Abstract
We previously identified an adenovirus (Ad) protein named U exon protein (UXP) encoded by a leftward-strand (l-strand) transcription unit. Here we identify and characterize the UXP promoter. Primer extension and RNase protection assays mapped the transcription initiation site at 32 nucleotides upstream of the UXP gene initiation codon. A series of viral mutants with mutations at two putative inverted CCAAT (I-CCAAT) boxes and two E2F sites were generated. With mutants lacking the proximal I-CCAAT box, the UXP mRNA level decreased significantly to 30% of the Ad type 5 (Ad5) mRNA level as measured by quantitative reverse transcription-PCR. Decreased UXP was also observed by immunoblotting and immunofluorescence. UXP mRNA and protein levels were similar to those of Ad5 for mutants lacking the distal I-CCAAT box or both putative E2F sites. Ad DNA levels were similar in mutant- and wild-type Ad5-infected cells during the late stage of infection, strongly suggesting that the decreased UXP mRNA and protein from mutants lacking the proximal I-CCAAT box was due to decreased promoter activity. Electrophoretic mobility shift assays (EMSA) indicated that a cellular factor binds specifically to the proximal I-CCAAT box of the UXP promoter. An in vitro luciferase reporter assay demonstrated that basal promoter activity lies between bp −158 and +30 of the transcription initiation site. No E1A-mediated promoter transactivation was observed in 293 cells compared with A549 cells. Thus, we propose that there is a previously unidentified Ad5 promoter that drives expression of the UXP transcription unit. This promoter is embedded within the gene for fiber, and it contains a proximal I-CCAAT box critical for UXP mRNA transcription.
Human adenoviruses (Ads) have been studied extensively as a model for eukaryotic gene regulation. The Ad transcription units are expressed in four temporal stages: immediate early, delayed early, intermediate, and late (1, 3, 4, 27). The assignment of genes to a particular stage is based on the time of appearance of the gene product. So far, 11 different promoters have been identified for initiation of Ad gene transcription at different stages of productive infection. The promoter of the immediate early E1A transcription unit becomes active upon Ad infection, and E1A is transcribed as soon as the viral genome enters the cell nucleus. The larger E1A protein activates transcription from other early transcription units, namely, E1B, E2E (E2 early), E3, and E4, via a variety of cellular transcriptional factors (3, 4). The major late promoter (MLP) is also active at a low level during this early stage, but transcription proceeds only as far as the L3 region, primarily producing the i-leader protein and L1-52/55K proteins (36). Proteins encoded from early transcription units modulate multiple cellular functions to facilitate Ad replication. E1B proteins inhibit apoptosis and regulate viral mRNA transport (in cooperation with E4 proteins) (3, 6). E3 proteins function to subvert the host cellular immunity (13, 16, 20, 44). E4 proteins facilitate viral mRNA metabolism (in association with E1B-55K), promote viral DNA replication by preventing a double-stranded DNA repair response, and induce the shutoff of host protein synthesis (12, 19, 42, 43). Efficient transcription from the E2 early promoter results in accumulation of the E2A DNA binding protein (DBP), E2B precursor terminal protein, and DNA polymerase, which set the stage for viral DNA replication to begin. At the initiation of viral genome replication, three intermediate viral promoters (pIX, IVa2, and E2 late) that are silent during the earliest phase of infection become active (3, 4). pIX, a virion structural protein, and IVa2 are involved in upregulating the transcriptional activity of the MLP during the early- to late-phase transition (22, 29, 30, 41). As viral DNA replication begins, L4-22K and L4-33K, transcribed from a novel L4 promoter (27), act as positive regulators for full activation of the MLP. Transcription proceeds through the full-length major late transcription unit to produce maximal expression of the late structural genes from all five subregions, L1 to L5, to supply the structural proteins for the packaging of newly replicated Ad genomes into mature infectious virions, and to express the Ad death protein (ADP) (39).
In an earlier study, we identified a previously unrecognized human Ad protein named U exon protein (UXP) (40). UXP is encoded from a late leftward-strand (l-strand) transcription unit, is first detected in the nucleoli and nuclei, and later is associated with Ad replication centers. UXP deletion mutants display aberrant DBP localization and have a modest growth defect. UXP is expressed abundantly at late stages of Ad infection. The regulation of expression of the UXP transcription unit is unknown. Here, we report experiments that map the UXP promoter.
MATERIALS AND METHODS
Cell lines.
The human lung carcinoma cell line A549 and cervical cancer cell line HeLa were obtained from the American Type Culture Collection (ATCC, Manassas, VA). HEK293 cells were obtained from Microbix (Ontario, Canada). All cells were grown in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS).
Viruses.
Wild-type Ad5 was obtained from the ATCC (VR-5), plaque purified three times, and expanded in KB cell spinner cultures (38). Construction of the UXP frameshift mutant (UXPFS) was as follows. (i) Ad5 HindIII B fragment (Ad5 bp 26328 to 31998) was cloned into the pGL3 plasmid (Promega, Madison, WI) at the HindIII site to generate the pGL3/H3B vector. (ii) The plasmid was then cut with XhoI to remove Ad5 bp 26328 to 29791, resulting in a smaller plasmid, pGL3-XH, containing Ad5 bp 29791 to 31998 at XhoI/HindIII sites. The pGL3-XH plasmid was used as the template in site-directed mutagenesis along with primers (forward, CTCCTGTTCCTGTCCGGATCCGCACCCACTAT; reverse ATAGTGGGTGCGGATCCGGACAGGAACAGGAG) to introduce an extra GG between Ad5 bp 31010 and 31011. The UXP frameshift occurs after seven amino acids. The mutagenesis was done using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). After mutagenesis in pGL3-XH, the previously removed DNA fragment containing Ad5 bp 26328 to 29791 was rebuilt back into pGL3-XH to reconstitute the pGL3-H3B plasmid. (iii) The reconstituted pGL3-H3B plasmid with the desired mutation was cotransfected into HEK293 cells with Ad5 virion DNA digested with EcoRI/SpeI to make the UXP frameshift mutant UXPFS. To make the UXP promoter mutants, two putative inverted CCAAT (I-CCAAT) (bp −86, −152) or E2F (bp −123, −387) sites located in the UXP putative promoter region were mutated individually or in combination by multiple rounds of PCR. Specific mutation changes to each CCAAT box and E2F site are shown in Fig. 2A. PCRs were performed according to the following protocol: 94°C for 2 min, followed by 35 cycles of 94°C for 25 s, 55°C for 30 s, and 72°C for 1 min 30 s, and a final extension at 68°C for 10 min. Each PCR fragment with the desired mutations was cloned into the pGL3-XH plasmid containing Ad5 bp 29791 to 31998 at NdeI/HindIII sites. The DNA fragment containing Ad5 bp 26328 to 29791 was subsequently rebuilt into pGL3-XH at the XhoI site to reconstitute the pGL3-H3B plasmid. The resulting pGL3-H3B plasmid with the desired mutation was cotransfected into HEK293 cells with Ad5 virion DNA digested with EcoRI/SpeI to make each of the promoter mutants.
All the mutations were verified by sequencing. The mutants were plaque purified three times on A549 cells and expanded in KB cell spinner cultures, and their titers were determined on A549 cells.
