Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) infection can cause the development of severe end-stage lung disease characterized by acute respiratory distress syndrome (ARDS) and pulmonary fibrosis. The mechanisms by which pulmonary lesions and fibrosis are generated during SARS-CoV infection are not known. Using high-throughput mRNA profiling, we examined the transcriptional response of wild-type (WT), type I interferon receptor knockout (IFNAR1−/−), and STAT1 knockout (STAT1−/−) mice infected with a recombinant mouse-adapted SARS-CoV (rMA15) to better understand the contribution of specific gene expression changes to disease progression. Despite a deletion of the type I interferon receptor, strong expression of interferon-stimulated genes was observed in the lungs of IFNAR1−/− mice, contributing to clearance of the virus. In contrast, STAT1−/− mice exhibited a defect in the expression of interferon-stimulated genes and were unable to clear the infection, resulting in a lethal outcome. STAT1−/− mice exhibited dysregulation of T-cell and macrophage differentiation, leading to a TH2-biased immune response and the development of alternatively activated macrophages that mediate a profibrotic environment within the lung. We propose that a combination of impaired viral clearance and T-cell/macrophage dysregulation causes the formation of prefibrotic lesions in the lungs of rMA15-infected STAT1−/− mice.
The severe acute respiratory syndrome coronavirus (SARS-CoV) is a highly pathogenic respiratory virus that emerged in the human population to cause a global epidemic in 2002 and 2003. The virus infected roughly 8,000 people and resulted in approximately 800 deaths, with high case mortality in populations of the elderly (9). In humans, SARS-CoV infection caused mortality due to development of acute respiratory distress syndrome (ARDS) (19, 23). The lung pathology in SARS-CoV-infected patients is believed to be caused by host immune dysregulation as well as by viral factors (1, 28, 63).
ARDS, with a mortality rate of approximately 50%, is the most severe form of acute lung injury and affects more than one million people per year worldwide (32, 77). No effective therapies for ARDS currently exist, making it a major challenge for critical-care medicine. The first phase of ARDS is the exudative phase, presenting with diffuse alveolar damage (DAD), inflammation, proteinaceous edema, hyaline membrane formation, and severe hypoxia. This may progress to a second phase of ARDS, the organizing phase, characterized by pulmonary fibrosis. In addition to SARS-CoV, viruses that can cause ARDS include influenza virus, respiratory syncytial virus, adenovirus, varicella zoster virus, human metapneumovirus, and hantavirus (5, 12, 34, 37, 38, 47, 69). Mice infected with select strains of SARS-CoV or influenza virus serve as models for the initial exudative phase of ARDS (63, 64, 80), but these animals do not develop pathology consistent with later ARDS phases, and there are few infection systems that serve as models of pulmonary fibrosis (75). Instead, most studies focused on pulmonary fibrosis utilize the bleomycin model, which does not recapitulate the effects of a viral inducer of the disease (50).
Several studies have shown the importance of interferon- and STAT1-dependent signaling for reducing viral replication as well as for limiting pathogenesis in animal models. Infection of type I interferon receptor knockout (IFNAR1−/−) animals with highly pathogenic H5N1 influenza virus results in accelerated mortality compared with wild-type (WT) animal results (71). Mouse hepatitis virus, another coronavirus, causes lethal disease in type I interferon receptor knockout mice but not in wild-type mice (7). Likewise, STAT1−/− mice exhibit dramatic defects in the immune response, leading to mortality in response to Listeria monocytogenes, vesicular stomatitis virus, and herpes simplex virus infections (46, 58). In vivo treatment with interferon reduces SARS-CoV replication and lung pathology in macaques, suggesting that the innate immune response does protect animals from SARS-CoV pathogenesis (29).
Previous gene expression studies of SARS-CoV in model systems have provided some insight into the mechanism of pathogenesis. Initial studies using macaques infected with sublethal inoculums revealed a strong innate immune response that corresponded with peak viral replication and a later proliferative response associated with increased expression of cell-division genes and the healing of the damaged tissue (17). Further studies of lethal SARS-CoV infection in aged mice revealed an accelerated upregulation of acute-phase response genes associated with ARDS, including the cytokines interleukin-1 beta (IL-1β), IL-6, and tumor necrosis factor alpha (TNF-α) (63). However, infection with clinical isolates of SARS-CoV did not cause significant lung disease in young mice and resulted in delayed upregulation of immune response genes compared with the results seen with aged mice. To enable routine studies of SARS-CoV pathogenesis, mouse-adapted SARS-CoV strains have been developed using serial passage techniques (16, 52, 62). The first of these strains, designated rMA15 (representing a recombinant mouse-adapted SARS-CoV), contains 6 mutations with respect to the Urbani clinical isolate of SARS-CoV (62). Both young and aged mice infected with rMA15 develop lung pathology, and several strains of mice succumb to infection. Using this virus, we have been able to infect young mice and elicit ARDS pathophysiology (61).
Our previous work (24) revealed that young type I interferon receptor knockout (IFNAR1−/−) mice did not exhibit increased pathogenesis as a result of rMA15 infection compared to wild-type (WT) mice. However, rMA15 infection of young STAT1−/− mice resulted in increased pathology, including evidence of pulmonary fibrosis-like lesions and mortality by 9 days postinfection (dpi). In this work, we extended the initial biological characterization of rMA15 infection in WT, IFNAR1−/−, and STAT1−/− mice by using microarray profiling, immunohistochemistry, and cell-sorting techniques to examine the molecular mechanisms governing severe end-stage lung disease. We found that STAT1−/− mice infected with rMA15 exhibited a shift to a TH2 response, with expansion and activation of alternatively activated M2 macrophages, resulting in fibrosis-like protein deposition in the lungs.
MATERIALS AND METHODS
Mouse infection.
129S6/SvEv (abbreviated here as “129”) wild-type and STAT1−/− mice (catalog number 002045-M-F) were obtained from Taconic Farms (Germantown, NY). IFNAR1−/− mice bred on a 129SvEv background were a gift from Mark Heise (University of North Carolina [UNC]-Chapel Hill) and were bred at the University of North Carolina mouse facility (Chapel Hill, North Carolina). All animal studies were conducted using animal biosafety level 3 laboratories and Sealsafe cages with HEPA filters, and personnel wore personal protective equipment, including Tyvek suits and hoods as well as positive-pressure air respirators with HEPA filters. Ten-week-old mice were anesthetized with a ketamine (1.3 mg/mouse)-xylazine (0.38 mg/mouse) mixture administered intraperitoneally in a 50-μl volume. Mice were infected intranasally with either phosphate-buffered saline (PBS) alone (Invitrogen, Carlsbad, CA) or 1 × 105 PFU rMA15-PBS. A total of 5 infected mice from each strain were euthanized at days 2, 5, and 9 postinfection (dpi) for virological, histological, and microarray analyses. Mock-infected animals from each of the strains were euthanized at 5 dpi. Samples from the right lung were reserved for viral titers and histology, and the entire left lung was reserved for microarray analyses. Animal housing, care, and experimental protocols were in accordance with all UNC-Chapel Hill Institutional Animal Care and Use Committee guidelines.
RNA preparation and oligonucleotide microarray processing.
The entire left lung of each mouse was dissected and homogenized using Trizol and a Magnalyser system (Roche) according to manufacturer instructions. RNA was further purified using RNeasy columns (Qiagen) according to manufacturer instructions. RNA samples were spectroscopically verified for purity, and the quality of the intact RNA was assessed using an Agilent 2100 Bioanalyzer. For each set of treatment conditions, 3 of the 5 RNA samples exhibiting the highest RNA integrity number (RIN) determined using the Bioanalyzer were used for microarray analysis. cRNA probes were generated from each sample by the use of an Agilent one-color Quick-Amp labeling kit. Individual cRNA samples were hybridized to Agilent mouse whole-genome oligonucleotide microarrays (4 × 44) according to manufacturer instructions. Samples from individual animals were not pooled to enable examination of animal-to-animal variations as part of the data analysis. Select samples were hybridized a second time (n = 2 technical replicates) to verify the quality of the process. Slides were scanned with an Agilent DNA microarray scanner, and the resulting images were analyzed using Agilent Feature Extractor software. Data were warehoused in a Katze Labkey system (Labkey, Inc., Seattle, WA) and analyzed using Resolver 7.1 software (Rosetta Biosoftware, Seattle, WA), and Spotfire DecisionSite for Functional Genomics 9.1 software (Tibco Spotfire, Somerville, MA). Primary data are available (http://viromics.washington.edu) in accordance with proposed Minimum Information About a Microarray Experiment (MIAME) standards.
Analysis of microarray data.
An initial analysis of gene expression in each mouse line was performed as a function of time (kinetic analysis). Genes were selected based on textbook analysis of variance (ANOVA) of microarray intensity scores, with a significance criterion of P < 0.05. The following infection groups (n = 3 per group) were compared for WT, IFNAR1−/−, and STAT1−/− mice, for a total of 9 gene lists: mock and 2 dpi, 2 dpi and 5 dpi, and 5 dpi and 9 dpi. These gene lists were reduced by requiring a change of at least 1.5-fold between the group averages. Genes that met these criteria were characterized using Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Redwood City, CA) functional annotations. Functional annotations were scored using the Fisher exact test. Subsequent analysis was performed to better understand the differences between strains. Textbook ANOVA was performed on the ratio data (i.e., ratios determined with respect to the averages of the strain-matched mock-infected sample results) at 2 dpi. The following comparisons of mouse groups were performed: WT versus IFNAR1−/−, WT versus STAT1−/−, and IFNAR1−/− versus STAT1−/−. As described above, a significance criterion of P < 0.05 was applied. The gene lists were trimmed by requiring that the change of at least one ratio comparing mock results was greater than 1.5-fold. The gene lists were then split based on the sample that exhibited the greater magnitude of change, yielding 6 gene lists based on both ANOVA and the relative differential regulation results for the mouse groups, as follows: genes that are differentially regulated more in WT than in IFNAR1−/−, more in IFNAR1−/− than in WT, more in WT than in STAT1−/−, and so forth. As described above, these gene lists were functionally annotated using IPA. This process was repeated for data obtained at 5 dpi. The resulting top 5 statistically significant canonical pathways for each gene list are shown (see Table 2). Additional analysis of interferon-stimulated genes (ISGs) was performed using a panel of genes induced in the lungs of WT mice upon intranasal stimulation with 10,000 units of interferon alpha for a 24-h time period (14). Gene lists characteristic of TH1 and TH2 immune responses were derived from annotations from the SABiosciences and NCBI Entrez Gene databases. Gene lists characteristic of M1 and M2 activated macrophages were derived from the work of Mantovani and coworkers (45) and NCBI Entrez Gene.
