Abstract
Penaeus stylirostris densovirus (PstDNV), a pathogen of penaeid shrimp, causes significant damage to farmed and wild shrimp populations. In contrast to other parvoviruses, PstDNV probably has only one type of capsid protein that lacks the phospholipase A2 activity that has been implicated as a requirement during parvoviral host cell infection. The structure of recombinant virus-like particles, composed of 60 copies of the 37.5-kDa coat protein, the smallest parvoviral capsid protein reported thus far, was determined to 2.5-Å resolution by X-ray crystallography. The structure represents the first near-atomic resolution structure within the genus Brevidensovirus. The capsid protein has a β-barrel “jelly roll” motif similar to that found in many icosahedral viruses, including other parvoviruses. The N-terminal portion of the PstDNV coat protein adopts a “domain-swapped” conformation relative to its twofold-related neighbor similar to the insect parvovirus Galleria mellonella densovirus (GmDNV) but in stark contrast to vertebrate parvoviruses. However, most of the surface loops have little structural resemblance to any of the known parvoviral capsid proteins.
The Parvoviridae family is a family of small DNA viruses that is divided into two subfamilies, the Parvovirinae that infect vertebrates and the Densovirinae that infect invertebrates. Penaeus stylirostris densovirus (PstDNV), also known as infectious hypodermal and hematopoietic necrosis virus (IHHNV), belongs to the Densovirinae subfamily and was first reported as a highly lethal disease of juvenile shrimp in 1983 (22). The virus has significant commercial impact on the shrimp farming industry, causing mass mortality and severe deformations in penaeid shrimp during catastrophic epidemics in marine aquaculture facilities worldwide (14). PstDNV is closely related to the mosquito brevidensoviruses (35), which have the potential to be used as biological control agents of mosquito-borne diseases, such as malaria (30), dengue, chikungunya, and yellow fever (8).
The single-stranded DNA genome of parvoviruses is encapsidated within a nonenveloped, icosahedral protein shell of less than 280 Å in external diameter. The capsid consists of 60 structurally equivalent subunits that are composed of the major viral coat protein and a few copies of N-terminally extended variants of the major capsid protein. A phospholipase A2 (PLA2) activity in the unique N-terminal extension of the largest minor capsid protein plays a crucial role during parvoviral host cell infection (7, 12, 13, 20, 46). The structures of the major capsid protein of several vertebrate parvoviruses have previously been determined to near-atomic resolution (1, 18, 23, 37, 41, 43, 44). However, the only high-resolution structure available for the invertebrate subfamily is that of the insect parvovirus Galleria mellonella densovirus (GmDNV) (36). The central motif of parvoviral capsid proteins is an eight-stranded, antiparallel β-barrel “jelly roll” fold. The surface of the virion, however, is formed by large insertions connecting the strands of the β-barrel, thereby creating features that govern antigenicity, receptor binding, and most intersubunit contacts. Surface characteristics common to most parvoviruses are protrusions at or around the icosahedral threefold axes, depressions on the twofold axes, and canyons surrounding the fivefold axes. At each fivefold apex, a cylindrical pore connects the interior of the virus particle with its exterior surroundings. In full virions, these pores are occupied by a glycine-rich motif in the N-terminal region of the major capsid protein, presumably positioning the N-terminal peptide for externalization. The general surface topology of GmDNV is smoother, probably due to smaller loop insertions. The structure of some of these insertions has diverged from vertebrate parvoviruses beyond recognition (4, 36). The N-terminal portions of twofold-related subunits in GmDNV have swapped their positions relative to those of the vertebrate parvoviruses. A cryo-electron microscopy (cryo-EM) study of Aedes albopictus densovirus, a brevidensovirus, has shown that its surface features are different from GmDNV and the mammalian parvoviruses, in particular in having prominent protrusions at the fivefold axes (9).
Although it has been reported that PstDNV contains four structural proteins, as determined by SDS-polyacrylamide gel electrophoresis (3), these data do not fit the coding sequence (35). The 4.1-kb DNA genome of PstDNV (3) encodes in the 3′ half of the plus strand just one structural protein of 329 amino acids, as of now the smallest reported parvoviral capsid protein, and in the 5′ half of the plus strand two nonstructural proteins (666 and 363 amino acids) (35). Having only a single type of capsid protein is an unusual feature for viruses in the Parvoviridae family, where capsids are generally reported to contain two or more coat protein variants. A stretch of 11 amino acids in the N-terminal region of the capsid protein (17-DAHNEDEEHAE-27) is reminiscent of the PLA2 catalytic site (35), but it lacks important conserved motifs of PLA2s. Consequently, but curiously, PstDNV does not have the enzymatic activity that has previously been described as a requirement for parvoviral infectivity.
We report here the three-dimensional (3D) crystal structure of recombinant, empty virus-like particles (VLPs) of the shrimp parvovirus PstDNV at 2.5-Å resolution. The loops connecting the strands of the structurally conserved jelly roll motif differ considerably in structure and length from other parvoviruses. The near-atomic resolution structure might provide the basis for the design of capsid binding antiviral compounds that may protect shrimp against parvoviral infection (16, 32, 42). Furthermore, the structure might aid the targeting of monoclonal antibodies to gain functional data about the role of the Brevidensovirus capsid protein during the infection cycle. Such information in turn may permit the design of densovirus-based delivery systems for drugs or pest control agents in aquacultural facilities. The small dimensions of PstDNV VLPs can be advantageous for their possible use as nanoparticles for antigen presentation and transport of immune stimulatory substances or interfering RNAs (21, 26). Additionally, the small size of the PstDNV capsid protein makes the system attractive as a model for studying assembly mechanisms of icosahedral virus capsids.