Primer extension analysis.
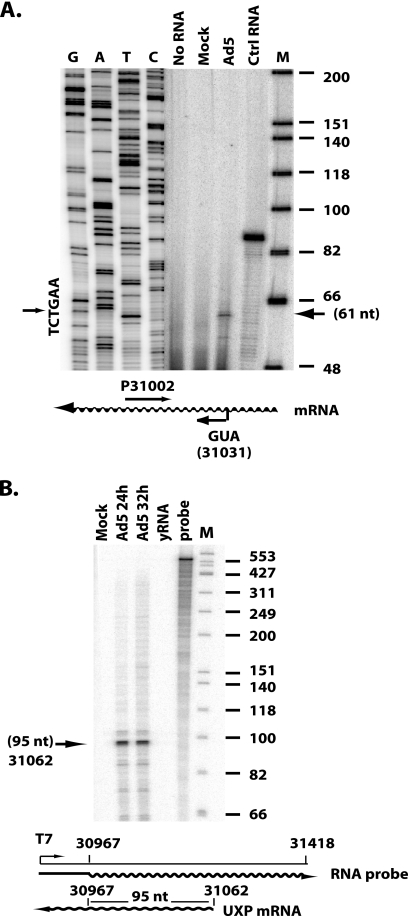
A549 cells were mock infected or infected with Ad5 at 20 PFU/cell. At 20 h postinfection (p.i.), cells were harvested, and total cytoplasmic RNA was isolated with the Qiagen RNeasy kit (Qiagen, Valencia, CA). Primer extension analysis was performed using the primer extension system kit (Promega). Briefly, equivalent amounts of total RNA (20 μg per sample) were hybridized to a 5′-end-labeled primer (TTCCTGTCCATCCGCACCCACTAT) at 58°C for 20 min, followed by cooling at room temperature for 10 min. The 5′ nucleotide of the primer is complementary to the first exon of UXP mRNA and is located 30 bp downstream from the initiation ATG codon (Fig. 1 A). The duplex was extended using avian myeloblastosis virus (AMV) reverse transcriptase at 42°C for 45 min. Extended DNA products were separated on a denaturing 6% polyacrylamide sequencing gel. The gel was dried and exposed to a PhosphorImager. The dideoxy sequence of a plasmid carrying Ad5 bp 30967 to 31418 with the same primer is shown as a size standard. 5′-end labeled φX174 DNA/HinfI was used as the molecular marker.
FIG. 1.
Determination of the transcription initiation site of the UXP pre-mRNA by primer extension analysis (A) and RNase protection assay (B). Total cytoplasmic RNAs isolated from mock- or Ad5-infected A549 cells at 24 or 32 h p.i. were used in the assays. (A) The primer used was a 5′-end-labeled oligonucleotide complementary to the first exon of the UXP mRNA. The 5′-end nucleotide of the primer is located 30 bp downstream of the initiation codon, ATG, of the UXP gene. The dideoxy sequence of a plasmid carrying Ad5 bp 30967 to 31418 with the same primer is shown as a size standard. The arrow on the right indicates the major extension product. The extension product in control (Ctrl) RNA was from kanamycin RNA, and its corresponding primer was included in the kit. 5′-end labeled φX174 DNA/HinfI was used as marker (lane M). (B) For RNase protection, the labeled riboprobe corresponding to Ad5 nt 30967 to 31418 was used. The arrow on the left indicates the major protected product. The number next to the arrow on the left indicates the size of the protected product and the location of the initiation site on the Ad5 genome. Torula yeast RNA (yRNA) was used as negative-control RNA. 5′-end-labeled φX174 DNA/HinfI was used as the marker (lane M).
RPA.
A549 cells were mock or Ad5 infected at 20 PFU/cell. At 24 or 32 h p.i., total cytoplasmic RNA was isolated as described for the primer extension assay. To prepare the RNA probe used for the RNase protection assay (RPA), a DNA fragment containing Ad5 bp 30967 to 31418 was PCR amplified and cloned into the pcDNA3.1/myc-His(−) A plasmid (Invitrogen, Carlsbad, CA) at BamHI/HindIII sites. The resulting plasmid was linearized with HindIII digestion, and an antisense probe was transcribed using the Ambion Maxscript in vitro transcription kit with T7 RNA polymerase. The probe was heated to 95°C for 3 min and run on a 5% acrylamide-8 M urea-1× Tris-borate-EDTA (TBE) gel and subsequently exposed to Kodak film to determine the band position within the gel. The full-length probe was excised from the gel and eluted in 0.35 ml of 0.5 M ammonium acetate-1 mM EDTA-0.2% SDS for 2 h at 37°C. The resulting probe (1.2 × 106 cpm) was used to hybridize to 20 μg total cytoplasmic RNA at 42°C overnight. The resulting hybrid was treated with RNase A and RNase T1 mixture for 30 min at 37°C using the RPA III RNase protection assay kit (Ambion, Austin, TX). The protected products were loaded on a 6% polyacrylamide sequencing gel and exposed on the PhosphorImager.
Quantitative PCR.
Subconfluent A549 cells in 100-mm tissue culture dishes were mock infected or infected with the UXP promoter mutants in triplicate at 10 PFU/cell. At 20 h p.i., cells were washed twice with phosphate-buffered saline (PBS) and trypsinized with trypsin-EDTA, gently pelleted, washed again with ice-cold PBS, resuspended in 1 ml ice-cold PBS, and aliquoted as follows: 500 μl for total RNA extraction and 500 μl for nuclear DNA isolation.
Total RNA were isolated with the RNeasy kit (Qiagen, Valencia, CA). RNA samples were treated with RNase-free DNase, followed by RNA Cleanup to eliminate DNA contamination according to the RNeasy minikit protocols (Qiagen). Two micrograms of RNA and 50 pmol of oligo(dT) primer were used for in vitro reverse transcription (RT) with SuperScript III reverse transcriptase. RT was performed as described in the manufacturer's instructions.
TaqMan-based quantitative reverse transcription-PCR (Q-RT-PCR) was used to specifically detect UXP mRNA. The primers and probe were designed using Primer Express software v2.0 (ABI, Foster City, CA) and synthesized by Integrated DNA Technologies (Coralville, IA). The probe was modified with the fluorophore 6-carboxyfluorescein (FAM) at the 5′ end and the quencher 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. The forward and reverse primers were designed to amplify a 76-bp segment of UXP cDNA. The sequences of the primers and probe are as follows: forward primer, CTGGGAGGAGGGCAAGGA; reverse primer, CGCGGCAAACGCTTTAAA; and probe, TTAGCAAATTTCTGTCCAGTTTATTCAGCAGCA. The reverse primer spans the junction of the UXP first and second exons. As a result, the assay preferentially detects UXP mRNA. The PCR was set up in a 50-μl volume containing 1× universal PCR master mix (ABI), 250 nM forward and reverse primers, 250 nM probe, and 5 μl of the diluted RT template. Quantification was done in triplicate for each sample using an ABI model 7500 genetic analyzer with the following cycling parameters: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. For absolute quantification, a plasmid containing full-length UXP cDNA was used to generate a standard curve. The mRNA copy number from each viral infection was normalized to that of wild-type Ad5 infection.