Masson's trichrome staining.
Lung tissues were fixed in PBS-4% paraformaldehyde (pH 7.3) and embedded in paraffin, and 5-μm-thick sections were prepared by the UNC histopathology core facility. Prior to use, sections were deparaffinized in xylene with 2 changes of 5 min each and washed with water. Slides were left to incubate overnight in picric acid at room temperature. Slides were then washed in running tap water for 10 min to remove the yellow color and rinsed with deionized water. The slides were then incubated in Weigert's iron hematoxylin solution (catalog number HT079; Sigma) for 10 min. After incubation, slides were washed in running water for 10 min and rinsed in deionized water. The slides were then incubated in Biebrich scarlet-acid fuschin solution (catalog number HT151; Sigma) for 5 min and washed in deionized water. After the washing was performed, the slides were incubated in phosphomolybdic-phosphotungstic acid (catalog number F9252; Sigma) for 10 min and then in aniline blue for 5 min (catalog number 415049; Sigma) before being rinsed in deionized water. The slides were then fixed in 1% acetic acid for 3 min followed by two rinses of 95% and 100% ethanol and a final xylene rinse.
Immunohistochemistry.
Lung tissues were fixed in PBS-4% paraformaldehyde (pH 7.3) and embedded in paraffin, and 5-μm-thick sections were prepared by the University of Maryland histopathology core facility. Sections were deparaffinized in xylene, with 2 washes of 5 min each. The slides were then placed in absolute alcohol and in 95% and 70% alcohol, with 2 washes of 2 min each. After 2 washes in PBS, the slides were immersed in methanol-0.3% H2O2 for 30 min to quench the endogenous peroxidase. The slides were then preincubated with 10% goat serum for 20 min at room temperature followed by incubation with primary antibody for 1 h at room temperature. Rabbit anti-arginase I (Santa Cruz Biotechnology, Santa Cruz, CA) was used at a 1:50 dilution, polyclonal rabbit anti-YM1 (StemCell Technologies, Vancouver, BC, Canada) was used at a 1:200 dilution, and monoclonal rat anti-Mac3 (Cedarlane Laboratories, Burlington, NC) was used at a 1:500 dilution. Slides were washed 3 times in PBS, and biotin secondary antibody was applied for 30 min at room temperature. The sections were washed in PBS 3 times and incubated with ABC reagent (Vector Laboratories Inc.). After 2 changes of PBS, DAB (3,3-diaminobenzidine) was used to develop the sections, which were then counterstained with Mayer's hematoxylin for 2 min. Slides were dehydrated in 2 changes of 95% and absolute alcohol and then mounted with coverslips.
Quantitative RT-PCR.
Relative quantitative reverse transcription-PCR (RT-PCR) was performed to validate a number of gene expression changes as detected with microarrays. A QuantiTect reverse transcription kit (Qiagen Inc., Valencia, CA) was used to generate cDNA, and quantitative RT-PCR was performed using an Applied Biosystems 7500 PCR system. The following TaqMan assays for mouse genes were obtained from Applied Biosystems: for Igf1, assay Mm00439560_m1; for Ccl11, assay Mm00441238_m1; for Arg1, assay Mm00475988_m1; for Il17a, assay Mm00439619_m1; and for Col3a1, assay Mm 01254476_m1. The TaqMan assay for SARS nucleoprotein (NP) was described previously (39). Loading variations were normalized using the invariant calibrator gene Mfap-1a according to the 2−ΔΔCT threshold cycle method (41). Differences in gene expression are represented as log ratios relative to cDNA samples from strain-matched mock-infected animals.
FACS analysis of alternatively activated macrophages (AAMs).
Ten-week-old 129SvEv and STAT1−/− mice (Taconic Laboratories) were infected with 105 PFU of rMA15 as described previously. Mice were euthanized at day 8 postinfection and their lungs dissected for fluorescence-activated cell sorter (FACS) analysis. Lungs of the mice were dissociated in collagenase and DNase, after which the cells were incubated with PerCP-Cy5.5-labeled anti-CD11b (eBioscience, San Diego, CA) and phycoerythrin-labeled anti-CD14 (eBioscience, San Diego, CA). Cells were permeabilized and stained with anti-Mac3 (Cedarlane Laboratories Ltd., Burlington, NC) and then with allophycocyanin-labeled goat anti-rat IgG (Biolegend, San Diego, CA). Samples were processed on a Becton Dickinson LSR II FACS machine and analyzed using FlowJo 7.6.
RESULTS
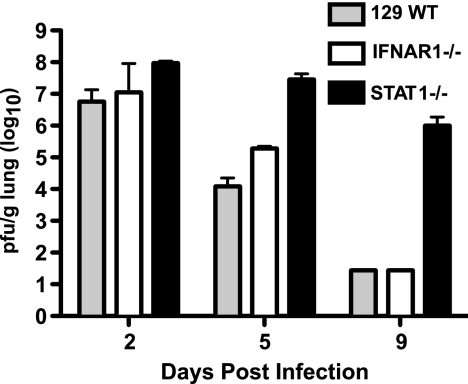
To better understand the contributions of specific signaling pathways and gene expression changes to SARS-CoV-induced lung disease, we examined the global transcriptional changes that occur during rMA15 infection of WT mice or mice containing genetic defects associated with innate immune signaling. WT, IFNAR1−/−, and STAT1−/− mice were infected with 1 × 105 PFU of rMA15. Five animals per group were sacrificed at 2, 5, and 9 dpi. Weight loss, tissue pathology, and viral titer data are shown in Fig. 1 and 2 and Table 1; a detailed description of the disease course was previously reported (24). Briefly, WT and IFNAR1−/− mice experienced similar disease courses based on viral titer, weight loss, and lung pathology results. WT and IFNAR1−/− mice lost 15% of their starting weight by 5 dpi but regained this weight by 9 dpi. Peak viral titers in the lung for both mouse strains were observed at 2 dpi and diminished over the course of the infection. At 9 dpi, virus was not detectable by plaque assay or by in situ hybridization.
FIG. 1.
Viral replication in the lungs of wild-type and knockout mice. Lung viral titer data from 5 mice infected with rMA15 per strain per time point are shown. The limit of detection for this assay is 5 × 102 PFU.
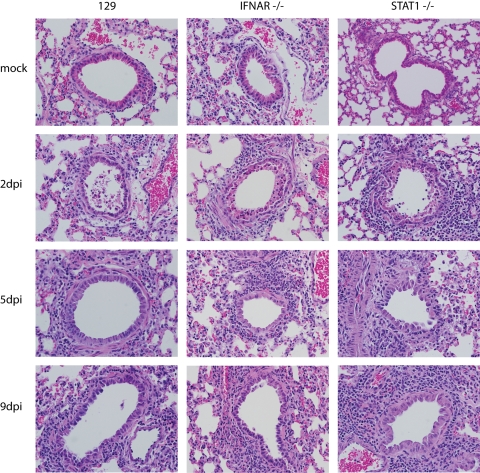
FIG. 2.
Sustained inflammation in the lungs of STAT1−/− mice infected with rMA15. Representative tissue samples from WT, IFNAR−/−, and STAT1−/− mice infected with rMA15 or mock-infected (2, 5, and 9 dpi) were stained with hematoxylin and an eosin counterstain. Images at lower magnification are available in Fig. S1 in the supplemental material. Magnification, ×40.
TABLE 1.
Summary of clinical and virological measures
| Characteristic | Value for indicated mouse straina |
||
|---|---|---|---|
| 129 | IFNAR1−/− | STAT1−/− | |
| Time of maximum weight loss (dpi) | 4 | 4 | 9 |
| Maximum wt loss (% of starting wt) | 14-16 | 14-16 | >20 |
| Peak viral titer (PFU/g) | 2.6-3.8 × 107 | 3-7.2 × 107 | 7.6 × 107-3.4 × 108 |
| Viral RNA level at 2 dpi | 6.4-16 × 104 | 1.4-2.8 × 105 | 8.9-23 × 104 |
| Viral RNA level at 5 dpi | 1.2-20 × 102 | 3.7-25 × 102 | 3.1-4.4 × 104 |
| Viral RNA level at 9 dpi | 1.0-7.5 | 5.2-84 | 5.5-9.5 × 103 |
| Alveolar protein deposition at 9 dpi | No | No | Yes |
| Endpoint | Recovery | Recovery | Death |
The ranges of viral RNA levels are shown on an arbitrary scale as determined by quantitative RT-PCR results determined using probes for the SARS nucleoprotein gene (39). Three animals were used per set of conditions and time point.
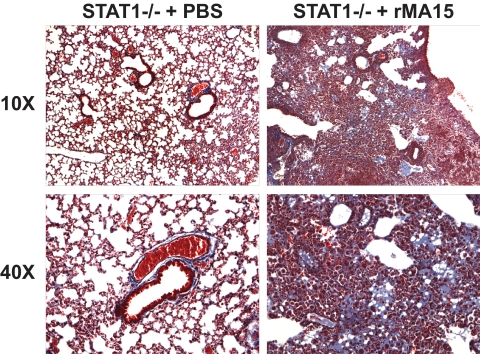
STAT1−/− mice exhibited weight loss and pathology similar to the results seen with the WT and IFNAR1−/− mice at 2 dpi but experienced increased weight loss and inflammatory cell infiltration into the lungs at 5 and 9 dpi. Notably, extensive collagen deposition was observed in the lungs of STAT1−/− mice by Frieman et al. (24) and in this work as judged by Masson's trichrome staining (Fig. 3), suggesting early-stage pulmonary fibrosis. STAT1−/− mice also exhibited higher viral titers in the lung throughout the time course, including 5 × 106 PFU per gram of tissue at 9 dpi. Gene expression profiling analyses of RNA samples from 3 of the 5 mice per group were used to identify mechanisms of protection against rMA15 exhibited by wild-type and IFNAR1−/− mice and to identify the pathological mechanisms that lead to fibrosis-like disease in the STAT1−/− mice, as described in the following sections.
