A. Muller. 2008. Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol. J. 5:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, K. E., N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman, and P. Daszak. 2008. Global trends in emerging infectious diseases. Nature 451:990-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 25.Lam, W. Y., A. C. Yeung, J. W. Tang, M. Ip, E. W. Chan, M. Hui, and P. K. Chan. 2007. Rapid multiplex nested PCR for detection of respiratory viruses. J. Clin. Microbiol. 45:3631-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 27.Lee, W. M., C. Kiesner, T. Pappas, I. Lee, K. Grindle, T. Jartti, B. Jakiela, R. F. Lemanske, Jr., P. A. Shult, and J. E. Gern. 2007. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One 2:e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibowitz, J. L., S. Perlman, G. Weinstock, J. R. DeVries, C. Budzilowicz, J. M. Weissemann, and S. R. Weiss. 1988. Detection of a murine coronavirus nonstructural protein encoded in a downstream open reading frame. Virology 164:156-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, D. X., and S. C. Inglis. 1992. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 66:6143-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIntosh, K., J. H. Dees, W. B. Becker, A. Z. Kapikian, and R. M. Chanock. 1967. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 57:933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connor, J. B., and D. A. Brian. 2000. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology 269:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyrc, K., B. Berkhout, and L. van der Hoek. 2007. Identification of new human coronaviruses. Expert Rev. Anti Infect. Ther. 5:245-253. [DOI] [PubMed] [Google Scholar]
- 35.Pyrc, K., B. Berkhout, and L. van der Hoek. 2007. The novel human coronaviruses NL63 and HKU1. J. Virol. 81:3051-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyrc, K., M. F. Jebbink, B. Berkhout, and L. van der Hoek. 2004. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyrc, K., M. F. Jebbink, B. Berkhout, and L. van der Hoek. 2008. Detection of new viruses by VIDISCA. Virus discovery based on cDNA-amplified fragment length polymorphism. Methods Mol. Biol. 454:73-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaad, M. C., W. Chen, S. A. Peel, and R. S. Baric. 1993. Studies into the mechanism for MHV transcription. Adv. Exp. Med. Biol. 342:85-90. [DOI] [PubMed] [Google Scholar]
- 41.Schildgen, O., M. F. Jebbink, M. de Vries, K. Pyrc, R. Dijkman, A. Simon, A. Muller, B. Kupfer, and L. van der Hoek. 2006. Identification of cell lines permissive for human coronavirus NL63. J. Virol. Methods 138:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scull, M. A., L. Gillim-Ross, C. Santos, K. L. Roberts, E. Bordonali, K. Subbarao, W. S. Barclay, and R. J. Pickles. 2009. Avian influenza virus glycoproteins restrict virus replication and spread through human airway epithelium at temperatures of the proximal airways. PLoS Pathog. 5:e1000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senanayake, S. D., and D. A. Brian. 1997. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 48:101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheahan, T., B. Rockx, E. Donaldson, A. Sims, R. Pickles, D. Corti, and R. Baric. 2008. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 82:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spaan, W., H. Delius, M. A. Skinner, J. Armstrong, P. Rottier, S. Smeekens, S. G. Siddell, and B. van der Zeijst. 1984. Transcription strategy of coronaviruses: fusion of non-contiguous sequences during mRNA synthesis. Adv. Exp. Med. Biol. 173:173-186. [DOI] [PubMed] [Google Scholar]
- 47.St-Jean, J. R., M. Desforges, F. Almazan, H. Jacomy, L. Enjuanes, and P. J. Talbot. 2006. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J. Virol. 80:3670-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talbot, H. K., J. E. Crowe, Jr., K. M. Edwards, M. R. Griffin, Y. Zhu, G. A. Weinberg, P. G. Szilagyi, C. B. Hall, A. B. Podsiad, M. Iwane, and J. V. Williams. 2009. Coronavirus infection and hospitalizations for acute respiratory illness in young children. J. Med. Virol. 81:853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel, V., and S. G. Siddell. 1994. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 75(Pt. 11):3041-3046. [DOI] [PubMed] [Google Scholar]
- 50.Tyrrell, D. A., and M. L. Bynoe. 1965. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1:1467-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vabret, A., T. Mourez, J. Dina, L. van der Hoek, S. Gouarin, J. Petitjean, J. Brouard, and F. Freymuth. 2005. Human coronavirus NL63, France. Emerg. Infect. Dis. 11:1225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallet, S., A. Gagneur, P. J. Talbot, M. C. Legrand, J. Sizun, and B. Picard. 2004. Detection of human coronavirus 229E in nasal specimens in large scale studies using an RT-PCR hybridization assay. Mol. Cell. Probes 18:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Hoek, L., G. Ihorst, K. Sure, A. Vabret, R. Dijkman, M. de Vries, J. Forster, B. Berkhout, and K. Uberla. 2010. Burden of disease due to human coronavirus NL63 infections and periodicity of infection. J. Clin. Virol. 48:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, G., C. Deering, M. Macke, J. Shao, R. Burns, D. M. Blau, K. V. Holmes, B. L. Davidson, S. Perlman, and P. B. McCray, Jr. 2000. Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol. 74:9234-9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, K., B. J. Zheng, R. Zeng, W. Lu, Y. P. Lin, L. Xue, L. Li, L. L. Yang, C. Xu, J. Dai, F. Wang, Q. Li, Q. X. Dong, R. F. Yang, J. R. Wu, and B. Sun. 2009. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology 388:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]