To isolate nuclear DNA, cells were lysed in 1 ml of Nonidet P-40 (NP-40) lysis buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1.5 mM MgCl2, 0.6% NP-40) for 1 h on ice (NP-40 is now available as Igepal CA-60 from Sigma-Aldrich [St. Louis, MO]). Nuclei were collected and washed once in 1 ml NP-40 lysis buffer and pelleted by centrifugation. Nuclear DNA was isolated with the Qiagen DNeasy kit with addition of RNase A in the first step of isolation.
The same amounts of the nuclear DNA samples were used in a Q-PCR to detect viral genome copy numbers. The primers, probe, and reaction conditions were as established previously, and assays were performed as described previously (45). For absolute quantification, 102 to 107 copies of purified Ad5 viral genomic DNA were used to generate a standard curve. The DNA copy number from each viral infection was normalized to that of wild-type Ad5 infection.
Immunoblotting.
A549 cells in 6-well plates were mock infected or infected with the viruses indicated in Fig. 3 at 25 PFU/cell. At 28 h p.i., cells were washed three times with PBS and lysed in lysis buffer (10 mM Tris-HCl [pH 7.4], 0.4% deoxycholic acid, 66 mM EDTA, 1.0% NP-40, 0.1% SDS), and the protein concentration was determined with the Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Hercules, CA). Twenty-five micrograms of each sample was electrophoresed on 15% SDS-polyacrylamide gels (SDS-PAGE) and transferred to an Immobilon-P membrane (Millipore, Bedford, MA). The blot was probed with a UXP-specific monoclonal antibody (40). The secondary antibody was horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG. Following UXP detection, the blot was stripped in stripping buffer containing 50 mM Tris-HCl [pH 6.8], 100 mM β-mercaptoethanol, and 2% SDS for 30 min at 50°C. The blot was subsequently washed twice with TBST (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.2% Tween 20) and reprobed with antiactin monoclonal antibody (Chemicon International, Temecula, CA). The bands were visualized by using the LumiGLO peroxidase chemiluminescent substrate kit (KPL, Inc., Gaithersburg, MD). The bands were quantified using ImageQuant software and normalized to actin. The data are represented after normalization to Ad5.
Indirect immunofluorescence.
A549 cells were plated onto number 1 glass coverslips in 6-well tissue culture plates. Cells (9.5 × 105 cells/well) were infected with 20 PFU/cell of the promoter mutant viruses or Ad5. At 28 h p.i., cells were fixed in 3.7% paraformaldehyde in PBS and subsequently permeabilized with methanol (−20°C) containing 4′,6-diamidino-2-phenylindole (DAPI) dihydrochloride. Cells were immunostained with a 1:1 mixture of two anti-UXP monoclonal antibodies. The secondary antibody was goat anti-mouse IgG (Molecular Probes Alexa Fluor 488 conjugate; Invitrogen Corp., Carlsbad, CA). Images were taken on a Nikon Optiphot microscope (Nikon, Melville, NY) equipped with a Nikon DXM1200 digital camera and ACT-1 software (Nikon). To assume uniformity in this analysis, the titers of all viruses were determined as a group at the same time. Also, all infections were done at the same time, identical conditions were used for immunostaining, and subsequent exposure times were identical.
Luciferase reporter assay.
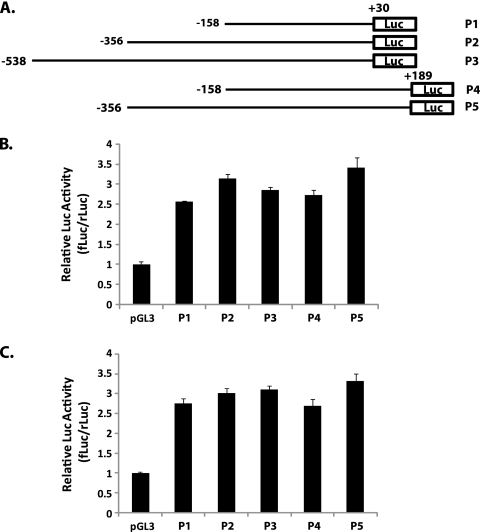
DNA fragments of different lengths spanning the transcription initiation site were amplified by PCR. The forward primers recognize sites located at bp 158, 356, or 538 upstream from the transcription initiation site, whereas reverse primers are located at bp 30 or 187 downstream of the transcription initiation site. PCR fragments were cloned into the pGL3 firefly luciferase (fLuc) vector (Promega) at BamHI/HindIII sites. HEK293 cells or A549 cells in 12-well tissue culture plates were cotransfected with 1 μg of the luciferase reporter construct and 5 ng of the phRL-TK plasmid. The phRL-TK plasmid contains Renilla luciferase (rLuc) cDNA under the control of a thymidine kinase (TK) promoter. All transfections were performed in triplicate. At 24 h posttransfection, cells were lysed in passive-lysis buffer. The activities of the firefly and Renilla luciferases were evaluated in 10 μl of cell lysate using the reagents in the dual-luciferase reporter assay system (Promega). The results are presented as ratios of firefly luciferase activity induced by the pGL3-derived promoter relative to the activity of the Renilla luciferase induced by the constitutively active phRL-TK vector.
Electrophoretic mobility shift assays (EMSA).
Nuclear extracts were prepared as follows. A549 cells (5 × 106) were mock infected or infected with Ad5 at 25 PFU/cell. At 24 h p.i., nuclear proteins were extracted using a nuclear extraction kit (Active Motif, Carlsbad, CA). Briefly, cells were washed twice with ice-cold PBS-phosphatase inhibitors, the cell pellet was resuspended in 500 μl 1× hypotonic buffer and incubated on ice for 15 min, and then 25 μl detergent was added and the solution was vortexed for 15 s. After the suspension was centrifuged at 14,000 × g for 30 s, the nuclear pellet was resuspended in 50 μl Complete lysis buffer and incubated on ice for 30 min. The supernatant was collected following centrifugation at 14,000 × g for 10 min. The nuclear protein concentration was determined with the Bio-Rad DC protein assay kit (Bio-Rad Laboratories).
For the EMSA, 6 μg of nuclear extract were added to 15 μl of reaction mixture containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 4% glycerol, 0.5 mM dithiothreitol (DTT), 50 μg/ml poly(dI-dC), and 150,000 cpm of the 32P-labeled probe. Double-stranded DNA containing the proximal I-CCAAT box of the UXP promoter was generated by annealing complementary pairs of oligonucleotides (Ad5 bp 31121 to 31168). The mutant I-CCAAT oligonucleotide used for competition carries the ATCAAAC sequence instead of the wild-type CCCCAAT sequence. The DNA fragment was subsequently end labeled with 5 U of T4 DNA kinase and 20 μCi of [γ-32P]ATP in 10 μl of buffer at 37°C for 15 min and then purified on a Sephadex G-25 spin column (Roche, Piscataway, NJ). The binding reaction mixtures were incubated at room temperature for 20 min and resolved in a 4% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer for 2 h at 200 V. Gels were dried and exposed to Kodak film at −80°C in the presence of intensifying screens. A competition experiment was carried out by preincubating the extract with an unlabeled competitor oligonucleotide for 15 min before addition of the probe. An unlabeled double-stranded oligonucleotide corresponding to wild-type I-CCAAT, the mutant I-CCAAT box, or AP1 binding sites in a 50-fold molar excess was added to the mixtures in the competition reaction.