FIG. 3.
Abnormal protein deposition in the lungs of STAT1−/− mice infected with rMA15. Representative tissue samples from STAT1−/− mice infected with rMA15 or subjected to mock infection (9 dpi) were stained with Masson's Trichrome. The presence of blue-purple staining indicates protein deposition in the alveolar spaces. Images at lower magnification are available in Fig. S2 in the supplemental material.
Immune and mitotic gene expression reveals common antiviral response and damage repair mechanisms.
We began our gene expression studies by performing ANOVA of gene expression data obtained from the lungs of WT, IFNAR1−/−, and STAT1−/− mice. ANOVA identifies genes for which the variation in expression levels between biological replicates is significantly smaller than the variation between different treatment groups (based on either time or mouse strain). We initially focused on identifying functional gene classes that were associated with the progression of rMA15 infection and pathology. To accomplish this, we performed three comparisons for each strain: (i) expression data from mock-infected animals were compared with expression data from infected animals sacrificed at 2 dpi; (ii) expression data from infected animals sacrificed at 2 dpi were compared with expression data from infected animals sacrificed at 5 dpi; and (iii) expression data from infected animals sacrificed at 5 dpi were compared with expression data from infected animals sacrificed at 9 dpi.
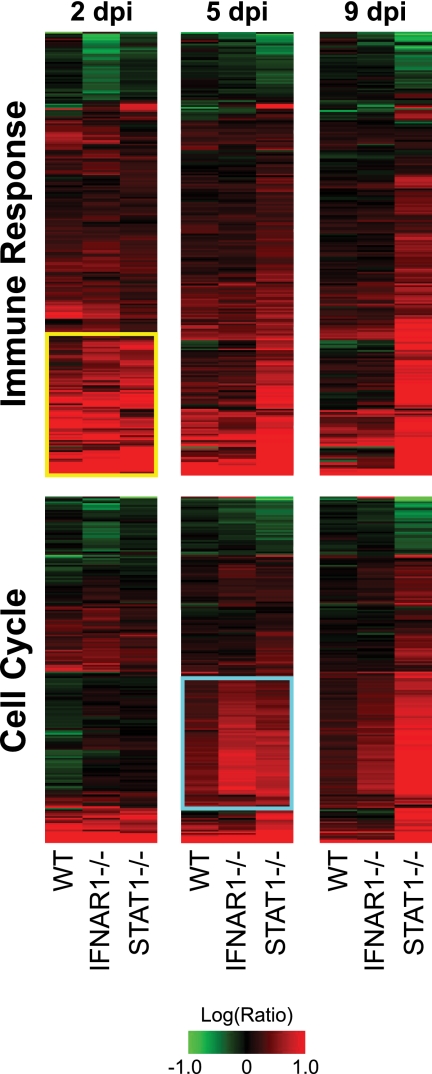
Using IPA to evaluate the gene sets determined using our ANOVA results, we identified responses common to rMA15-infected WT, IFNAR1−/−, and STAT1−/− mice relative to mock-infected animals. The commonly identified response at 2 dpi consisted of a set of genes involved in immunity and the inflammatory response to infection that were annotated by the IPA “immunological disease” functional category. WT mice differentially regulated 183 genes (P = 1.85 × 10−36), IFNAR1−/− mice differentially regulated 230 genes (P = 4.36 × 10−11), and STAT1−/− mice differentially regulated 202 genes (P = 8.30 × 10−45) in this category. A union of these genes is shown as a heat map in Fig. 4 (top panel). Increased expression of a subset of these genes at 2 dpi, including genes encoding cytokines (Il1a, Il1b, Il6, Il10, Il12b, and Tnf), chemokines (Ccl2, Ccl3, Ccl4, Ccl7, Ccl20, Cxcl2, Cxcl3, and Cxcl10), and immune sensor molecules (Tlr1, Tlr2, and Tlr3), suggested a common initial immune and inflammatory response in all three strains of mice (Fig. 4, top panel [yellow box]) (individual genes are listed in Table S1 in the supplemental material).
FIG. 4.
Kinetics of the transcriptional response to infection correlate with pathological findings. Expression profiles of 261 immune-related genes (top) and 204 cell cycle-related genes (bottom) selected by ANOVA are shown. Each column represents averages of data from the results of 3 replicate animal experiments performed using WT, IFNAR1−/−, or STAT1−/− mice at 2, 5, or 9 dpi compared to averages of data from experiments performed using mock-infected mice of the same strain. Genes shown in red were upregulated, genes shown in green were downregulated, and genes shown in black indicate no change in expression relative to mock-infected mice. Genes in the top panel were identified as part of the IPA “immunological disease” functional category in at least one ANOVA comparison between mock-infected animals and animals sacrificed at 2 dpi. A yellow box highlights 81 genes similarly upregulated by the 3 strains at 2 dpi. The identities of these genes are listed in Table S1 in the supplemental material. Genes in the lower panel were identified as part of the IPA “cell stage” functional category in at least one ANOVA comparison between mice sacrificed at 2 and at 5 dpi. A cyan box highlights 80 genes commonly upregulated by all strains of mice by 5 dpi. The identities of these genes are listed in Table S2 in the supplemental material.
By 5 dpi, the mRNA abundance of these same immune-related genes was reduced in WT and IFNAR1−/− mice, which correlated with the reduced pathology seen with these animals. In contrast, expression of this gene set in STAT1−/− mice was increased at 5 dpi, reflecting an observed increase in inflammation. At 9 dpi, expression of immune-related genes was further reduced in WT and IFNAR1−/− mice whereas expression of these genes was still significantly increased in STAT1−/− mice. This may reflect the extensive immune cell infiltration observed in the lungs of these animals (24).
Our ANOVA and IPA results also suggested the presence of extensive cell proliferation by 5 dpi. In particular, the IPA “cell stage” functional category was strongly associated with this time point. WT mice differentially regulated 171 genes (P = 1.05 × 10−30), IFNAR1−/− mice differentially regulated 185 genes (P = 1.82 × 10−19), and STAT1−/− mice differentially regulated 160 genes (P = 4.71 × 10−14) in this category. Expression of a union of these genes is shown in Fig. 4 (lower panel). Of particular interest are members of a subset of these genes that exhibit low expression at 2 dpi and high expression at 5 dpi, with the results being independent of the mouse strain (Fig. 4, cyan box). These genes include DNA damage repair-associated genes (Brca1, Rad51, and Rad54l), cyclins (Ccna2, Ccnb1, Ccne1, Ccne2, and Ccnf), cell division cycle genes (Cdc6, Cdc20, Cdc25c, Cdca5, and Cdca8), and genes associated with DNA replication initiation (Mcm10 and Pole). A full list of these genes is included in Table S2 in the supplemental material. In particular, IFNAR1−/− mice exhibited the strongest expression of these genes, suggesting that more cell division occurs in these animals at 5 dpi. By 9 dpi, the expression of these genes was reduced in WT and IFNAR1−/− mice, suggesting a decrease in cell proliferation as the virus was cleared. In contrast, STAT1−/− mice exhibited a further increase in gene expression associated with the cell cycle at 9 dpi. This may have been due to proliferation of fibroblasts or cell division of infiltrating immune cells.
Differential host response patterns suggest mechanisms of pathogenesis and survival.
In order to reveal the differences in the host response that underlie the inability of STAT1−/− mice to clear the virus and the subsequent development of pulmonary fibrosis-like lesions, we performed ANOVA to identify genes that were differentially expressed based on the mouse strain. These genes were then functionally annotated using pathway information from IPA. Table 2 contains the pathways most significantly enriched in the ANOVA gene lists from 2 dpi. Genes associated with the interferon signaling pathway, including Ifi35, Ifit1, Ifit3, Irf1, Irf9, Jak2, Med14, Oas1, Psmb8, Socs3, Stat1, Stat2, and Tap1, were robustly upregulated in WT or IFNAR1−/− mice compared with STAT1−/− mice (Table 2).
TABLE 2.
Top differentially expressed pathways at 2 dpi
| Gene regulation comparison for indicated strainsa | Canonical pathwayb | No. of genes | Scorec |
|---|---|---|---|
| 129 more regulated than IFNAR1−/− | Acute-phase response signaling | 15 | 4.3 |
| LXR/RXR activation | 9 | 4.1 | |
| FXR/RXR activation | 10 | 4.0 | |
| TREM1 signaling | 8 | 4.0 | |
| Hepatic cholestasis | 11 | 3.1 | |
| 129 more regulated than STAT1−/− | Pattern recognition receptors in recognition of bacteria and viruses | 21 | 7.6 |
| Interferon signaling | 11 | 5.9 | |
| Allograft rejection signaling | 12 | 5.3 | |
| Antigen presentation pathway | 11 | 5.3 | |
| Graft-vs-host disease signaling | 12 | 5.2 | |
| IFNAR1−/− more regulated than 129 | Valine, leucine, and isoleucine degradation | 25 | 7.4 |
| Propanoate metabolism | 21 | 5.9 | |
| LPS/IL-1-mediated inhibition of RXR function | 43 | 4.9 | |
| β-alanine metabolism | 18 | 4.8 | |
| Pyruvate metabolism | 22 | 4.8 | |
| IFNAR1−/− more regulated than STAT1−/− | Valine, leucine, and isoleucine degradation | 28 | 8.9 |
| Propanoate metabolism | 23 | 6.1 | |
| Pyruvate metabolism | 23 | 5.5 | |
| β-alanine metabolism | 18 | 4.4 | |
| Interferon signaling | 12 | 4.1 | |
| STAT1−/− more regulated than 129 | Arginine and proline metabolism | 9 | 4.3 |
| Urea cycle and metabolism of amino groups | 6 | 4.2 | |
| Bile acid biosynthesis | 7 | 4.0 | |
| IL-10 signaling | 7 | 3.2 | |
| Complement system | 5 | 2.9 | |
| STAT1−/− more regulated than IFNAR1−/− | LXR/RXR activation | 9 | 6.2 |
| Acute-phase response signaling | 12 | 5.2 | |
| IL-10 signaling | 7 | 4.6 | |
| Cytokine mediation of communication between immune cells | 6 | 3.9 | |
| Hepatic cholestasis | 9 | 3.8 |
Lists of differentially expressed genes were determined as described in Materials and Methods and were investigated using IPA. The 5 top-scoring canonical pathways are shown.