MATERIALS AND METHODS
Preparation of virus-like particles and crystallization.
The complete coding sequence of the PstDNV viral coat protein (VP) was amplified by PCR using the primers IHVPF1 (5′-TTTGGATCCA TGTGCGCCGA TTCAACAAGA ACAAGC-3′) and IHVPR1 (5′-TTTTCTAGAT TAGTTAGTAT GCATAATATA ACATTTG-3′) and cloned into the pFastbac1 vector (Invitrogen) using the BamHI and XbaI restriction sites. The insert was sequenced and identical to the sequence that has been assigned GenBank accession no. AF273215. The recombinant plasmid was then transfected into Sf9 insect cells to obtain recombinant baculovirus for the heterologous expression of PstDNV VP (329 residues, 37.5 kDa).
Self-assembled, recombinant, empty PstDNV VLPs were purified from baculovirus-infected insect cells by gradient density centrifugation. In short, capsids were concentrated from the cell culture supernatant by centrifugation (4.5 h, 150,000 × g, 10°C) through a CsCl cushion (0.6 g/ml in 50 mM Tris-HCl [pH 8.7], 25 mM EDTA, 0.5% Triton X-100). The particles were further purified by iodixanol gradient centrifugation (4.2 to 24% iodixanol in 10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM MgCl2, 1 mM CaCl2; 1.5 h, 160,000 × g, 10°C). The purity and quality of the final virus preparation was estimated by SDS-polyacrylamide gel electrophoresis with Coomassie blue staining. Two protein bands were observed. The major protein band corresponded to a slightly lower molecular mass of ∼36 kDa than the expected 37.5 kDa (minor band), probably due to proteolytic processing.
The purified protein capsids were crystallized using the hanging drop vapor diffusion technique. Aliquots (2 μl) of sample in 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM MgCl2, 1 mM CaCl2 (protein concentration, 5 to 14 mg/ml) were mixed with an equal volume of reservoir solution (1.5 to 1.75% polyethylene glycol 8000 [PEG 8000] in 10 mM Tris-HCl [pH 7.5] containing 5% glycerol or 100 mM NaCl), and the droplets were equilibrated against 1 ml reservoir solution at 20°C. The crystals were soaked for 5 min in the presence of 35% glycerol as a cryoprotectant prior to flash freezing for X-ray data collection.
Cryo-EM and 3D image reconstruction.
Micrographs of the frozen-hydrated PstDNV particles were recorded on Kodak SO-163 films with a Philips CM300 FEG transmission electron microscope (Philips, Eindhoven, Netherlands) (see Fig. S1A in the supplemental material). Images were taken at a calibrated magnification of 47,010 and at a total electron dose of 18.4 e−/Å2. The cryo-EM micrographs were digitized at 7-μm intervals with a Zeiss SCAI scanner, resulting in a sampling of the specimen at 1.49-Å intervals. A total of 18,014 particles from 9 micrographs were boxed and processed using the RobEM program (2). The defocus levels were determined by fitting the theoretical microscope contrast transfer functions (CTF) to the incoherent average of the Fourier transforms of all particle images from each micrograph and ranged from 3.06 to 1.36 μm. A rough 3D model of PstDNV was generated with the starticos program of the EMAN software package (25) by identifying particles with the best fivefold, threefold, or twofold rotational symmetry from a subset of about 2,400 particle images. The refined 3D model calculated from the class averages for each of these three views was used to initiate the reconstruction from all available data. The orientations and origins of all the particles were determined by comparing the CTF-corrected particle images against reference projections of the current 3D model using the AUTO3DEM automated image reconstruction system (45). The procedure was repeated at progressively higher spatial frequencies until no further increase in the correlation coefficients could be obtained. To improve the reliability of the orientation refinement, the image data were band pass filtered to reduce low and high frequency noise. A total of 9,009 particles were selected on the basis of correlation between the observed image and the density projection of the current model oriented in the presumed orientation to calculate the final 3D electron density map. The resolution of the resulting map was estimated by comparing structure factors of the virus shell computed from two independent half-data sets (see Fig. S1B in the supplemental material). The Fourier shell correlation coefficient was 0.5 at a resolution of 6.17 Å. For the final three-dimensional reconstruction, data were included to a resolution of 6.1 Å, where the correlation between the two independent data sets was 0.2 (see Fig. S1C in the supplemental material).
X-ray structure determination.