RESULTS
Determination of the transcription initiation site of UXP RNA.
Guided by the work of Chow et al. (10), we cloned the mRNA (as cDNA) that encodes the protein named UXP. The UXP mRNA corresponds to the mRNA “2c” described by Chow et al. The 5′-end exon of UXP is located on the l strand between the early E3 region and the fiber gene (map unit [m.u.] 86.7); this exon is spliced to the second exon of DBP mRNA (m.u. 68.6) and further spliced to the main exon of DBP mRNA (m.u. 66.6), but the protein is translated in a reading frame different from that of DBP (40). Although the cloned mRNA covered the full-length sequence encoding UXP, the 5′-end transcription initiation site of the UXP pre-mRNA has not been defined. To determine the UXP pre-mRNA transcription initiation site, primer extension analysis and RNase protection assay were performed. For primer extension analysis, an oligonucleotide complementary to the first exon of UXP mRNA was end labeled with 32P, hybridized to total cytoplasmic RNA, and extended by reverse transcription. RNA from Ad5-infected A549 cells directed the synthesis of a major extension product of 61 nucleotides (nt) (Fig. 1A). No band was detected with mock-infected RNA, identifying the 61-nt product as defining the probable major (if not exclusive) transcription start site. The 5′ end of this product mapped to Ad5 bp 31062 in the Ad5 genome, 32 bp upstream of the UXP initiation codon.
Similar results were obtained in a RNase protection assay using an antisense riboprobe. When this probe was hybridized to cytoplasmic RNA and digested with RNase A/T1, a major fragment of 95 nt was protected with RNA from Ad5-infected cells (Fig. 1B). No band was detected with the RNA from the mock infection or a Torula yeast RNA negative control (Fig. 1B). The 5′ end of the major protected band mapped to around Ad5 bp 31062 in the genome, agreeing with the major transcription initiation site identified by the primer extension shown in Fig. 1A.
Identification of the proximal I-CCAAT box as a cis-acting element.
Identification of cis-acting elements of the promoter is crucial for our understanding of the regulation of UXP transcription. To examine whether potential cis-acting elements that control UXP transcription exist, computational cis-regulatory analysis of the regions flanking the transcription initiation site was performed using the transcription factor-binding site database TRANSFAC (25) via the Match platform. Because many promoters contain functionally important cis-regulatory elements downstream of the transcription initiation site and such downstream elements have been found in both TATA-containing and TATA-less promoters, we conducted the computational analysis downstream (up to bp +62) of the UXP transcription initiation site in order to cover any potential downstream promoter elements (DPE). The DPE is a core promoter element usually located at bp +28 to +32 from the initiation site. No DPE was found downstream of the UXP initiation site.
On the upstream site of the transcription initiation site, a typical eukaryotic promoter element, the TATA box, was not found in the region immediately 5′ to the UXP pre-mRNA initiation site. However, we identified several potential transcription factor binding sites (Fig. 2 A), including two CCAAT boxes in the inverted orientation (5′-ATTGG-3′ at bp −86 and −152), one E2F site in the opposite orientation (5′-GGCGCAAA-3′ at bp −123), and one E2F site in the sense orientation (5′-TTAGCGGG-3′ at bp −387). We decided to test whether these predicted inverted CCAAT (I-CCAAT) boxes and E2F sites function in the control of UXP transcription in vivo for three reasons. First, the CCAAT box is one of the most common elements in eukaryotic promoters. Second, E2F regulation of the Ad E2 early promoter through cooperative binding to a pair of E2F sites upstream of the transcription initiation site has been well documented. Third, E2F DNA-binding activity is induced in Ad-infected cells. Therefore, we mutated the I-CCAAT boxes and the E2F sites either individually or in combination. The promoter constructs containing mutations in the corresponding elements were rebuilt into the Ad5 genome so that the UXP promoter is situated in its natural context. These sites are located within the Ad fiber gene; the mutations were done in such a way that the amino acid sequence of fiber is preserved, as follows. Many CCAAT motifs that bind to different CCAAT box binding proteins with different binding specificities and affinities have been identified. The core CCAAT pentanucleotide (positions +1 to +5) is almost invariably conserved, but high variation is found in the flanking sequences. A T residue is seldom found in close proximity (positions −3 to −1 and +6 to +9); therefore, we generated our I-CCAAT box mutants by replacing the bases inside the core I-CCAAT pentanucleotide. To preserve the corresponding amino acid sequence of fiber, nucleotides 32 C at position +1 and A at +3 and +4 had to be maintained. The C at position +2 could be changed to G, A, or T, but base T at +5 could be changed only to C in order to preserve the coding asparagine of fiber. The above changes result in mutation of the proximal I-CCAAT box to CGAAC and the distal I-CCAAT box to CAAAC. Similar considerations applied in making mutations at the E2F sites; the mutations were made so as to change the E2F site as much as possible away from the E2F site consensus sequence but not change the amino acid sequence of fiber.
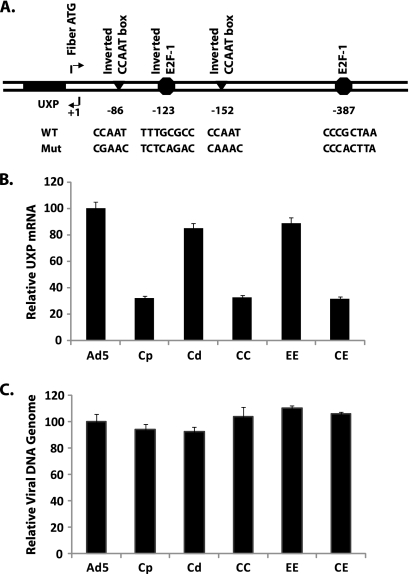
FIG. 2.
(A) Schematic diagram of putative transcriptional control elements of the Ad5 UXP promoter by TRANSFAC analysis. Triangles represent CCAAT boxes; octagons represent E2F sites. Numbers indicate the location of each element relative to the transcription initiation site (+1). Details of nucleotide changes at each site are shown underneath. WT, wild type; Mut, mutant. (B) Effects of putative cis-element mutations on UXP transcription. (C) Quantification of Ad DNA template. A549 cells were infected in triplicate with wild-type Ad5 or the indicated promoter mutants at 10 PFU/cell. At 20 h p.i., total RNA and nuclear DNA were isolated and analyzed by Q-RT-PCR and Q-PCR, respectively. Values are normalized to that for Ad5. Data represent means ± standard deviations (SD) of results from triplicate cultures assayed in triplicate. (C) The P values of results for Cp, CC, or CE versus Ad5 are 0.095, 0.272, and 0.068, respectively. Abbreviations correspond to mutations at the following sites: Cp-proximal I-CCAAT box; Cd-distal I-CCAAT box; CC-double I-CCAAT boxes; EE-double E2F mutations (sites at bp −123 and −387); CE-double I-CCAAT box plus double E2F site mutations (sites at bp −123 and −387).