LXR, liver X receptor; RXR, retinoid X receptor; FXR, farsenoid X receptor; LPS, lipopolysaccharide.
Enrichment scores are calculated as −log(P value). The P value is derived from Fisher's exact test as described in Materials and Methods.
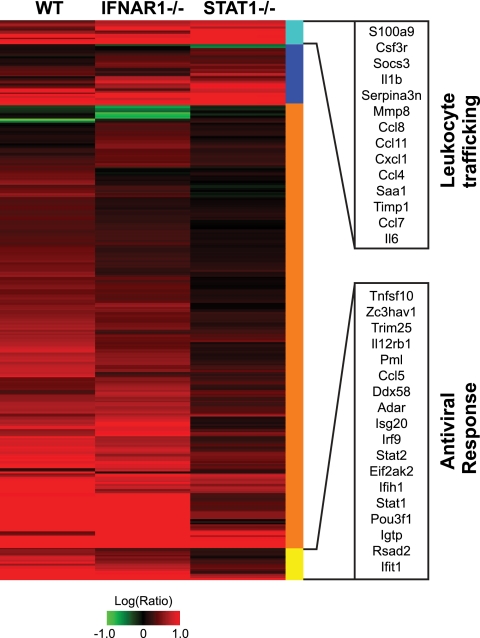
Since most of the genes noted above are induced in response to interferon stimulation, we compared our ANOVA gene list with a more comprehensive list of genes that have been shown to be upregulated in mice treated intranasally with interferon alpha (see Materials and Methods and reference 14). We identified 268 interferon-stimulated genes that were more strongly induced or repressed in WT or IFNAR1−/− mice than in STAT1−/− mice (Fig. 5, orange and yellow bars). Genes annotated with an antiviral function were most significantly enriched in this gene set (Fig. 5, yellow bar), suggesting that signaling independent of the type I interferon receptor, but dependent on STAT1, activates interferon-stimulated gene (ISG) expression during rMA15 infection, including expression of genes required for clearance of rMA15. Such signaling could be the result of interferon lambda production. Il28b (interferon lambda 3) is modestly induced at 2 dpi, averaging 4.5-fold induction in 129 mice, 7.4-fold induction in IFNAR1−/− mice, and 10.2-fold induction in STAT1−/− mice but not at later time points (data not shown). This level of induction is modest compared with the more than 10-fold induction seen with a number of type I interferons at 2 dpi (data not shown).
FIG. 5.
Deviant interferon response in STAT1−/− mice. Expression profiles are shown for interferon-stimulated genes (identified by intranasal administration of 10,000 units of interferon alpha for 24 h) (see details in Materials and Methods and reference 14) that were significantly different between STAT1−/− mice and WT or IFNAR1−/− mice as judged by ANOVA. Each column represents gene expression data from an average of 3 animal replicate experiments compared to mock-infected animal results. Genes shown in red were upregulated, genes shown in green were downregulated, and genes shown in black indicate no change in expression relative to mock-infected animal results. Genes are marked according to whether greater differential regulation was observed in STAT1−/− mice (blue and cyan) or in WT and IFNAR1−/− mice (orange and yellow). Genes marked with a cyan bar were functionally annotated as involved with “cell movement of leukocytes” by IPA (14 genes; P = 8.61 × 10−11). Genes marked with a yellow bar were functionally annotated as “antiviral response” genes by IPA (18 genes; P = 8.58 × 10−12). Genes in these functionally annotated categories are listed by Entrez Gene name. Genes comprising this heat map are listed in Table S3 in the supplemental material.
In contrast, only 48 ISGs were identified as being more strongly induced in STAT1−/− mice than in WT or IFNAR1−/− mice (Fig. 5, blue and cyan bars). Many of these genes are involved in leukocyte trafficking (Fig. 5, cyan bar). In particular, S110a9, Csf3r, Mmp8, Cxcl1, Saa1, Cxcl10, Ccl7, and Il6 gene expression is associated with neutrophil infiltration (4, 10, 22, 48, 55, 74), Ccl8 and Ccl11 are associated with eosinophilia (10), and Ccl4 is associated with macrophage activation (20). Notably, levels of neutrophils, eosinophils, and macrophages were elevated in the lungs of STAT1−/− mice infected with rMA15 (24). These results suggest that leukocyte infiltration of the lung is the main antiviral response available to STAT1−/− mice in the absence of effective ISG induction. Although these genes were identified as stimulated by interferon, many of these genes are also activated by other proinflammatory signaling pathways, most notably AP-1 and NFκB (18, 54, 72).
STAT1−/− mice exhibit induction of genes associated with leukocyte recruitment, TH2 bias, and fibrosis.
In order to understand the regulatory patterns associated with the increased pathogenesis of rMA15 in STAT1−/− mice, we examined those genes selected by ANOVA with the STAT1−/− and WT or IFNAR1−/− datasets (described above) that exhibited greater differential regulation in STAT1−/− mice. At 2 dpi, this analysis revealed a significant enrichment for genes associated with leukocyte trafficking (Table 2). In addition, eosinophilic chemokine Ccl24 and TH2 signaling transcripts (Csf3, Il5, and Il9) were identified. The involvement of TH2-associated genes was unexpected in the context of an acute viral infection of naïve animals. TH2 responses are associated with humoral immunity to bacteria and viruses, wound healing, and helminth infection, whereas clinical and experimental SARS infections are generally characterized by a TH1 immune response (40, 63). In addition, STAT1−/− mice exhibited increased gene expression associated with the arginine-proline metabolism pathway at 2 dpi (Table 2), including inducible arginase (Arg1), a marker for profibrotic “alternatively activated macrophages,” as described in detail below.
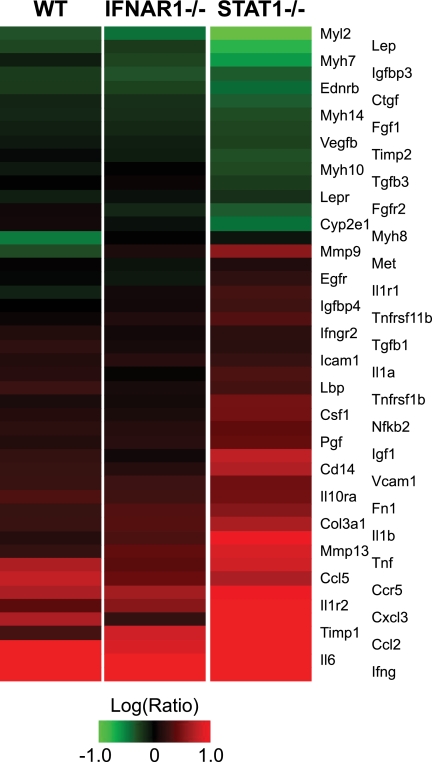
Of particular significance, numerous genes associated with the development of fibrosis were differentially induced by STAT1−/− mice compared with WT and IFNAR1−/− mice at 5 dpi (Fig. 6), including mediators of inflammation (Ccl2, Cxcl3, Ifng, Il1a, Il1b, Il6, Nfkb2, and Tnf) and tissue remodeling (Igf1, Igfbp4, Igfbp3, Timp1, Timp2, Mmp9, Mmp13, Fn1, and Col3a1). These results suggested that we were able to detect the activation of profibrotic gene expression several days prior to observing a fibrotic phenotype in the infected lung; we previously observed a similar phenomenon in the context of hepatic fibrosis caused by hepatitis C virus (68). RT-PCR validation performed using Col3a1 and Igf1 confirmed this pattern of gene expression (data not shown). Fibrosis genes included extracellular matrix proteins (Col3a1 and Fn1), metalloprotease inhibitor Timp1, and insulin-like growth factor (Igf1), which have previously been implicated as hallmarks of pulmonary fibrosis progression in the bleomycin model (11, 43, 70).
FIG. 6.
Increased fibrotic gene expression in STAT1−/− mice. Expression profiles are shown for 48 genes associated with the IPA “hepatic fibrosis” canonical pathway. The heat map is colored according to the ratio of gene expression at 5 dpi to that seen with mock-infected mice. Red indicates upregulation, green indicates downregulation, and black indicates no change in expression upon infection.
Knockout mice exhibit a bias in T-helper and macrophage differentiation.
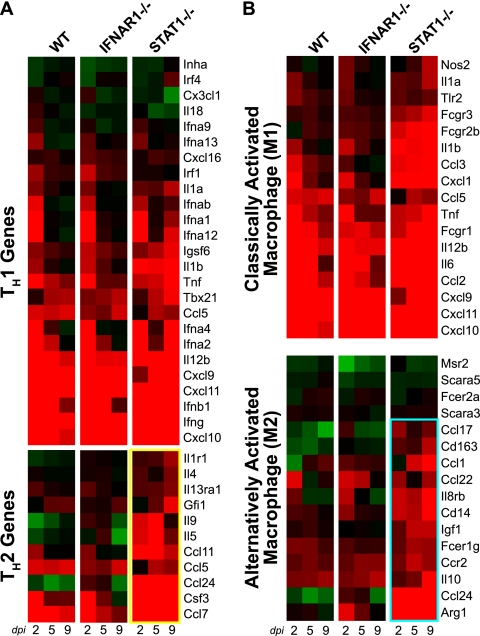
Because several genes identified in the time-matched ANOVA were associated with a TH2 immune response, we investigated gene regulation associated with the TH1/TH2 balance in greater detail. Expression levels for TH1-associated genes (Fig. 7 A) were similar in all mouse strains at 2 dpi. However, the STAT1−/− mice exhibited stronger induction of TH1 genes at 5 and 9 dpi compared with WT and IFNAR1−/− mice. This may reflect the ongoing viral replication and activation of the TH1 master regulator T-bet in the lungs of STAT1−/− mice. In contrast, whereas WT and IFNAR1−/− mice exhibited little regulation of TH2-associated gene expression, STAT1−/− mice exhibited significant induction of several TH2 genes, including Il5, Il9, Ccl7, Ccl11, Ccl24, and Csf3 (Fig. 7A, yellow box). RT-PCR performed on Ccl11 confirmed this pattern of gene regulation (data not shown). These data suggest that STAT1−/− animals experience a TH2 bias compared with WT mice.