X-ray diffraction data were collected from a single frozen crystal at the Advanced Photon Source (beamline GM/CA-CAT 23-ID-B, Argonne National Laboratory) (Table 1). Data collection was stopped after 450 films because of noticeable radiation damage. The crystal diffracted to 2.2-Å resolution, although data beyond 2.5 Å were weak. The diffraction data were integrated and scaled using the HKL data processing package (28) (Table 1). Only the first 250 films were further processed for structure determination to ensure high-quality data. The orthorhombic space group P21221 was assigned on the basis of symmetry and systematic absences. Assuming two virus particles per unit cell, the Matthews coefficient VM was 3.46 Å3/Da, corresponding to a solvent content of 64.5%. Thus, the particle is located on a crystallographic twofold axis parallel to b, with half a particle per asymmetric unit. The particle orientation about the b axis was determined by a self-rotation function calculated with the GLRF program (39) using data between 5- and 3.5-Å resolution. Packing consideration using spheres with a diameter between 215 and 250 Å suggested a rough position of the capsid center at (0, 1/4, 0) for maximum separation within the unit cell. The cryo-EM density map of PstDNV was used as a phasing model for molecular replacement. Structure factors between 20- and 7.5-Å resolution were derived from this model and used to calculate cross-rotation and translation functions with the AMoRe program (27, 40), verifying the rotation and approximate center parameters. The particle center and rotation matrix were refined several times during phase extension using the climb function of the real-space averaging program envelp (34). Averaging was performed within a spherical mask between the radii of 60 and 130 Å from the particle center. The initial phases from the model were extended from 8 to 2.5 Å in resolution steps of one reciprocal lattice point (1/c) with 10 cycles of 30-fold noncrystallographic symmetry (NCS) averaging per extension step. An additional 30 cycles of density averaging at 2.5-Å resolution were performed to achieve maximum convergence between observed and calculated structure factors. Averaging was finalized with 70 cycles during which calculated structure factors were used and weighted with the mean figure of merit for the given resolution in place of unobserved reflections. The final center position of the particle was (0.000, 0.256, 0.000) with an orientation given by
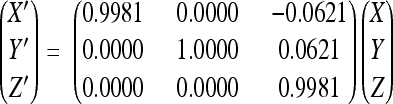 |
relative to the standard icosahedral orientation and position of the search model, where (X Y Z) are the coordinates of a position in the standard particle.
TABLE 1.
Data collection and refinement statistics
| Parameter or statistic | Value(s) for parameter or statistica |
|---|---|
| Data collection parameters | |
| Wavelength (Å) | 1.0332 |
| CCD detectorb | Rayonix MarMosaic 300 |
| Distance from sample to detector (mm) | 400 |
| Exposure time per frame (s) | 3 |
| Oscillation angle per frame (°) | 0.2 |
| No. of frames collected | 450 |
| Data reduction and refinement statistics | |
| Resolution range (Å) | 50-2.5 (2.59-2.50) |
| Space group | P21221 |
| No. of frames used | 1-250 |
| Cell dimensions (a, b, c) (Å) | 235.9, 245.5, 268.5 |
| Mosaicity (°) | 0.42 |
| No. of observed reflections | 896,125 |
| No. of unique reflections | 338,268 (23,750) |
| Redundancy | 2.6 (1.5) |
| Completeness (%) | 64.4 (45.5) |
| 〈I〉/〈σ(I)〉c | 8.10 (1.35) |
| Rsym = ∑‖(I − 〈I〉)∑‖(I) | 0.107 (0.598) |
| No. of residues/no. of atoms | 299/2,396 |
| No. of metal ions | 3 |
| No. of water molecules | 159 |
| Final R-factor | 0.2778 (0.3758) |
| Rfreed | 0.2851 (0.3764) |
| RMSD bond lengths (Å) | 0.0063 |
| RMSD bond angles (°) | 1.4116 |
| Residues in most favored regions (%)/residues in additionally allowed | |
| regions (%)e | 89.5/10.5 |
Values in parentheses refer to the highest-resolution shell.
CCD detector, charge-coupled-device detector.
I is the intensity of a reflection, and 〈I〉 is the average intensity.
The test set contains 1.3% of observed unique reflections.
The protein structure (residues 31 to 329) was built into the electron density map using the Coot program (10) (Fig. 1; see Fig. S2 in the supplemental material). The initial R-factor for the manually built model was 36.0% (46.2% in the highest resolution shell between 2.59 and 2.50 Å). Atomic positions were refined with the CNS program package (5, 6) applying strict icosahedral NCS constraints (simulated annealing refinement, group B-factor refinement, restrained individual B-factor refinement, crystallographic conjugate gradient minimization, automated and manual water picking) alternated with model rebuilding into NCS-averaged 2Fo-Fc and Fo-Fc electron density maps. Additionally, 159 water molecules and three divalent metal ions (1 Ca2+ and 2 Mg2+) were tentatively placed into the density. The identity of the metal ions, present in equimolar ratio in the sample buffer, was tentatively assigned according to density height and coordination. The final R-factor for the model was 27.8% (Table 1). Structure analysis with the PROCHECK program (19) showed that 89.5% of the nonglycine, nonproline residues were within the most favored regions and 10.5% within additionally allowed regions of the Ramachandran plot. The bond lengths and angles deviated from idealized values by 0.006 Å and 1.412°, respectively. Secondary structure elements were defined by using the PROCHECK program (17, 19) (Table 2).
FIG. 1.