A549 cells were infected with wild-type Ad5 or the promoter mutants. At 20 h p.i., RNA was prepared and quantified by Q-RT-PCR. The reverse primer in the PCR spans the junction of the UXP first and second exons, so the assay preferentially detects UXP RNA. The relative levels of UXP RNA in cells infected with the mutants indicated here were measured. The EE mutant, which abolishes both of the putative E2F sites, did not cause a significant decrease (10% decrease) in the UXP RNA level compared with that of wild-type Ad5 (Fig. 2B). The Cd mutant, which alters only the distal I-CCAAT box, also had little effect on the UXP RNA level. However, with the Cp mutant, in which only the proximal I-CCAAT box is mutated, the UXP RNA level was decreased markedly (70% decrease) compared with that in Ad5. The CC mutant (both I-CCAAT boxes mutated) and CE mutant (both I-CCAAT boxes and both E2F sites mutated) showed approximately the same level of UXP RNA as did the Cp mutant, providing further evidence that the I-CCAAT box at position −84 represents a cis-acting element that is vital for UXP promoter activity but that the distal I-CCAAT box and two E2F sites are not critical.
Decreased UXP RNA obtained with the promoter mutants is not due to decreased DNA template number.
To determine whether the decreased UXP RNA was due to a difference in DNA template number through reduced DNA replication by the Cp, CC, and CE mutants compared to that of wild-type Ad5, nuclear DNA was isolated at 20 h p.i. after a 10-PFU/cell infection, and the DNA copy number was quantified by Q-PCR. As shown in Fig. 2C, the amount of nuclear DNA template present with each mutant was similar to the amount present after Ad5 infection. The differences in DNA copy number from that in Ad5 were not significant by a two-tailed Student t test (P > 0.05). This result ruled out the possibility that differences in DNA template number account for decreased expression of UXP RNA in mutants with the proximal I-CCAAT site mutation. Therefore, we conclude that the decreased transcript numbers of the UXP gene in cells infected with mutants (Cp, CC, CE) is due to decreased promoter activity.
Decreased UXP mRNA results in decreased levels of UXP.
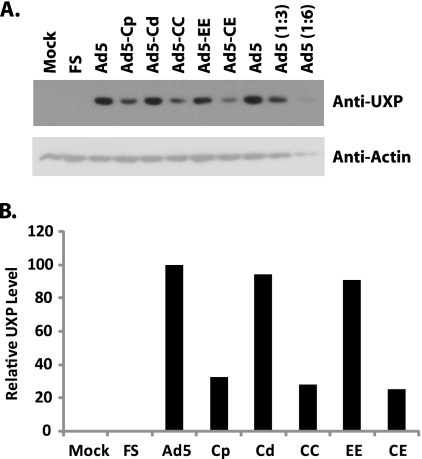
Expression of UXP in cells infected with promoter mutants was examined further at the protein level by Western blotting and immunofluorescence assays for UXP. A549 cells were infected with wild-type Ad5 or the promoter mutants indicated in Fig. 3. At 28 h p.i., cell extracts were prepared and 25 μg of each sample was assayed by immunoblotting for UXP (Fig. 3 A, upper panel) and then reprobed with antibody against actin (Fig. 3A, bottom panel). The UXP bands were quantified and normalized to actin (Fig. 3B). For the Cp mutant, in which the proximal I-CCAAT box is mutated, the UXP level was down to 30% of the Ad5 level. Mutations of either the distal I-CCAAT box (Cd) or double E2F (EE) sites had little effect on UXP expression (90% of that of wild-type Ad5). Mutations of the two I-CCAAT boxes together (CC) or mutations of the two I-CCAAT boxes as well as the two E2F sites (CE) showed decreased levels of UXP similar to that in the Cp mutant. The extent of the effect of each mutation on the UXP level is comparable to the effect for RNA levels seen from Q-RT-PCR.
FIG. 3.
Detection of UXP expression by immunoblotting. A549 cells were mock infected or infected with 25 PFU/cell of the indicated viruses. At 28 h p.i., the cells were harvested and proteins were extracted. (A) Samples containing 25 μg of protein were electrophoresed by 15% SDS-PAGE and immunoblotted for UXP (top panel) and reprobed for actin (bottom panel). (B) The bands were quantified using ImageQuant software and normalized to the corresponding actin. The data are presented after normalization to Ad5.
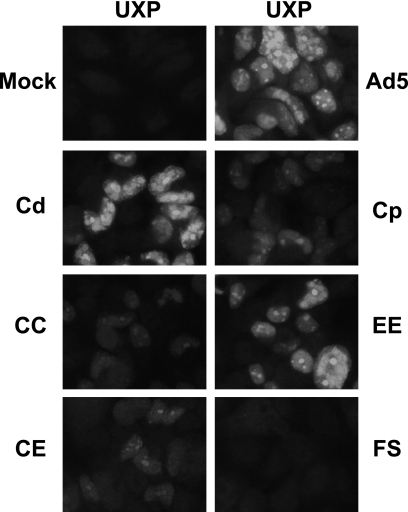
Indirect immunofluorescence of UXP in cells infected by the mutants further verified the results from Q-RT-PCR and the Western blot. As shown in Fig. 4 and compared to that of wild-type Ad5, no expression was detected with the UXP frameshift mutant (UXPFS), and markedly less UXP was detected with mutants containing the proximal I-CCAAT box mutation (Cp, CC, CE). Mutations of either the distal I-CCAAT site (Cd) or the double E2F (EE) sites had little effect on UXP expression (Fig. 4). Mutations of the two I-CCAAT boxes together (CC) or mutation of the two I-CCAAT boxes and two E2F sites combined (CE) did not result in further decreased UXP compared to the single mutation at the proximal I-CCAAT site (Cp). Taken together, these results strongly support our conclusion that the proximal I-CCAAT box is a cis-acting element in the control of UXP expression but that the distal I-CCAAT box and two E2F sites are not critical.
FIG. 4.
Immunofluorescence of UXP in cells infected with the indicated viruses. A549 cells were infected at 20 PFU/cell. At 28 h p.i., cells were fixed and immunostained with UXP-specific monoclonal antibodies.
Analysis of the promoter activity of the UXP 5′-end-flanking region.
To test whether the region surrounding the UXP pre-mRNA initiation site has promoter activity in vitro, a series of DNA fragments of different lengths spanning the transcription initiation site were ligated upstream of the luciferase reporter gene in pGL3, a promoter-free nonreplicating vector (Fig. 5 A). The plasmids were transiently transfected into A549 or HeLa cells, and luciferase activity was measured at 24 h posttransfection. To control for transfection efficiency, cells were cotransfected with a TK-driven Renilla luciferase plasmid. Firefly luciferase activity was normalized to Renilla luciferase activity and is presented as the ratio of fLuc to rLuc (fLuc/rLuc). As shown in Fig. 5B, the region between bp −538 and bp +30 (P3) or that between bp −356 and bp +30 (P2) supported an ∼3-fold-higher level of reporter expression than did the promoter-free pGL3 vector. The region from bp −158 to +30 (P1) showed luciferase activity similar to that of the longer constructs (P3 and P2). Extending the downstream region to +187 (P4 and P5) had little effect on promoter activity compared to ending the downstream region at +30. These results suggest that the cis-acting elements residing in the region between bp −158 and bp +30 of the UXP pre-mRNA initiation site are necessary for UXP basal promoter activity. This region seems to contain all the elements that are necessary and sufficient in this assay. Similar results were seen in HeLa cells transfected with the constructs (data not shown).