FIG. 7.
Increased TH2- and M2-associated gene expression in STAT1−/− mice. (A and B) Expression profiles are shown for genes associated with TH1 and TH2 responses (A) and for M1 classical macrophage activation and M2 alternative macrophage activation (45) (B). Individual genes are identified by the mouse Entrez Gene name. TH2 responses and M2 responses were upregulated only in STAT1−/− mice and are highlighted by yellow and cyan boxes, respectively.
Because STAT1−/− mice exhibited dysregulation of the TH1 and TH2 responses, we examined the possibility that the recently described TH17 lineage could also be perturbed in these animals. These cells contribute to pathogenesis in asthma and chronic inflammation (49, 73, 81) and could be relevant to the ongoing pathogenesis observed in STAT1−/− mice. Although induction of TH17 cytokine gene Il17a was negligible in WT and IFNAR1−/− mice, expression of this gene was strongly induced at 9 dpi in STAT1−/− mice (averaging 86-fold by microarray and 1,600-fold by RT-PCR) (data not shown). Likewise, the expression of Il23a, a cytokine responsible for survival of TH17 cells, was induced 4-fold (based on microarray data) in STAT1−/− animals at 9 dpi. These results suggest that activation of the TH17 pathway occurs concurrently with the development of fibrosis in STAT1−/− mice.
Effect of the TH2 bias on macrophage phenotype.
Previous studies have suggested that macrophage phenotypes play a role in TH2-mediated fibrosis (2, 25, 51). In response to viral or bacterial pathogens, macrophages become activated by gamma interferon along a classical pathway (M1 macrophages). These cells are characterized by production of proinflammatory cytokines as well as nitric oxide synthase (45). Gene expression for individual M1-associated genes was strong at 2 dpi in all mouse strains studied (Fig. 7B). This gene expression decreased in the WT and IFNAR1−/− mice at 5 and 9 dpi as these strains cleared the virus. In contrast, M1 gene expression remained elevated in STAT1−/− mice, reflecting sustained infection in these animals.
Under certain circumstances, such as response to helminth infections, macrophages (specifically, M2 macrophages) can become activated by Il4 along an alternative pathway. These AAMs are characterized by production of arginase I, Il10, eosinophilic chemokines, and insulin-like growth factor-1. Increased expression of individual M2-associated genes, including arginase I, Ccl24, Cd14, and Il8rb by 2 dpi and Ccl1, Igf1, and Ccr2 by 5 dpi, revealed strong activation of the M2 response only in STAT1−/− animals (Fig. 7B, cyan box). This pattern of gene expression was also confirmed using RT-PCR for Arg1 and Igf1 (data not shown).
Lungs of STAT1−/− mice exhibit macrophage infiltration and activation along the alternative pathway.
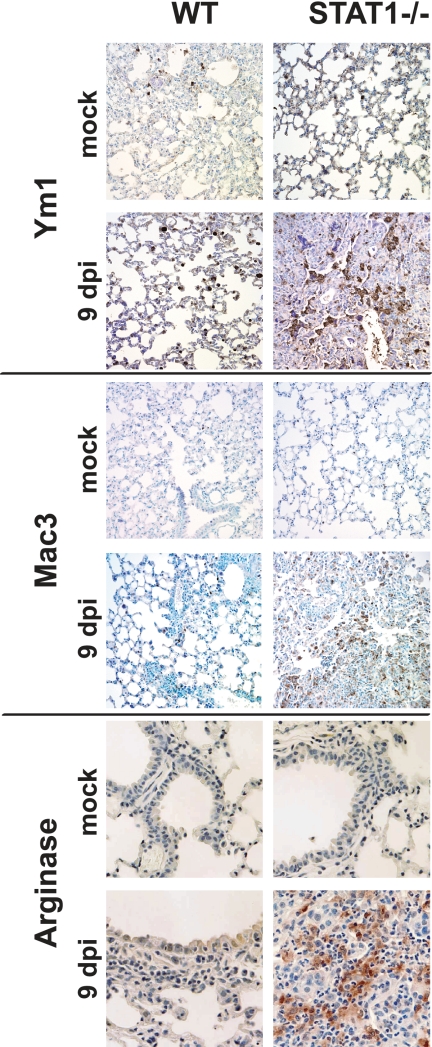
To confirm the production of arginase I by M2 macrophages in STAT1−/− mice, we performed immunohistochemistry to detect several protein markers of alternative macrophage activation: arginase I, Ym1, and Mac3. As shown in Fig. 8, minimal staining was detected in the lungs of mock-infected animals. By 9 dpi, both the number of cells staining for these proteins and the density of the staining had increased in STAT1−/− mice. Increased staining was also observed at 2 and 5 dpi (data not shown). Of particular interest, the cells exhibiting high levels of staining exhibited morphology characteristics consistent with macrophages. These results extend our gene expression data, suggesting that, in STAT1−/− mice, arginase I-expressing M2 macrophages infiltrate the lung in large numbers during rMA15 infection.
FIG. 8.
Detection of AAM markers in lung tissue of infected STAT1−/− mice by the use of immunohistochemistry. Lung sections were stained for the AAM markers arginase I, Ym1, and Mac3 with a hematoxylin counterstain. At 9 dpi, prominent staining is observed in STAT1−/− mice infected with rMA15 compared with uninfected animals and WT animals infected with rMA15. Magnification, ×40.
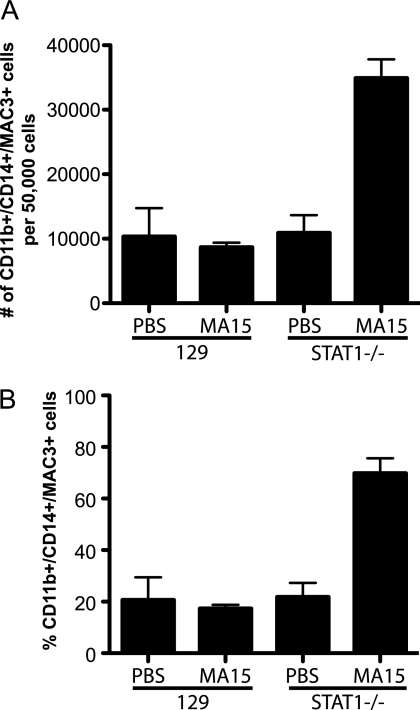
To confirm that lung macrophages were being activated along the alternative pathway, we performed flow cytometry using antibodies to CD11b and CD14 to identify macrophages and Mac3 to identify AAMs (A. Q. Ford, P. Dasgupta, E. A. Smith, N. Noben-Trauth, and A. D. Keegan, submitted for publication). Numbers of AAMs remained low in strain 129 mice and uninfected STAT1−/− mice. However, STAT1−/− mice infected with rMA15 exhibited a dramatic increase in the population of AAMs (Fig. 9 A) as well as in the number of AAMs as a percentage of total macrophages (Fig. 9B). Based on these data, we conclude that AAMs are disproportionately activated in STAT1−/− mice infected with rMA15.
FIG. 9.
Enrichment of alternatively activated macrophages in STAT1−/− mice infected with SARS-CoV. Ten-week-old strain 129 WT and STAT1−/− mice were infected with MA15, and their induction of AAMs was analyzed at 8 days postinfection. A total of 50,000 lung cells were sorted for the total macrophage population according to the presence of CD11b and CD14 double positives. Of these cells, AAMs were identified by the presence of Mac3 in the cells by intracellular staining. (A) The numbers of CD11b+, CD14+, and MAC3+ cells are graphed. (B) The numbers of AAMs are graphed as percentages of the total number of macrophages (CD11b+/CD14+ cells).
DISCUSSION
Previous studies have demonstrated that SARS-CoV infection of mice mirrors the acute exudative phase of ARDS (63, 64). More recently, STAT1−/− mice were shown to be highly susceptible to rMA15 infection, whereas disruption of interferon signaling at the level of the type I, II, or III interferon receptors did not significantly alter the disease course from that observed in WT mice (24). Our current gene expression analyses further demonstrate that although there were differences in the transcriptional profiles of WT and IFNAR1−/− mice, signaling through the type I interferon receptor was not necessary for a protective antiviral transcriptional response during rMA15 infection. Other studies performed by our group have revealed that IFNAR1−/− mice, and mouse embryonic fibroblasts derived from these animals, also induce ISG expression in response to infection with influenza virus (14, 27), suggesting that there exists a common mechanism of compensation for the loss of type I interferon receptor signaling. Our results also reveal molecular mechanisms by which STAT1−/− mice progress to fibrosis-like disease, mirroring the exudative and fibrotic phases of ARDS.
Compensatory gene expression in IFNAR1−/− mice.
Although WT and IFNAR1−/− mice exhibited similar weight loss and pathology characteristics, absence of the type I interferon receptor was associated with changes in host gene expression with respect to the immune response and healing. For example, at 2 dpi, a large number of genes associated with metabolism exhibited decreased expression in IFNAR1−/− mice (Table 2). Similar metabolic changes have been observed in response to a variety of viral infections, suggesting that this is an additional protective response to viral infection (3, 8, 13). Although genes associated with mitosis were induced in WT and IFNAR1−/− mice at 5 dpi, IFNAR1−/− mice exhibited greater induction of these genes, particularly those associated with DNA damage repair. In addition, we observed a slight attenuation of ISG expression in IFNAR1−/− mice compared with WT mice (Fig. 5). This attenuation may be responsible for the larger viral load observed in the lungs of IFNAR1−/− mice at 5 dpi compared with that observed in WT mice at the same time point (24). This compensatory ISG activation was potentially the result of interferon lambda signaling, but only modest induction of Il28b (interferon lambda 3) was observed in the lungs of all strains at 2 dpi. However, Frieman et al. showed that mice with defects in both type I and type III interferon signaling cleared rMA15 infection, suggesting the presence of additional compensatory pathways (24). We also cannot rule out the possibility that induction of some ISGs was the result of secondary signaling through other pathways, including that involving interferon gamma. However, Frieman et al. showed that interferon gamma signaling was also dispensable for clearing rMA15 (24). Through redundant pathways, some defects in innate immunity can be overcome, including removal of type I interferon signaling. By comparison, there was a dramatic reduction of ISG expression in rMA15-infected STAT1−/− mice; this reduction correlated with increased pathology and death for these animals. Since many immune response pathways, including the interferon response, signal through STAT1, the STAT1−/− mice appear to be unable to mount an effective antiviral response to rMA15 infection.