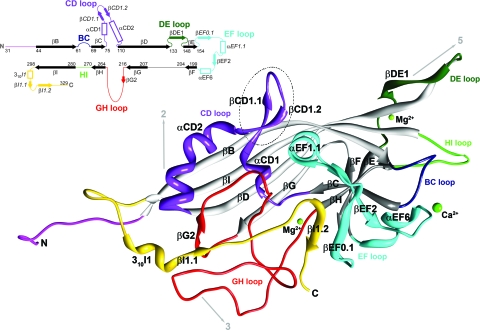
Ribbon diagram of the crystal structure of the PstDNV capsid protein. The BIDG and CHEF sheets of the eight-stranded β-barrel are shown in light gray and dark gray, respectively. The surface loops connecting the strands of the β-barrel are labeled by color as follows: BC loop, dark blue; CD, purple; DE, dark green; EF, cyan; GH, red; and HI, green. The N-terminal region upstream of βB is shown in magenta, and the C-terminal portion downstream of βI is shown in gold. The positions of icosahedral symmetry elements and their direction from the viral center are indicated by arrows. Between the conserved helical elements αCD1 and αCD2 (αA), the dashed circle highlights a hairpin loop insertion that is unique to the PstDNV structure. The position of secondary structural elements along the primary sequence is also presented in a schematic shown in the top left corner. Numbering corresponds to the starting residues of the β-strands (solid arrows), shown black in the conserved jelly roll β-barrel and colored in connecting loops. Helical elements are depicted as open rectangles. Secondary structure elements are named as defined by Xie and Chapman (44). Additional secondary structure elements designated for PstDNV (Table 3) are labeled in italic font. The N terminus (N) and C terminus (C) are indicated.
TABLE 2.
Sequential position of secondary structure elements in the PstDNV capsid proteina
| SSECPVb | SSEPSTc | Amino acid positions in PstDNV |
|---|---|---|
| βB | 44-60 | |
| βC | 69-74 | |
| αCD1 | 79-83 | |
| βCD1.1 | 84-87 | |
| βCD1.2 | 92-95 | |
| αCD2d | 96-110 | |
| βD | 111-132 | |
| βDE1 | 136-142 | |
| βE | 148-153 | |
| βEF0.1 | 160-165 | |
| αEF1.1 | 170-177 | |
| βEF2 | 179-183 | |
| αEF6e | 191-197 | |
| βF | 199-203 | |
| βG | 207-215 | |
| βG2 | 220-222 | |
| βH | 264-269 | |
| βI | 280-297 | |
| 310I1 | 306-310 | |
| βI1.1 | 311-313 | |
| βI1.2 | 321-325 |
Conserved parvoviral secondary elements (β-barrel and αA) are indicated by boldface type.
SSECPV, secondary structure elements as defined for CPV by Xie and Chapman (44) that structurally align with PstDNV.
SSEPST, additional secondary structure elements designated for PstDNV.
Also referred to as α-helix A.
Also referred to as α-helix B.
Structure comparison and evolutionary analysis.
The structure of PstDNV was compared with the insect parvovirus GmDNV (Protein Data Bank [PDB] accession number 1DNV; 3.7 Å) and the mammalian parvoviruses canine parvovirus (CPV) (PDB accession number 4DPV; 2.9 Å), adeno-associated virus 2 (AAV-2) (PDB accession number 1LP3; 3.0 Å), and human parvovirus B19 (PDB accession number 1S58; 3.5 Å). Secondary structure elements were designated as defined by the PROCHECK program (17, 19) (see Table S1 in the supplemental material).
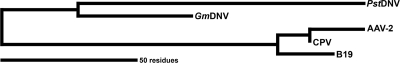
As an exploratory evolutionary analysis, the differences in the lengths of the secondary structural elements as extracted from atomic models (Table 3) were encoded as weighted additive binary characters and then analyzed using PAUP* (38). An unrooted phylogenetic tree was obtained with a consistency index (CI) of 0.8222 using Wagner parsimony. When calculating a Hamming or edit distance between these characters and then applying a least-squares method (unweighted or Fitch Margoliash [FM]), the tree remained the same, with a reported percentage of standard deviation per distance (%SD) of 3.8 and 3.1, respectively. Bootstrapping of the data showed a split between mammalian and nonmammalian viruses with 74% confidence. CPV and AAV-2 were grouped together with 63% confidence. These confidence values are relatively low, most likely because only one character, the GH loop, strongly separates the mammalian viruses from the nonmammalian viruses and seems to be the defining feature for determining the evolutionary separation of the different viruses (11). Consideration of only the loops resulted in the same tree with a slightly better fit (CI of 0.8667; %SD FM of 2.8), while consideration of just the strands of the β-barrel showed no consistent trend in distinguishing these viruses. The latter was to be expected, as the β-barrel is a conserved structural feature among all parvoviruses. Analysis of the most conserved sequence elements suggests that the human parvoviruses B19 and AAV-2 might be closer relatives, but the results were inconclusive.
TABLE 3.