FIG. 5.
Transient-transfection analysis of promoter activity at 5′-end-flanking regions of UXP. (A) Schematic representation of the 5′-end-flanking region relative to the transcription initiation site; (B) luciferase assay on A549 cells; (C) luciferase assay on HEK293 cells. (B, C) Reporter plasmids containing the firefly luciferase cDNA linked with 5′-end-flanking regions of UXP were cotransfected with a control plasmid expressing Renilla luciferase. Transfection was performed in triplicate, and individual samples were analyzed using the dual-luciferase kit. Promoter activity is presented as the ratio of fLuc to rLuc (fLuc/rLuc). Data are means ± SD of results from two independent experiments.
Ad E1A transactivates a variety of different promoters, including all other Ad early promoters and major late promoters (3, 4). To address whether E1A transactivates UXP transcription, the reporter assays were performed on HEK293 cells that constitutively express Ad5 E1A gene products (Fig. 5C). Similar patterns emerged in HEK293 cells, and no increased promoter activity was detected compared with that in A549 cells, suggesting that E1A is not involved in UXP trans-activation. The result indirectly implied that the E2F sites in the 5′-end-flanking region appear not to be critical for UXP promoter activity.
A cellular factor binds specifically to the proximal I-CCAAT site of the UXP promoter.
The observation that the UXP promoter requires the proximal I-CCAAT site for efficient transcription prompted us to investigate the formation of an I-CCAAT complex in vitro. A DNA fragment containing the wild-type I-CCAAT box proximal to the UXP promoter (bp −59 to −106 relative to the transcription initiation site) was end labeled and used in gel shift assays with nuclear extracts prepared from mock- or Ad5-infected A549 cells. A double-stranded oligonucleotide corresponding to the wild-type sequence and an oligonucleotide in which the sequence is mutated were used in competition experiments. The mutant oligonucleotide has the same core mutation as that of the Cp virus, plus it has two additional residue changes. We included these two additional changes because the mutation in the Cp mutant reduced transcription to only 30% of the wild-type level, not completely. Inasmuch as transcription was not abrogated completely in the Cp mutant, we thought that there still might be binding affinity between the mutated CGAAC box and its transcription factor. Since we wanted to ensure as much as possible that the cold mutant oligonucleotide would not have any factor binding activity, we introduced two AT mutations at the −2 and −1 positions in addition to the CGAAC core mutation. Specifically, we mutated the wild-type CCC+1CAAT sequence to ATC+1AAAC (in which boldface indicates the core mutation); the T at position −1 is rarely found in close proximity to a CCAAT box.
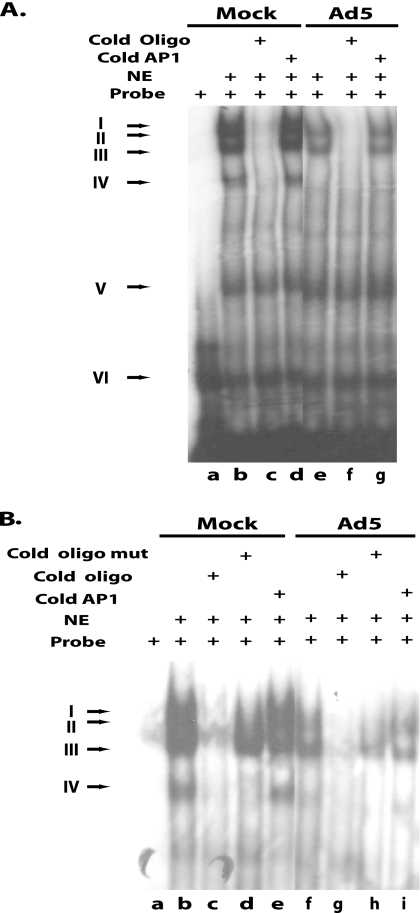
Several slower-moving bands representing DNA-protein complexes were observed in both mock- and Ad5-infected cell extracts (Fig. 6 A, lanes b and e). Bands V and VI in Fig. 6A are nonspecific, since they appear in all reactions and could not be abolished with excess cold oligonucleotides containing the I-CCAAT or AP1 sequence. The DNA-protein complex formed in band IV was present in mock-infected nuclear extracts (Fig. 6A, lanes b and d) but was absent in nuclear extracts from Ad5 infection, so protein in this band could not be responsible for binding the proximal I-CCAAT site of the UXP promoter. Even though bands I and II were competed out by the cold unlabeled oligonucleotide carrying the I-CCAAT site (Fig. 6A, lanes c and f, and B, lanes c and g), they were competed out by the oligonucleotide carrying mutations at the I-CCAAT site (Fig. 6B, lanes d and h). Therefore, the protein in this complex could bind to sequence outside the I-CCAAT sequence in the oligonucleotide instead of the core I-CCAAT site. The nuclear factor in band III appears to bind specifically to the proximal I-CCAAT site, inasmuch as the band was competed out by an unlabeled oligonucleotide carrying the wild-type I-CCAAT site of the UXP promoter (Fig. 6A, lanes c and f, and B, lanes c and g) but not by an oligonucleotide carrying a I-CCAAT mutation or AP1 site (Fig. 6B, lanes d and h). There are several nuclear transcriptional factors known to bind to the I-CCAAT box sequence (23). It remains to be determined which I-CCAAT box binding proteins are involved in the binding of this I-CCAAT box of the UXP promoter.
FIG. 6.
Recognition of the proximal I-CCAAT box of the UXP promoter by a cellular factor. Nuclear extracts (NE) prepared from mock- or Ad5-infected A549 cells were used in an EMSA. Double-stranded DNA corresponding to the proximal I-CCAAT box of the UXP promoter was end labeled as the probe in the EMSA. In the competition reaction, the unlabeled wild-type double-stranded DNA containing an I-CCAAT box (Cold Oligo), a mutant I-CCAAT box (Cold Oligo Mut), or an unrelated DNA fragment (Cold Ap1) in a 50-fold molar excess was used in the reaction. The mutant I-CCAAT oligonucleotide carries the ATCAAAC mutation instead of the wild-type CCCCAAT sequence. Components in the binding reaction mixtures are shown at the top of the figure. Arrows at the left indicate the positions of DNA-protein complexes. The cold mutant I-CCAAT oligonucleotide was included in lanes d and h in panel B.