Macrophage dysregulation in STAT1−/− mice.
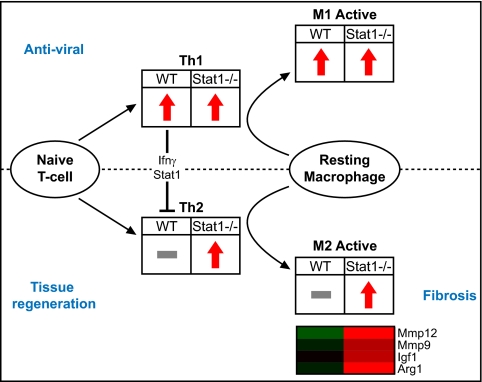
Our gene expression data suggest that infection of STAT1−/− mice with rMA15 dysregulates TH1/TH2 and macrophage host responses, resulting in a profibrotic phenotype. A model consistent with our data is shown in Fig. 10. In WT and IFNAR1−/− mice (but not STAT1−/− mice), T cells that differentiate with a TH1 program repress TH2 differentiation via gamma interferon signaling through STAT1 (82). Gamma interferon also stimulates the differentiation of macrophages along a classical activation pathway (M1) characterized by production of interleukins 1 and 6, tumor necrosis factor, and Cxcr3 ligands (Cxcl9, Cxcl10, and Cxcl11) (45); upregulation of these genes was observed in WT and IFNAR1−/− mice at 2 dpi.
FIG. 10.
Dysregulation of T-helper and macrophage differentiation in STAT1−/− mice. A functional network that depicts the effects of T-helper differentiation on macrophage phenotype and disease resolution is shown. The regulation of genes associated with TH1, TH2, M1, and M2 is represented with red arrows to indicate upregulation or with a gray bar to indicate no significant change from mock-infected animal results. A heat map is shown for select macrophage-associated genes that contribute to fibrosis and eosinophilia. Genes are colored according to differential regulation at 9 dpi.
In contrast, we found that STAT1−/− mice exhibited a TH2-biased immune response to infection with rMA15. TH2 cells produce Il4 (production of which was transcriptionally increased in STAT1−/− mice infected with rMA15), which triggers the differentiation of macrophages along an alternative activation pathway (M2). Alternatively activated macrophages were observed at 2 dpi in STAT1−/− mice, and their numbers increased at 5 and 9 dpi. The TH2/M2 response is associated with a variety of biological functions, including wound healing, chronic inflammation, and fibrotic immune responses to helminths (15, 66, 78). Notably, similarities with respect to TH2/M2 bias and pulmonary fibrosis have been observed in gamma interferon receptor knockout (IFNGR−/−) mice infected with murine gammaherpesvirus 68 (51). In contrast, we did not observe fibrosis in IFNGR−/− mice infected with rMA15 (24). This could have been due either to inhibition of uncontrolled cell proliferation by STAT1 (76) or to effective clearance of the virus in these mice.
Thus, a signaling pathway that promotes healing under favorable conditions may instead promote fibrosis in the context of continuing immune dysregulation, uncontrolled fibroblast proliferation, and/or persistent viral replication (Fig. 10). One hallmark of M2 macrophages is the production of arginase I (31), which regulates the first committed step in the conversion of arginine to proline (42). Proline is an important building block for collagen production, so increases in proline concentrations are considered profibrotic. It is interesting that, although arginase I was strongly induced transcriptionally at all time points in STAT1−/− mice, we observed a lag in the production of arginase I protein in the lung (Fig. 8). Likewise, genes in a panel associated with fibrosis progression (Fig. 6) were differentially induced in STAT1−/− mice prior to the appearance of protein deposition in the lungs. Similar results have also been found in the context of hepatic cell apoptosis and fibrosis (68). These results suggest the potential for diagnostic tests to detect gene expression changes predictive of the initiation of fibrosis.
STAT1−/− mice also exhibited increased expression of additional M2 macrophage-associated genes that contribute to fibrosis and eosinophilia. For example, we observed increased expression of Igf1, an important regulator of cell proliferation (65) that promotes fibroblast survival and increased extracellular matrix protein deposition (79). It is negatively regulated by several IGF-binding proteins (Igfbp1 through Igfbp6), and expression of genes encoding these proteins was decreased in STAT1−/− mice in response to rMA15 infection (data not shown). In addition, we observed increased expression of Mmp9 and Mmp12, which are expressed by M2 macrophages to promote wound healing (56) and tissue remodeling (33), respectively.
Contribution of eosinophils and TH17 cells.
TH2-mediated inflammation causes eosinophil infiltration, which is generally not observed during acute viral infection. Mmp12 is an important contributor to airway eosinophilia in mice (60), and eosinophilia is associated with pulmonary fibrosis in humans (21) and with ARDS (35) and is also observed in some cases of persistent inflammation. Expression of genes encoding eosinophil attractants, including Ccl11, Ccl24, and Mmp12, was observed in STAT1−/− mice as early as 2 dpi, and significant eosinophilia was observed at 8 dpi (24).
TH17 cells are a recently described T-helper population distinct from TH1 and TH2 cells (57) and are associated with asthma, chronic obstructive pulmonary disease, and autoimmune inflammation (30, 36, 53, 73, 81). As with eosinophilia, the presence of TH17 cells, which mediate inflammation through the production of Il17 and Il23, is not characteristic of most acute viral infections. However, gene expression characteristic of TH17 cells was observed in STAT1−/− mice at 9 dpi. As with TH2, TH17 differentiation is repressed by gamma interferon signaling through STAT1 (57), suggesting that removal of this block in STAT1−/− mice enables the expansion of TH17 cells. The occurrence of eosinophils and TH17 cell markers in the lungs of STAT1−/− mice at 8 to 9 dpi suggests that these cells may play a role in the later progression rather than the initiation of fibrosis, although this has not been directly examined.
Comparison of rMA15-infected STAT1−/− mice with other SARS-CoV models.
This is the first report of a study examining the host transcriptional responses elicited by the mouse-adapted rMA15 variant of SARS-CoV in young mice. The disease course caused by rMA15 infection in young strain 129 mice was similar to that observed in aged BALB/c mice infected with SARS-CoV Urbani (63). Of particular interest, a comparison of TH2 and M2 gene expression characteristics in aged and young BALB/c mice (63) reveals greater induction of Il13ra1, Ccl5, Ccl7, Fcer1g, and Igf1 in aged mice, suggesting that TH2 and M2 induction upon viral infection is a common feature of aged BALB/c and STAT1−/− mice. In contrast, induction of TH17-associated genes was not observed in aged BALB/c mice infected with SARS-CoV. In addition, previously described SARS-CoV mouse models that used stain rMA15 or Urbani (62-64, 67) exhibited either mortality or recovery from infection within 6 dpi. In contrast, STAT1−/− mice did not succumb to the infection until approximately 10 dpi and exhibited protein deposition in the lungs consistent with fibrosis. This suggests that rMA15-infected STAT1−/− mice represent a model of both the exudative and the organizing (fibrotic) phases of ARDS pathophysiology (61).
Previous transcriptome analysis of blood samples from patients suffering from SARS-CoV infection suggested that sustained innate immune activation and a failure to switch to humoral immunity are associated with a severe disease course (6). STAT1−/− mice exhibited a similar sustained, yet ineffective, immune response. However, these clinical cases exhibited gene expression more characteristic of the type I interferon response and not of the TH2, M2, and TH17 cell expression observed in STAT1−/− mice. These differences may reflect the choice of tissue used for microarray analysis (Cameron and coworkers used blood from patients whereas our analysis used lung tissue), but they could also be the result of the profound dysregulation associated with the loss of STAT1-dependent transcription. However, similar TH2 bias and macrophage dysregulation characteristics have been observed in aged human populations (26, 59), members of which are more susceptible to pulmonary fibrosis (44) and exhibited greater mortality rates during the SARS outbreak (9). Based on these common factors, STAT1−/− mice may be a good model for investigation of the SARS-dependent initiation of fibrosis observed in aged populations.
In conclusion, rMA15 infection of STAT1−/− mice resulted in an aberrant TH2 response, sustained viral infection, inflammation, and a profibrotic phenotype. This infection system serves as a model for the early and intermediate stages of ARDS and should be of value for the testing of therapeutic interventions to alleviate virus-induced ARDS. In addition, this model may be useful for studying the role of eosinophils and TH17 cells in mediating persistent inflammation in response to viral infection.
Supplementary Material
Acknowledgments
We thank Elizabeth Smith (University of Maryland School of Medicine) for her help with immunohistochemistry. We thank Sean Proll, Sarah Belisle, and Chester Ni for assistance with statistical analysis and figure design.
This work was supported by Public Health Service grants AI66542 to M.B.F., AI075297 and AI059443 to R.S.B., and HL080621 to M.G.K. Additional SARS-CoV work in the Katze laboratory is supported by U.S. Public Health Service grant U54 AI081680 and by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272200800060C.
Footnotes
Published ahead of print on 11 August 2010.
Supplemental material for this article is available at http://jvi.asm.org/.