Length comparison between structural elements (strands of β-barrel and connecting loops) of different parvoviruses
| Element | Length comparison (%)a |
No. of residues in the structural element in CPV | |||
|---|---|---|---|---|---|
| PstDNV | GmDNV | B19 | AAV-2 | ||
| βB | 106.3 | 93.8 | 75.0 | 75.0 | 16 |
| BC loop | 22.9 | 65.7 | 111.4 | 82.9 | 35 |
| βC | 150.0 | 100.0 | 125.0 | 100.0 | 4 |
| CD loop | 180.0 | 120.0 | 120.0 | 100.0 | 20 |
| βD | 100.0 | 63.6 | 81.8 | 90.9 | 22 |
| DE loop | 88.2 | 100.0 | 111.8 | 94.1 | 17 |
| βE | 85.7 | 71.4 | 42.9 | 100.0 | 7 |
| EF loop | 61.6 | 138.4 | 89.0 | 75.3 | 73 |
| βF | 125.0 | 100.0 | 100.0 | 75.0 | 4 |
| FG loop | 60.0 | 220.0 | 80.0 | 100.0 | 5 |
| βG | 112.5 | 37.5 | 87.5 | 87.5 | 8 |
| GH loop | 21.1 | 42.1 | 96.5 | 100.4 | 228 |
| βH | 85.7 | 85.7 | 42.9 | 100.0 | 7 |
| HI loop | 55.6 | 94.4 | 100.0 | 116.7 | 18 |
| βI | 94.7 | 84.2 | 94.7 | 89.5 | 19 |
| C terminus | 72.7 | 104.6 | 138.6 | 104.6 | 44 |
| Total proteinb | 54.3 | 76.3 | 98.7 | 94.5 | 527 |
| Total jelly roll | 102.3 | 77.0 | 80.5 | 88.5 | 87 |
| Total loops | 44.8 | 76.1 | 102.3 | 95.7 | 440 |
Length is given as percentage relative to the number of residues in the equivalent element in CPV. Strands of the jelly roll β-barrel are indicated by boldface type.
Total protein length is considered starting from strand B of the β-barrel.
Figure preparation.
Molecular graphics images were produced using the Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (29).
Structure accession numbers.
The cryo-EM density map was deposited in the EM data bank under accession number EMD-1624. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under PDB accession number 3N7X.
RESULTS AND DISCUSSION
The crystal structure of virus-like particles of PstDNV, the first near-atomic structure of a Brevidensovirus, has been solved using diffraction data to 2.5-Å resolution (Fig. 1 and Table 1; see Fig. S2 in the supplemental material). Most of the 329 residues of the viral capsid protein were traceable in the icosahedrally averaged electron density, except for 30 N-terminal residues. Such lack of ordered structure in the N-terminal region of the major coat proteins was consistently observed for all parvoviral structures thus far. For PstDNV, this is most likely due to a positional variation, incompatible with the icosahedral symmetry averaging imposed during structure determination, and to the partial proteolytic cleavage of the N-terminal region (see Materials and Methods).
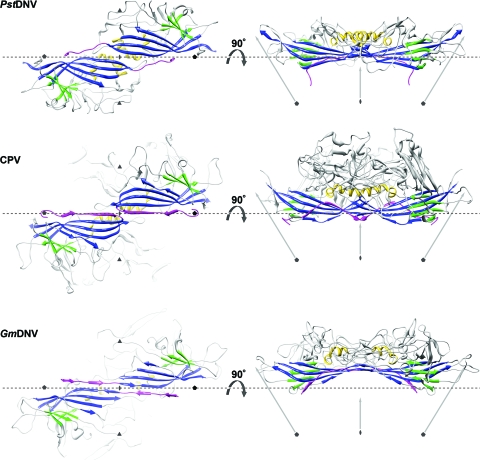
The PstDNV capsid protein contains an eight-stranded, antiparallel β-barrel jelly roll motif, similar to that found in many other viral capsid proteins, consisting of two β-sheets in the BIDG and CHEF arrangement (for nomenclature, see Rossmann and Johnson [33]) (Fig. 1 and 2 and Table 2). The β-barrel of PstDNV superimposed on the same motif in GmDNV with a root mean square deviation (RMSD) of 1.6 Å for 72 equivalenced Cα atoms (80.9% of Cα atoms in the β-barrel) and on CPV with an RMSD of 1.4 Å (72 Cα atoms). The β-barrel motif is in approximately the same position relative to the icosahedral symmetry axes as in other parvoviruses, but in comparison with the mammalian counterparts, it is laterally slightly shifted toward an imaginary line connecting two neighboring fivefold axes (Fig. 2). In the vertebrate parvoviruses, such as CPV, the β-BIDG sheet is extended to an antiparallel β-ABIDG sheet by the backfolded β-strand A (βA) that is located in the N-terminal portion of the protein sequentially upstream of βB (Fig. 2). In contrast, in the densovirus GmDNV, βB is essentially the linear extension of βA (36) (Fig. 2). In this “domain-swapped” organization, βA engages in hydrogen bonding with βB of a neighboring twofold-related subunit, thereby possibly contributing to capsid stability. Similarly, the N terminus of the PstDNV coat protein is pointing toward a fivefold axis in a domain-swapped fashion (Fig. 2). The N-terminal region upstream of βB in PstDNV is spatially farther removed from the respective neighboring BIDG β-sheet than is the case in GmDNV, thus making hydrogen bonding with the symmetry-related β-BIDG unfeasible. Capsid stability, however, seems to be promoted by coordinated divalent metal ions that cross-link neighboring fivefold-symmetry-related subunits. No intra- or intermolecular disulfide bonds within the ordered part of the structure were found that had been suggested as being essential for the structural integrity of PstDNV VLPs (15).
FIG. 2.
Spatial arrangement of the parvoviral core jelly roll and the N-terminal region for the capsid proteins of PstDNV, CPV, and GmDNV. (Left) Two twofold-related symmetry mates are shown, viewed from the viral center along an icosahedral twofold axis. (Right) The side view of the dimer, turned by 90° around the broken-line axis, demonstrates the spatial outward displacement of the β-barrel in GmDNV. Conserved secondary structure elements of each protein subunit are colored blue (β-BIDG), green (β-CHEF), and gold (helical elements). The N-terminal region of the capsid protein, upstream of βB, including βA, is shown in magenta. The positions of icosahedral symmetry axes are indicated by polygonal symbols and arrows. The spatial position of the broken line connecting two neighboring fivefold axes is the same for the three viruses to allow positional comparison of the structures.