DISCUSSION
Ad UXP is encoded from an l-strand transcription unit in the late stage of infection. To begin to characterize this transcription unit, we mapped the UXP mRNA start site using traditional primer extension and RNase protection assays. Both assays identified the major transcription initiation site as being at bp 32 upstream of the UXP initiation codon, with a C at nt −1 and an A at the +1 position of the transcription start site. The C-A dinucleotide is in accordance with the pyrimidine-purine preference at positions −1 and +1 in eukaryotic transcription initiation sites. Eukaryotic transcription mediated by RNA polymerase II starts with a purine at position +1, with a preference for pyrimidine at position −1 (7, 31, 33, 35). This pyrimidine-purine dinucleotide corresponds in part to the consensus initiator element Inr (pyrimidine, pyrimidine, A+1, X, T/A, pyrimidine, pyrimidine, where X is any nucleotide) (7, 33). Further examination of the surrounding sequence of the UXP transcription initiation site (TCA+1GACG) revealed that it is in good (but not exact) agreement with the consensus Inr element, consistent with our conclusion that this site is the major initiation site. However, no Inr element was identified in our computational analysis of the UXP promoter. Currently, the many computer programs and databases developed to search for cis-acting elements that control transcription rely on motif searches and/or comparative techniques to search for transcription factor binding sites. However, the prediction of core promoter Inr elements and the localization of the transcription initiation site remain challenging and unreliable, primarily because the defined Inr consensus element (Py Py A+1 X T/A Py Py) is loose and highly degenerate, leading to a high false-discovery rate. Thus, any initiation site detected by computational analysis still needs to be experimentally verified by traditional methods, such as primer extension and RNase protection assays, as we have done.
The Inr element is defined as a discrete core promoter element that is functionally similar to the TATA box and can function independently of a TATA box to initiate transcription (32, 33). Inrs have been identified in a variety of mammalian and viral promoters, including the TATA-containing Ad MLP (8, 9). It has been demonstrated that the Inr in the Ad MLP or IVa2 promoter is able to direct initiation of transcription independently of a TATA box (8, 9, 21). Further studies are needed to determine the role of the putative UXP Inr element in the specificity and efficiency of UXP transcription initiation.
Q-RT-PCR of UXP RNA from the I-CCAAT box mutants demonstrated that the proximal I-CCAAT box is critical for the promoter to function. This I-CCAAT box is located at nt 31148 in the Ad5 genome (GenBank accession number M73260). The proximal I-CCAAT box is well conserved among different serotypes of species C human Ads, suggesting the importance of the proximal I-CCAAT box in controlling UXP expression. The distal I-CCAAT box sequence appears not to be required for UXP transcription. This sequence is not conserved among species C Ad serotypes (ATTGG in Ad5 [GenBank accession number M73260.1] versus GTGGG in Ad1 [NCBI reference sequence AC_000017.1] and Ad2 [NCBI reference sequence AC_000007.1] and GTGTG in Ad6 [GenBank accession number AB108424.1]). Regarding the putative E2F sites, our data indicate that neither site is required for UXP transcription. Despite this result, these two sites fit the consensus sequence for E2F binding sites, and their position relative to the transcription initiation site is conserved in species C serotypes.
The presence of promoter activity in the upstream region of the UXP first exon was further verified in the transient-transfection reporter assay with both the A549 and HEK293 cell lines. However, the luciferase activity detected in the cells transfected with plasmids containing sequences upstream of UXP was only moderately increased (∼3-fold) compared to the activity of those transfected with the parental pGL3 plasmid. Several possibilities can be considered for the relatively low luciferase activity. First, the low activity may simply be due to the weakness of the promoter. However, we have not encountered any difficulty in detecting UXP in Ad-infected cells. Second, the promoter may require DNA replication to activate maximal transcription. The pGL3-derived plasmids used in the reporter assay do not contain a replication origin, so presumably no replication of the plasmids occurs in the transfected cells. It has been shown that when a nonreplicating plasmid DNA containing the Ad pIX transcription unit was introduced into HeLa cells, no pIX transcript was detected (24, 28). In contrast, efficient transcription was detected in cells transfected with a replicating plasmid containing the pIX gene. The same phenomenon was demonstrated for the Ad IVa2 promoter. The IVa2 promoter was unable to drive efficient synthesis of GFP in the absence of a simian virus 40 (SV40) origin of replication in a reporter assay in a transient-transfection system (17). Furthermore, it has been well documented in superinfection experiments that replication of the DNA template is required for maximal activation of expression of pIX, IVa2, and late-region L2 to L5 proteins (11, 24, 37). Expression of pIX, IVa2, and late genes of Ad is detected only after Ad DNA synthesis has occurred. It has been shown that IVa2 transcription is regulated by a cellular repressor that is titrated out upon Ad genome replication (17, 18), and MLP is regulated by IVa2 in conjunction with the late L4-22K and -33K proteins (2, 26). Our previous study had shown that UXP expression is detected only in cells at the late stage of infection; no UXP was detected in the presence of cytosine arabinoside (an inhibitor of DNA replication). This observation led us to speculate that UXP expression, like that of pIX, pIVa2, and MLP, might require DNA replication to achieve maximal active transcription. However, the mechanism underlying this restriction of UXP to late-phase infection is not understood.
As discussed, our mutation data clearly showed that the proximal I-CCAAT box is critical for the promoter to function. The CCAAT box is one of the most common elements in eukaryotic promoters, present in more than 30% of all promoters in the forward or reverse orientation (7). The Ad MLP contains an I-CCAAT box which is highly conserved in Ads (34). The frequency of CCAAT boxes appears to be relatively higher in TATA-less promoters, particularly in the inverted (ATTGG) orientation. The CCAAT pentanucleotide is typically present at bp −60 to −100 upstream of transcription start sites (23). An I-CCAAT box (positions −72 and −135 relative to the E2 late cap site) has been demonstrated in the regulation of the activity of the Ad E2L promoter, a promoter with a poor TATA consensus that is activated after Ad DNA replication (5, 14). The Y box protein YB-1 has been shown to bind to the proximal I-CCAAT box of E2L to control E2 gene expression at late stages of infection (15). We have observed some similarities between the E2L and UXP transcription units: both are transcribed from the l strand and are active at late stages of infection, both mRNAs are spliced to the DBP mRNA leader and DBP mRNA main-body exons, both DBP and UXP are localized in Ad DNA replication centers, both promoters are regulated by an I-CCAAT box located upstream of the transcription initiation site, and neither promoter has a good TATA consensus. It remains to be determined whether YB1 binds to the proximal I-CCAAT box in the UXP promoter. The CCAAT box is known to bind to a plethora of proteins, including CCAAT/enhancer binding protein (c/EBP), CCAAT transcription factor (CTF), CCAAT displacement protein (CDP), Y box factors, and NF-Y (23). Our EMSA has indicated that a cellular factor binds specifically to the proximal I-CCAAT site of the UXP promoter. Further experiments are needed to identify the underlying transcription factor(s) in the control of UXP transcription.
In conclusion, this communication is the first definitive identification of a promoter in the Ad5 genome that is active at late stages of infection and that is embedded in the fiber gene and drives transcription off the l strand. Further, we report the first mapping of the transcription initiation site of the UXP pre-mRNA and of the control elements of the UXP promoter. Our data support the requirement of the proximal I-CCAAT box in the control of UXP transcription and indicate that the E2F sites are not critical. This analysis has shed new light on the transcriptional regulation of this novel promoter.
Acknowledgments
This research was supported by grant RO1 CA118022 to W.S.M.W. from the National Institutes of Health.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Akusjarvi, G. 2008. Temporal regulation of adenovirus major late alternative RNA splicing. Front. Biosci. 13:5006-5015. [DOI] [PubMed] [Google Scholar]
- 2.Ali, H., G. LeRoy, G. Bridge, and S. J. Flint. 2007. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late-phase-specific stimulation of transcription. J. Virol. 81:1327-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673-7685. [DOI] [PubMed] [Google Scholar]
- 4.Berk, A. J. 2007. Adenoviridae: the viruses and their replication, p. 2355-2394. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams, & Wilkins, Philadelphia, PA.