REFERENCES
- 1.Baas, T., A. Roberts, T. H. Teal, L. Vogel, J. Chen, T. M. Tumpey, M. G. Katze, and K. Subbarao. 2008. Genomic analysis reveals age-dependent innate immune responses to severe acute respiratory syndrome coronavirus. J. Virol. 82:9465-9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belperio, J. A., M. Dy, L. Murray, M. D. Burdick, Y. Y. Xue, R. M. Strieter, and M. P. Keane. 2004. The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J. Immunol. 173:4692-4698. [DOI] [PubMed] [Google Scholar]
- 3.Billharz, R., H. Zeng, S. C. Proll, M. J. Korth, S. Lederer, R. Albrecht, A. G. Goodman, E. Rosenzweig, T. M. Tumpey, A. García-Sastre, and M. G. Katze. 2009. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 83:10557-10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björkman, L., J. Karlsson, A. Karlsson, M. Rabiet, F. Boulay, H. Fu, J. Bylund, and C. Dahlgren. 2008. Serum amyloid A mediates human neutrophil production of reactive oxygen species through a receptor independent of formyl peptide receptor like-1. J. Leukoc. Biol. 83:245-253. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Côté, T. C. T. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 6.Cameron, M. J., L. Ran, L. Xu, A. Danesh, J. F. Bermejo-Martin, C. M. Cameron, M. P. Muller, W. L. Gold, S. E. Richardson, S. M. Poutanen, B. M. Willey, M. E. DeVries, Y. Fang, C. Seneviratne, S. E. Bosinger, D. Persad, P. Wilkinson, L. D. Greller, R. Somogyi, A. Humar, S. Keshavjee, M. Louie, M. B. Loeb, J. Brunton, A. J. McGeer, and D. J. Kelvin. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81:8692-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cervantes-Barragan, L., R. Züst, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, E. Y., J. N. Sutton, J. M. Jacobs, A. Bondarenko, R. D. Smith, and M. G. Katze. 2009. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J. Virol. 83:9283-9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan-Yeung, M., and R. Xu. 2003. SARS: epidemiology. Respirology 8:S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, K., Y. Wei, A. Alter, G. C. Sharp, and H. Braley-Mullen. 2005. Chemokine expression during development of fibrosis versus resolution in a murine model of granulomatous experimental autoimmune thyroiditis. J. Leukoc. Biol. 78:716-724. [DOI] [PubMed] [Google Scholar]
- 11.Choi, J., S. Lee, D. A. Sunde, I. Huizar, K. L. Haugk, V. J. Thannickal, R. Vittal, S. R. Plymate, and L. M. Schnapp. 2009. Insulin-like growth factor-I receptor blockade improves outcome in mouse model of lung injury. Am. J. Respir. Crit. Care Med. 179:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, D. W., C. H. Lee, C. W. Chen, H. Y. Chang, and T. R. Hsiue. 1999. Varicella pneumonia complicated by acute respiratory distress syndrome in an adult. J. Formos. Med. Assoc. 98:778-782. [PubMed] [Google Scholar]
- 13.Chow, E. K., A. Castrillo, A. Shahangian, L. Pei, R. M. O'Connell, R. L. Modlin, P. Tontonoz, and G. Cheng. 2006. A role for IRF3-dependent RXRα repression in hepatotoxicity associated with viral infections. J. Exp. Med. 203:2589-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cilloniz, C., M. J. Pantin-Jackwood, C. Ni, A. G. Goodman, X. Peng, S. C. Proll, V. S. Carter, E. R. Rosenzweig, K. J. Szretter, J. M. Katz, M. J. Korth, D. E. Swayne, T. M. Tumpey, and M. G. Katze. 2010. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 84:7613-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho, H. M., L. P. Acosta, H. W. Wu, S. T. McGarvey, L. Su, G. C. Langdon, M. A. Jiz, B. Jarilla, R. M. Olveda, J. F. Friedman, and J. D. Kurtis. 2007. Th2 cytokines are associated with persistent hepatic fibrosis in human Schistosoma japonicum infection. J. Infect. Dis. 195:288-295. [DOI] [PubMed] [Google Scholar]
- 16.Day, C. W., R. Baric, S. X. Cai, M. Frieman, Y. Kumaki, J. D. Morrey, D. F. Smee, and D. L. Barnard. 2009. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology 395:210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lang, A., T. Baas, T. Teal, L. M. Leijten, B. Rain, A. D. Osterhaus, B. L. Haagmans, and M. G. Katze. 2007. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 3:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dendorfer, U., P. Oettgen, and T. A. Libermann. 1994. Multiple regulatory elements in the interleukin-6 gene mediate induction by prostaglandins, cyclic AMP, and lipopolysaccharide. Mol. Cell. Biol. 14:4443-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding, Y., H. Wang, H. Shen, Z. Li, J. Geng, H. Han, J. Cai, X. Li, W. Kang, D. Weng, Y. Lu, D. Wu, L. He, and K. Yao. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200:282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorner, B. G., A. Scheffold, M. S. Rolph, M. B. Huser, S. H. E. Kaufmann, A. Radbruch, I. E. A. Flesch, and R. A. Kroczek. 2002. MIP-1α, MIP-1β, RANTES, and ATAC/lymphotactin function together with IFN-γ as type 1 cytokines. Proc. Natl. Acad. Sci. U. S. A. 99:6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emad, A., and Y. Emad. 2007. Relationship between eosinophilia and levels of chemokines (CCL5 and CCL11) and IL-5 in bronchoalveolar lavage fluid of patients with mustard gas-induced pulmonary fibrosis. J. Clin. Immunol. 27:605-612. [DOI] [PubMed] [Google Scholar]
- 22.Fielding, C. A., R. M. McLoughlin, L. McLeod, C. S. Colmont, M. Najdovska, D. Grail, M. Ernst, S. A. Jones, N. Topley, and B. J. Jenkins. 2008. IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J. Immunol. 181:2189-2195. [DOI] [PubMed] [Google Scholar]
- 23.Franks, T. J., P. Y. Chong, P. Chui, J. R. Galvin, R. M. Lourens, A. H. Reid, E. Selbs, C. P. L. McEvoy, C. D. L. Hayden, J. Fukuoka, J. K. Taubenberger, and W. D. Travis. 2003. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34:743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frieman, M. B., J. Chen, T. E. Morrison, A. Whitmore, W. Funkhouser, J. M. Ward, E. W. Lamirande, A. Roberts, M. Heise, K. Subbarao, and R. S. Baric. 2010. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6:e1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gangadharan, B., M. A. Hoeve, J. E. Allen, B. Ebrahimi, S. M. Rhind, B. M. Dutia, and A. A. Nash. 2008. Murine gammaherpesvirus-induced fibrosis is associated with the development of alternatively activated macrophages. J. Leukoc. Biol. 84:50-58. [DOI] [PubMed] [Google Scholar]
- 26.Glaser, R., R. C. MacCallum, B. F. Laskowski, W. B. Malarkey, J. F. Sheridan, and J. K. Kiecolt-Glaser. 2001. Evidence for a shift in the Th-1 to Th-2 cytokine response associated with chronic stress and aging. J. Gerontol. A Biol. Sci. Med. Sci. 56:M477-M482. [DOI] [PubMed] [Google Scholar]
- 27.Goodman, A. G., H. Zeng, S. C. Proll, X. Peng, C. Cillóniz, V. S. Carter, M. J. Korth, T. M. Tumpey, and M. G. Katze. 2010. The α/β interferon receptor provides protection against influenza virus replication but is dispensable for inflammatory response signaling. J. Virol. 84:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu, J., and C. Korteweg. 2007. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 170:1136-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haagmans, B. L., T. Kuiken, B. E. Martina, R. A. M. Fouchier, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, T. de Jong, S. Itamura, K. Chan, M. Tashiro, and A. D. M. E. Osterhaus. 2004. Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10:290-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto, T., K. Akiyama, N. Kobayashi, and A. Mori. 2005. Comparison of IL-17 production by helper T cells among atopic and nonatopic asthmatics and control subjects. Int. Arch. Allergy Immunol. 137(Suppl. 1):51-54. [DOI] [PubMed] [Google Scholar]
- 31.Hesse, M., M. Modolell, A. C. La Flamme, M. Schito, J. M. Fuentes, A. W. Cheever, E. J. Pearce, and T. A. Wynn. 2001. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J. Immunol. 167:6533-6544. [DOI] [PubMed] [Google Scholar]
- 32.Hudson, L. D., J. A. Milberg, D. Anardi, and R. J. Maunder. 1995. Clinical risks for development of the acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 151:293-301. [DOI] [PubMed] [Google Scholar]
- 33.Kahnert, A., P. Seiler, M. Stein, S. Bandermann, K. Hahnke, H. Mollenkopf, and S. H. E. Kaufmann. 2006. Alternative activation deprives macrophages of a coordinated defense program to Mycobacterium tuberculosis. Eur. J. Immunol. 36:631-647. [DOI] [PubMed] [Google Scholar]
- 34.Klinger, J. R., M. P. Sanchez, L. A. Curtin, M. Durkin, and B. Matyas. 1998. Multiple cases of life-threatening adenovirus pneumonia in a mental health care center. Am. J. Respir. Crit. Care Med. 157:645-649. [DOI] [PubMed] [Google Scholar]
- 35.Kollef, M. H., and D. P. Schuster. 1995. The acute respiratory distress syndrome. N. Engl. J. Med. 332:27-37. [DOI] [PubMed] [Google Scholar]
- 36.Komiyama, Y., S. Nakae, T. Matsuki, A. Nambu, H. Ishigame, S. Kakuta, K. Sudo, and Y. Iwakura. 2006. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 177:566-573. [DOI] [PubMed] [Google Scholar]
- 37.Ksiazek, T. G., C. J. Peters, P. E. Rollin, S. Zaki, S. Nichol, C. Spiropoulou, S. Morzunov, H. Feldmann, A. Sanchez, A. S. Khan, et al. 1995. Identification of a new North American hantavirus that causes acute pulmonary insufficiency. Am. J. Trop. Med. Hyg. 52:117-123. [DOI] [PubMed] [Google Scholar]
- 38.Ku, A. S., and L. T. Chan. 1999. The first case of H5N1 avian influenza infection in a human with complications of adult respiratory distress syndrome and Reye's syndrome. J. Paediatr. Child Health 35:207-209. [DOI] [PubMed] [Google Scholar]