The first few ordered N-terminal residues of PstDNV are pointing away from the fivefold channel slightly toward the interior of the virus particle, as has been observed in VLPs of human parvovirus B19, and slightly sideward toward some weak additional density nestled below the neighboring threefold-related subunit. This weak density is most likely due to the remaining N-terminal sequence but is too disordered and discontinuous to allow building of further residues. It has been suggested that the N terminus of the PstDNV capsid protein is located on the outer surfaces of VLPs expressed in Escherichia coli (15). However, in this work, no density that might be due to an externalized N-terminal sequence was found in the fivefold pore. As in GmDNV, the PstDNV fivefold channel is about 20 Å long and is lined by large, mostly hydrophobic residues (132Val, 138Phe, 130Leu, 140Thr, 141Leu, and 143Phe). The pore is closed at the outside of the particle where the pore diameter is 5.1 Å compared with 14 Å in CPV and 8.3 Å in GmDNV. The pore structure is formed by the DE loop. The tip of the DE loop contains a Ser-Gly-Gly sequence (residues 133 to 135), similar to a glycine-rich motif found in parvovirus B19 VLPs in which the pore is also closed (5.2 Å) (18). This region has the highest temperature factor of the observed structure, suggesting that the flexibility at the outside tip of the DE loop allows the fivefold channel to be switched from closed to open, as might be required for externalization of the N termini or packaging/release of DNA.
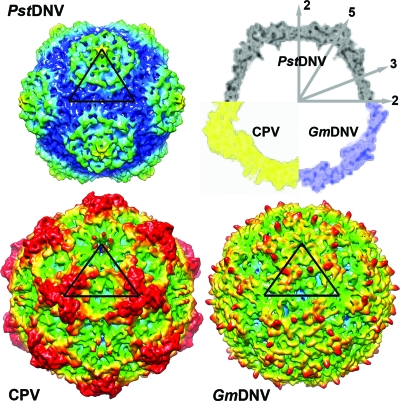
The majority of any parvoviral capsid structure is made up of loops connecting the strands of the β-barrel (see Table 2 for nomenclature) that form most intersubunit contacts and define the outer surface of the virus. Regions of the PstDNV structure with the highest temperature factors coincide with exposed surface loops. The protein shell of the shrimp parvovirus PstDNV has a significantly smaller diameter and is thinner than mammalian parvoviruses, such as CPV (Fig. 3). The virus capsid with an outer diameter of only 215 to 250 Å (mean diameter of 233 Å) does not show significant threefold-proximal projections as observed in members of the Parvovirinae subfamily, but the most protruding feature is located at the icosahedral fivefold apexes. The lack of a projecting structure close to the threefold axes and a raised canyon rim are consistent with the significantly shorter loops connecting the strands of the β-barrel in PstDNV (Table 3). Compared to PstDNV, the capsid surface of GmDNV is relatively featureless, presumably because most of its loops, even though for the most part shorter than in mammalian parvoviruses, are significantly longer than in PstDNV, making the void around the threefold axes shallower (Fig. 3). Because the β-barrel of GmDNV is slightly rotated and displaced outward from the viral center (Fig. 2), there is a difference in the inner diameter of PstDNV and GmDNV (Fig. 3). The resulting larger inner capsid volume likely allows GmDNV to accommodate its genome (6 kb), which is about 1.5 times larger than that of PstDNV.
FIG. 3.
Topographic comparison of the PstDNV protein capsid with other parvoviruses. Surface renderings of 3D density maps of PstDNV, GmDNV, and CPV at 8-Å resolution, generated from atomic coordinates. One icosahedral asymmetric unit is indicated by a black triangle. The surface is colored according to the distance from the viral center (100 Å [blue], 107.5 Å [cyan], 115 Å [green], 122.5 Å [yellow], and 130 Å[red]). In the top right corner of the figure, a combination display of the equatorial slices of the density maps depicts the differences in thickness and expansion of the protein shells. The positions of icosahedral symmetry axes are indicated by the numbered gray arrows.