- 5.Bhat, G., L. SivaRaman, S. Murthy, P. Domer, and B. Thimmappaya. 1987. In vivo identification of multiple promoter domains of adenovirus EIIA-late promoter. EMBO J. 6:2045-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackford, A. N., and R. J. Grand. 2009. Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J. Virol. 83:4000-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucher, P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563-578. [DOI] [PubMed] [Google Scholar]
- 8.Carcamo, J., L. Buckbinder, and D. Reinberg. 1991. The initiator directs the assembly of a transcription factor IID-dependent transcription complex. Proc. Natl. Acad. Sci. U. S. A. 88:8052-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., and S. J. Flint. 1992. Mutational analysis of the adenovirus 2 IVa2 initiator and downstream elements. J. Biol. Chem. 267:25457-25465. [PubMed] [Google Scholar]
- 10.Chow, L. T., T. R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. [DOI] [PubMed] [Google Scholar]
- 11.Crossland, L. D., and H. J. Raskas. 1983. Identification of adenovirus genes that require template replication for expression. J. Virol. 46:737-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25-54. [DOI] [PubMed] [Google Scholar]
- 13.Fessler, S. P., F. Delgado-Lopez, and M. S. Horwitz. 2004. Mechanisms of E3 modulation of immune and inflammatory responses. Curr. Top. Microbiol. Immunol. 273:113-135. [DOI] [PubMed] [Google Scholar]
- 14.Goding, C. R., S. M. Temperley, and F. Fisher. 1987. Multiple transcription factors interact with the adenovirus-2 EII-late promoter: evidence for a novel CCAAT recognition factor. Nucleic Acids Res. 15:7761-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, P. S., S. Bergmann, K. Jurchott, H. Lage, K. Brand, A. Ladhoff, K. Mantwill, D. T. Curiel, M. Dobbelstein, M. Dietel, B. Gansbacher, and H. D. Royer. 2002. YB-1 relocates to the nucleus in adenovirus-infected cells and facilitates viral replication by inducing E2 gene expression through the E2 late promoter. J. Biol. Chem. 277:10427-10434. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz, M. S. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gen. Med. 6(Suppl. 1):S172-S183. [DOI] [PubMed] [Google Scholar]
- 17.Huang, W., J. Kiefer, D. Whalen, and S. J. Flint. 2003. DNA synthesis-dependent relief of repression of transcription from the adenovirus type 2 IVa(2) promoter by a cellular protein. Virology 314:394-402. [DOI] [PubMed] [Google Scholar]
- 18.Iftode, C., and S. J. Flint. 2004. Viral DNA synthesis-dependent titration of a cellular repressor activates transcription of the human adenovirus type 2 IVa2 gene. Proc. Natl. Acad. Sci. U. S. A. 101:17831-17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppard, K. N. 1997. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J. Gen. Virol. 78:2131-2138. [DOI] [PubMed] [Google Scholar]
- 20.Lichtenstein, D. L., K. Toth, K. Doronin, A. E. Tollefson, and W. S. M. Wold. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23:75-111. [DOI] [PubMed] [Google Scholar]
- 21.Lu, H., M. D. Reach, E. Minaya, and C. S. Young. 1997. The initiator element of the adenovirus major late promoter has an important role in transcription initiation in vivo. J. Virol. 71:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutz, P., M. Rosa-Calatrava, and C. Kedinger. 1997. The product of the adenovirus intermediate gene IX is a transcriptional activator. J. Virol. 71:5102-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantovani, R. 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui, T., M. Murayama, and T. Mita. 1986. Adenovirus 2 peptide IX gene is expressed only on replicated DNA molecules. Mol. Cell. Biol. 6:4149-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris, S. J., and K. N. Leppard. 2009. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 83:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris, S. J., G. E. Scott, and K. N. Leppard. 2010. Adenovirus late phase infection is controlled by a novel L4 promoter. J. Virol. 84:7096-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan, V., and N. P. Salzman. 1985. Cis and trans activation of adenovirus IVa2 gene transcription. Nucleic Acids Res. 13:4067-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardo-Mateos, A., and C. S. Young. 2004. Adenovirus IVa2 protein plays an important role in transcription from the major late promoter in vivo. Virology 327:50-59. [DOI] [PubMed] [Google Scholar]
- 30.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smale, S. T. 1997. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta 1351:73-88. [DOI] [PubMed] [Google Scholar]
- 32.Smale, S. T., and D. Baltimore. 1989. The “initiator” as a transcription control element. Cell 57:103-113. [DOI] [PubMed] [Google Scholar]
- 33.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 34.Song, B., and C. S. Young. 1998. Functional analysis of the CAAT box in the major late promoter of the subgroup C human adenoviruses. J. Virol. 72:3213-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, Y., H. Taira, T. Tsunoda, J. Mizushima-Sugano, J. Sese, H. Hata, T. Ota, T. Isogai, T. Tanaka, S. Morishita, K. Okubo, Y. Sakaki, Y. Nakamura, A. Suyama, and S. Sugano. 2001. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Symington, J. S., L. A. Lucher, K. H. Brackmann, A. Virtanen, U. Pettersson, and M. Green. 1986. Biosynthesis of adenovirus type 2 i-leader protein. J. Virol. 57:848-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, G. P., and M. B. Mathews. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22:523-533. [DOI] [PubMed] [Google Scholar]
- 38.Tollefson, A. E., M. Kuppuswamy, E. V. Shashkova, K. Doronin, and W. S. M. Wold. 2007. Preparation and titration of CsCl-banded adenovirus stocks. Methods Mol. Med. 130:223-235. [DOI] [PubMed] [Google Scholar]
- 39.Tollefson, A. E., A. Scaria, S. K. Saha, and W. S. M. Wold. 1992. The 11,600-MW protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J. Virol. 66:3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tollefson, A. E., B. Ying, K. Doronin, P. D. Sidor, and W. S. Wold. 2007. Identification of a new human adenovirus protein encoded by a novel late l-strand transcription unit. J. Virol. 81:12918-12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weitzman, M. D. 2005. Functions of the adenovirus E4 proteins and their impact on viral vectors. Front. Biosci. 10:1106-1117. [DOI] [PubMed] [Google Scholar]
- 43.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686-7696. [DOI] [PubMed] [Google Scholar]
- 44.Windheim, M., A. Hilgendorf, and H. G. Burgert. 2004. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr. Top. Microbiol. Immunol. 273:29-85. [DOI] [PubMed] [Google Scholar]
- 45.Ying, B., K. Toth, J. F. Spencer, J. Meyer, A. E. Tollefson, D. Patra, D. Dhar, E. V. Shashkova, M. Kuppuswamy, K. Doronin, M. A. Thomas, L. A. Zumstein, W. S. M. Wold, and D. L. Lichtenstein. 2009. INGN 007, an oncolytic adenovirus vector, replicates in Syrian hamsters but not mice: comparison of biodistribution studies. Cancer Gene Ther. 16:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]