- 39.Kuiken, T., R. A. M. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. S. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. Manuguerra, K. Stöhr, J. S. M. Peiris, and A. D. M. E. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam, C. W. K., M. H. M. Chan, and C. K. Wong. 2004. Severe acute respiratory syndrome: clinical and laboratory manifestations. Clin. Biochem. Rev. 25:121-132. [PMC free article] [PubMed] [Google Scholar]
- 41.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 42.Maarsingh, H., T. Pera, and H. Meurs. 2008. Arginase and pulmonary diseases. Naunyn Schmiedebergs Arch. Pharmacol. 378:171-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda, A., K. Hiyama, H. Yamakido, S. Ishioka, and M. Yamakido. 1996. Increased expression of platelet-derived growth factor A and insulin-like growth factor-I in BAL cells during the development of bleomycin-induced pulmonary fibrosis in mice. Chest 109:780-786. [DOI] [PubMed] [Google Scholar]
- 44.Mallick, S. 2008. Outcome of patients with idiopathic pulmonary fibrosis (IPF) ventilated in intensive care unit. Respir. Med. 102:1355-1359. [DOI] [PubMed] [Google Scholar]
- 45.Mantovani, A., A. Sica, S. Sozzani, P. Allavena, A. Vecchi, and M. Locati. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25:677-686. [DOI] [PubMed] [Google Scholar]
- 46.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 47.Merk, J., F. X. Schmid, M. Fleck, S. Schwarz, C. Lehane, S. Boehm, B. Salzberger, and D. E. Birnbaum. 2005. Fatal pulmonary failure attributable to viral pneumonia with human herpes virus 6 (HHV6) in a young immunocompetent woman. J. Intensive Care Med. 20:302-306. [DOI] [PubMed] [Google Scholar]
- 48.Michalec, L., B. K. Choudhury, E. Postlethwait, J. S. Wild, R. Alam, M. Lett-Brown, and S. Sur. 2002. CCL7 and CXCL10 orchestrate oxidative stress-induced neutrophilic lung inflammation. J. Immunol. 168:846-852. [DOI] [PubMed] [Google Scholar]
- 49.Molet, S., Q. Hamid, F. Davoine, E. Nutku, R. Taha, N. Pagé, R. Olivenstein, J. Elias, and J. Chakir. 2001. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J. Allergy Clin. Immunol. 108:430-438. [DOI] [PubMed] [Google Scholar]
- 50.Moore, B. B., and C. M. Hogaboam. 2008. Murine models of pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 294:L152-L160. [DOI] [PubMed] [Google Scholar]
- 51.Mora, A. L., E. Torres-González, M. Rojas, C. Corredor, J. Ritzenthaler, J. Xu, J. Roman, K. Brigham, and A. Stecenko. 2006. Activation of alveolar macrophages via the alternative pathway in herpesvirus-induced lung fibrosis. Am. J. Respir. Cell Mol. Biol. 35:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagata, N., N. Iwata, H. Hasegawa, S. Fukushi, M. Yokoyama, A. Harashima, Y. Sato, M. Saijo, S. Morikawa, and T. Sata. 2007. Participation of both host and virus factors in induction of severe acute respiratory syndrome (SARS) in F344 rats infected with SARS coronavirus. J. Virol. 81:1848-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakae, S., A. Nambu, K. Sudo, and Y. Iwakura. 2003. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J. Immunol. 171:6173-6177. [DOI] [PubMed] [Google Scholar]
- 54.Németh, J., I. Stein, D. Haag, A. Riehl, T. Longerich, E. Horwitz, K. Breuhahn, C. Gebhardt, P. Schirmacher, M. Hahn, Y. Ben-Neriah, E. Pikarsky, P. Angel, and J. Hess. 2009. S100A8 and S100A9 are novel nuclear factor kappa B target genes during malignant progression of murine and human liver carcinogenesis. Hepatology 50:1251-1262. [DOI] [PubMed] [Google Scholar]
- 55.Nicola, N. A., and D. Metcalf. 1985. Binding of 125I-labeled granulocyte colony-stimulating factor to normal murine hemopoietic cells. J. Cell. Physiol. 124:313-321. [DOI] [PubMed] [Google Scholar]
- 56.Okuma, T., Y. Terasaki, K. Kaikita, H. Kobayashi, W. A. Kuziel, M. Kawasuji, and M. Takeya. 2004. C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases. J. Pathol. 204:594-604. [DOI] [PubMed] [Google Scholar]
- 57.Park, H., Z. Li, X. O. Yang, S. H. Chang, R. Nurieva, Y. Wang, Y. Wang, L. Hood, Z. Zhu, Q. Tian, and C. Dong. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pasieka, T. J., T. Baas, V. S. Carter, S. C. Proll, M. G. Katze, and D. A. Leib. 2006. Functional genomic analysis of herpes simplex virus type 1 counteraction of the host innate response. J. Virol. 80:7600-7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plackett, T. P., E. M. Schilling, D. E. Faunce, M. A. Choudhry, P. L. Witte, and E. J. Kovacs. 2003. Aging enhances lymphocyte cytokine defects after injury. FASEB J. 17:688-689. [DOI] [PubMed] [Google Scholar]
- 60.Pouladi, M. A., C. S. Robbins, F. K. Swirski, M. Cundall, A. N. J. McKenzie, M. Jordana, S. D. Shapiro, and M. R. Stämpfli. 2004. Interleukin-13-dependent expression of matrix metalloproteinase-12 is required for the development of airway eosinophilia in mice. Am. J. Respir. Cell Mol. Biol. 30:84-90. [DOI] [PubMed] [Google Scholar]
- 61.Puneet, P., S. Moochhala, and M. Bhatia. 2005. Chemokines in acute respiratory distress syndrome. Am. J. Physiol. Lung Cell. Mol. Physiol. 288:L3-L15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rockx, B., T. Baas, G. A. Zornetzer, B. Haagmans, T. Sheahan, M. Frieman, M. D. Dyer, T. H. Teal, S. Proll, J. van den Brand, R. Baric, and M. G. Katze. 2009. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 83:7062-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rockx, B., T. Sheahan, E. Donaldson, J. Harkema, A. Sims, M. Heise, R. Pickles, M. Cameron, D. Kelvin, and R. Baric. 2007. Synthetic reconstruction of zoonotic and early human severe acute respiratory syndrome coronavirus isolates that produce fatal disease in aged mice. J. Virol. 81:7410-7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rom, W. N., P. Basset, G. A. Fells, T. Nukiwa, B. C. Trapnell, and R. G. Crysal. 1988. Alveolar macrophages release an insulin-like growth factor I-type molecule. J. Clin. Invest. 82:1685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shao, D. D., R. Suresh, V. Vakil, R. H. Gomer, and D. Pilling. 2008. Pivotal advance: Th-1 cytokines inhibit, and Th-2 cytokines promote fibrocyte differentiation. J. Leukoc. Biol. 83:1323-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheahan, T., T. E. Morrison, W. Funkhouser, S. Uematsu, S. Akira, R. S. Baric, and M. T. Heise. 2008. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 4:e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith, M. W., K. Walters, M. J. Korth, M. Fitzgibbon, S. Proll, J. C. Thompson, M. M. Yeh, M. C. Shuhart, J. C. Furlong, P. P. Cox, D. L. Thomas, J. D. Phillips, J. P. Kushner, N. Fausto, R. L. J. Carithers, and M. G. Katze. 2006. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterology 130:179-187. [DOI] [PubMed] [Google Scholar]
- 69.Spelman, D. W., and P. A. Stanley. 1983. Respiratory syncytial virus pneumonitis in adults. Med. J. Aust. 1:430-431. [DOI] [PubMed] [Google Scholar]
- 70.Swiderski, R. E., J. E. Dencoff, C. S. Floerchinger, S. D. Shapiro, and G. W. Hunninghake. 1998. Differential expression of extracellular matrix remodeling genes in a murine model of bleomycin-induced pulmonary fibrosis. Am. J. Pathol. 152:821-828. [PMC free article] [PubMed] [Google Scholar]
- 71.Szretter, K. J., S. Gangappa, J. A. Belser, H. Zeng, H. Chen, Y. Matsuoka, S. Sambhara, D. E. Swayne, T. M. Tumpey, and J. M. Katz. 2009. Early control of H5N1 influenza virus replication by the type I interferon response in mice. J. Virol. 83:5825-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tian, B., D. E. Nowak, M. Jamaluddin, S. Wang, and A. R. Brasier. 2005. Identification of direct genomic targets downstream of the nuclear factor-kappaB transcription factor mediating tumor necrosis factor signaling. J. Biol. Chem. 280:17435-17448. [DOI] [PubMed] [Google Scholar]
- 73.Traves, S. L., and L. E. Donnelly. 2008. Th17 cells in airway diseases. Curr. Mol. Med. 8:416-426. [DOI] [PubMed] [Google Scholar]
- 74.Vandal, K., P. Rouleau, A. Boivin, C. Ryckman, M. Talbot, and P. A. Tessier. 2003. Blockade of S100A8 and S100A9 suppresses neutrophil migration in response to lipopolysaccharide. J. Immunol. 171:2602-2609. [DOI] [PubMed] [Google Scholar]
- 75.Vannella, K. M., and B. B. Moore. 2008. Viruses as co-factors for the initiation or exacerbation of lung fibrosis. Fibrogenesis Tissue Repair 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walters, D. M., A. Antao-Menezes, J. L. Ingram, A. B. Rice, A. Nyska, Y. Tani, S. R. Kleeberger, and J. C. Bonner. 2005. Susceptibility of signal transducer and activator of transcription-1-deficient mice to pulmonary fibrogenesis. Am. J. Pathol. 167:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ware, L. B., and M. A. Matthay. 2000. The acute respiratory distress syndrome. N. Engl. J. Med. 342:1334-1349. [DOI] [PubMed] [Google Scholar]
- 78.Wills-Karp, M., J. Luyimbazi, X. Xu, B. Schofield, T. Y. Neben, C. L. Karp, and D. D. Donaldson. 1998. Interleukin-13: central mediator of allergic asthma. Science 282:2258-2261. [DOI] [PubMed] [Google Scholar]
- 79.Wynes, M. W., S. K. Frankel, and D. W. H. Riches. 2004. IL-4-induced macrophage-derived IGF-I protects myofibroblasts from apoptosis following growth factor withdrawal. J. Leukoc. Biol. 76:1019-1027. [DOI] [PubMed] [Google Scholar]
- 80.Xu, T., J. Qiao, L. Zhao, G. Wang, G. He, K. Li, Y. Tian, M. Gao, J. Wang, H. Wang, and C. Dong. 2006. Acute respiratory distress syndrome induced by avian influenza A (H5N1) virus in mice. Am. J. Respir. Crit. Care Med. 174:1011-1017. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, Z., M. Zheng, J. Bindas, P. Schwarzenberger, and J. K. Kolls. 2006. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel Dis. 12:382-388. [DOI] [PubMed] [Google Scholar]
- 82.Zhuang, Y., Z. Huang, J. Nishida, L. Zhang, and H. Huang. 2009. Signaling pathways that lead to the silencing of the interleukin-4-producing potential in Th1 cells. J. Interferon Cytokine Res. 29:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.