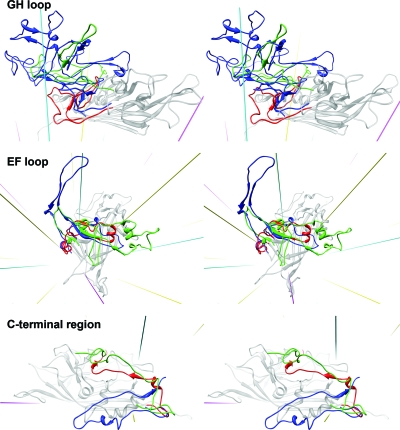
The backbone of PstDNV superimposes on GmDNV with an RMSD of 2.04 Å for 128 equivalenced Cα atoms (42.8% of all Cα atoms) and on CPV with an RMSD of 1.96 Å (144 Cα atoms or 48.2%). Most of these equivalenced atoms (79 and 85 residues, respectively) are part of the conserved β-barrel. The GH loop that forms the characteristic raised structures at and around the threefold apexes of the mammalian parvovirus capsid and a β-annulus-type structure in GmDNV is only ∼21% and 50% as long in PstDNV as the respective loops in CPV and GmDNV (Fig. 4 and Table 3). Similarly, the BC loop, one of the loops that contribute to the outer rim of the canyon, is ∼77% shorter than in CPV and ∼65% shorter than in GmDNV (Table 3). The EF loop also differs greatly between PstDNV, CPV, and GmDNV (Fig. 4 and Table 3). An insertion before αEF6 that contributes to the projections at the threefold axes in CPV is completely missing in PstDNV. In the latter, the CHEF β-sheet of the β-barrel is extended on the outer surface of the particle by a fifth strand, βEF2, and an additional β-structure is formed by hydrogen bonding between strands βEF0.1 and βI1.2 (Fig. 1 and Table 2). Interestingly, there is a loop insertion, a β-hairpin formed by βCD1.1 and βCD1.2, between the conserved helical elements αCD1 and αCD2 (Fig. 1). This structural element is unique to PstDNV with no equivalent in the other parvoviruses, and its functional role is unknown. It is possible that this hairpin contributes to capsid integrity by contacting the EF loop of a neighboring fivefold-related subunit. This might be necessary because no intertwining of the threefold-proximal loops occurs in PstDNV as observed in vertebrate parvoviruses. In mammalian parvoviruses, such as human parvovirus B19, the spatial position of the hairpin is occupied by a part of the C-terminal portion of the capsid protein. The structure of the C-terminal portions downstream of βI in PstDNV and GmDNV is different from mammalian parvoviruses (Fig. 4). Less significant structural differences exist in loops DE and HI which comprise the cylindrical structure around the fivefold axis.
FIG. 4.
Structure comparison of the GH and EF loops and the C-terminal region of PstDNV (red) with GmDNV (green) and CPV (blue). The gray-colored background structure is the PstDNV capsid protein. Icosahedral symmetry axes are shown in magenta (fivefold), cyan (threefold), and yellow (twofold).
The structure of the GH loop of PstDNV is very different from that of other known parvovirus structures. This loop is very long (>200 residues) in CPV, minute virus of mice (23), and porcine parvovirus (37) and is divided into 2 subloops (loop 3 and 4) that interdigitate with the neighboring subunit to create trimers. The major capsid protein would be transported to the nucleus only after correct cytoplasmic folding of the trimer yielded a nuclear localization signal (24, 31). The GH loop thus constitutes a significant feedback control in assembly. The GH loop plays a similar role in GmDNV, although it is much shorter (lacking loop 4) and is responsible for a relatively simple interdigitation in the trimer around the threefold axis (36). Surprisingly, the GH loop of PstDNV does not intertwine with the neighboring subunit (although interacting at the surface) to provide a similarly strong feedback control in assembly.
In summary, the near-atomic resolution structure of PstDNV is compatible with the low-resolution 3D structure features of Aedes albopictus densovirus and underscores their joint classification within the genus Brevidensovirus. However, the structure and length of the loop regions in PstDNV differ greatly from the loop regions in the vertebrate parvoviruses and in the insect parvovirus GmDNV (Table 3 and Fig. 4), suggesting a lengthy divergence in the evolutionary development between these viruses (Fig. 5).
FIG. 5.
Unrooted parsimony phylogenetic tree between different parvoviruses based on the length of loops connecting the strands of the β-barrel. In the bottom left corner, the length of the scale bar represents a 50-amino-acid distance or total character difference.
Supplementary Material
Acknowledgments
We thank Sheryl Kelly for assistance in the preparation of this paper. We are grateful for the support we received from the staff of the GM/CA CAT beamline.
GM/CA CAT has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract DE-AC02-06CH11357. The Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, was supported by NIH P41 RR-01081. This work was supported by NIH grants AI 33468 to Colin R. Parrish (Cornell University), AI 11219 to M.G.R., and an operating grant to P.T. from the Natural Sciences and Engineering Research Council of Canada. P.J.W. was supported by grant 5R01LM008626.
We have no conflict of interest.
Footnotes
Published ahead of print on 11 August 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S. 2004. RobEM—image processing and visualization program. http://cryoem.ucsd.edu/programDocs/runRobem.txt.
- 3.Bonami, J. R., B. Trumper, J. Mari, M. Brehelin, and D. V. Lightner. 1990. Purification and characterization of the infectious hypodermal and haematopoietic necrosis virus of penaeid shrimps. J. Gen. Virol. 71:2657-2664. [DOI] [PubMed] [Google Scholar]
- 4.Bruemmer, A., F. Scholari, M. Lopez-Ferber, J. F. Conway, and E. A. Hewat. 2005. Structure of an insect parvovirus (Junonia coenia densovirus) determined by cryo-electron microscopy. J. Mol. Biol. 347:791-801. [DOI] [PubMed] [Google Scholar]
- 5.Brünger, A. T. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2:2728-2733. [DOI] [PubMed] [Google Scholar]
- 6.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Canaan, S., Z. Zadori, F. Ghomashchi, J. Bollinger, M. Sadilek, M. E. Moreau, P. Tijssen, and M. H. Gelb. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502-14508. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, J., E. Suchman, and L. Buchatsky. 2006. Densoviruses for control and genetic manipulation of mosquitoes. Adv. Virus Res. 68:361-392. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S., L. Cheng, Q. Zhang, W. Lin, X. Lu, J. Brannan, Z. H. Zhou, and J. Zhang. 2004. Genetic, biochemical, and structural characterization of a new densovirus isolated from a chronically infected Aedes albopictus C6/36 cell line. Virology 318:123-133. [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 11.Eventoff, W., M. L. Hackert, and M. G. Rossmann. 1975. A low-resolution crystallographic study of porcine heart lactate dehydrogenase. J. Mol. Biol. 98:249-258. [DOI] [PubMed] [Google Scholar]
- 12.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U. S. A. 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 14.Harvell, C. D., K. Kim, J. M. Burkholder, R. R. Colwell, P. R. Epstein, D. J. Grimes, E. E. Hofmann, E. K. Lipp, A. D. Osterhaus, R. M. Overstreet, J. W. Porter, G. W. Smith, and G. R. Vasta. 1999. Emerging marine diseases—climate links and anthropogenic factors. Science 285:1505-1510. [DOI] [PubMed] [Google Scholar]
- 15.Hou, L., H. Wu, L. Xu, and F. Yang. 2009. Expression and self-assembly of virus-like particles of infectious hypodermal and hematopoietic necrosis virus in Escherichia coli. Arch. Virol. 154:547-553. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, K. N., M. C. van Hulten, and A. C. Barnes. 2008. “Vaccination” of shrimp against viral pathogens: phenomenology and underlying mechanisms. Vaccine 26:4885-4892. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann, B., A. A. Simpson, and M. G. Rossmann. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. U. S. A. 101:11628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 20.Li, Y., Z. Zadori, H. Bando, R. Dubuc, G. Fediere, J. Szelei, and P. Tijssen. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82:2821-2825. [DOI] [PubMed] [Google Scholar]
- 21.Li, Z., J. He, X. Huang, A. Dai, L. Cheng, D. Shao, and J. Zhang. 2008. The truncated virus-like particles of C6/36 cell densovirus: implications for the assembly mechanism of brevidensovirus. Virus Res. 132:248-252. [DOI] [PubMed] [Google Scholar]
- 22.Lightner, D. V., R. M. Redman, and T. A. Bell. 1983. Infectious hypodermal and hematopoietic necrosis, a newly recognized virus disease of penaeid shrimp. J. Invertebr. Pathol. 42:62-70. [DOI] [PubMed] [Google Scholar]
- 23.Llamas-Saiz, A. L., M. Agbandje-McKenna, W. R. Wikoff, J. Bratton, P. Tattersall, and M. G. Rossmann. 1997. Structure determination of minute virus of mice. Acta Crystallogr. D Biol. Crystallogr. 53:93-102. [DOI] [PubMed] [Google Scholar]
- 24.Lombardo, E., J. C. Ramirez, M. Agbandje-McKenna, and J. M. Almendral. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 26.Manolova, V., A. Flace, M. Bauer, K. Schwarz, P. Saudan, and M. F. Bachmann. 2008. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 38:1404-1413. [DOI] [PubMed] [Google Scholar]
- 27.Navaza, J. 1994. Amore—an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 28.Otwinowski, Z., W. Minor, and W. C. Charles. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 30.Ren, X., E. Hoiczyk, and J. L. Rasgon. 2008. Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog. 4:e1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riolobos, L., J. Reguera, M. G. Mateu, and J. M. Almendral. 2006. Nuclear transport of trimeric assembly intermediates exerts a morphogenetic control on the icosahedral parvovirus capsid. J. Mol. Biol. 357:1026-1038. [DOI] [PubMed] [Google Scholar]
- 32.Rollinger, J. M., and M. Schmidtke. 27 August 2009. The human rhinovirus: human-pathological impact, mechanisms of antirhinoviral agents, and strategies for their discovery. Med. Res. Rev. doi: 10.1002/med.20176. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 33.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533-573. [DOI] [PubMed] [Google Scholar]
- 34.Rossmann, M. G., R. McKenna, L. Tong, D. Xia, J. B. Dai, H. Wu, H. K. Choi, and R. E. Lynch. 1992. Molecular replacement real-space averaging. J. Appl. Crystallogr. 25:166-180. [Google Scholar]
- 35.Shike, H., A. K. Dhar, J. C. Burns, C. Shimizu, F. X. Jousset, K. R. Klimpel, and M. Bergoin. 2000. Infectious hypodermal and hematopoietic necrosis virus of shrimp is related to mosquito brevidensoviruses. Virology 277:167-177. [DOI] [PubMed] [Google Scholar]
- 36.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 38.Swofford, D. L., P. J. Waddell, J. P. Huelsenbeck, P. G. Foster, P. O. Lewis, and J. S. Rogers. 2001. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst. Biol. 50:525-539. [PubMed] [Google Scholar]
- 39.Tong, L. A., and M. G. Rossmann. 1990. The locked rotation function. Acta Crystallogr. A 46:783-792. [DOI] [PubMed] [Google Scholar]
- 40.Trapani, S., and J. Navaza. 2008. AMoRe: classical and modern. Acta Crystallogr. D Biol. Crystallogr. 64:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, and C. R. Parrish. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456-1464. [DOI] [PubMed] [Google Scholar]
- 42.Witteveldt, J., J. M. Vlak, and M. C. van Hulten. 2006. Increased tolerance of Litopenaeus vannamei to white spot syndrome virus (WSSV) infection after oral application of the viral envelope protein VP28. Dis. Aquat. Organ. 70:167-170. [DOI] [PubMed] [Google Scholar]
- 43.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]
- 45.Yan, X., R. S. Sinkovits, and T. S. Baker. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.