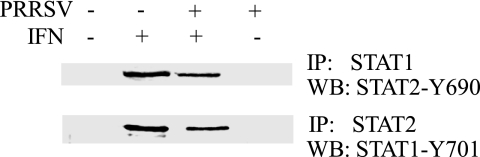Abstract
Type I interferons (IFNs) IFN-α/β play an important role in innate immunity against viral infections by inducing antiviral responses. Porcine reproductive and respiratory syndrome virus (PRRSV) inhibits the synthesis of type I IFNs. However, whether PRRSV can inhibit IFN signaling is less well understood. In the present study, we found that PRRSV interferes with the IFN signaling pathway. The transcript levels of IFN-stimulated genes ISG15 and ISG56 and protein level of signal transducer and activator of transcription 2 (STAT2) in PRRSV VR2385-infected MARC-145 cells were significantly lower than those in mock-infected cells after IFN-α treatment. IFN-induced phosphorylation of both STAT1 and STAT2 and their heterodimer formation in the PRRSV-infected cells were not affected. However, the majority of the STAT1/STAT2/IRF9 (IFN regulatory factor 9) heterotrimers remained in the cytoplasm of PRRSV-infected cells, which indicates that the nuclear translocation of the heterotrimers was blocked. Overexpression of NSP1β of PRRSV VR2385 inhibited expression of ISG15 and ISG56 and blocked nuclear translocation of STAT1, which suggests that NSP1β might be the viral protein responsible for the inhibition of IFN signaling. PRRSV infection in primary porcine pulmonary alveolar macrophages (PAMs) also inhibited IFN-α-stimulated expression of the ISGs and the STAT2 protein. In contrast, a licensed low-virulence vaccine strain, Ingelvac PRRS modified live virus (MLV), activated expression of IFN-inducible genes, including those of chemokines and antiviral proteins, in PAMs without the addition of external IFN and had no detectable effect on IFN signaling. These findings suggest that PRRSV interferes with the activation and signaling pathway of type I IFNs by blocking ISG factor 3 (ISGF3) nuclear translocation.
Porcine reproductive and respiratory syndrome (PRRS) is an economically important disease, causing an estimated loss of $560 million per year to the swine industry in the United States (24). The causative agent, PRRS virus (PRRSV), is a positive-sense single-stranded RNA virus belonging to the family Arteriviridae (20). The genome of PRRSV is about 15 kb in length, with nine open reading frames (ORFs) (7, 22). ORF1a and -1b comprise 80% of the viral genome and are predicted to encode viral enzymes for RNA synthesis. ORF2, -2a, -3, and -4 of PRRSV encode minor membrane-associated proteins GP2, E, GP3, and GP4, respectively. ORF5, -6, and -7 encode major structural proteins, i.e., a major envelope glycoprotein (GP5), a membrane protein (M), and a nucleocapsid protein (N), respectively (18, 21). PRRSV can be propagated in vitro in the epithelial cell-derived monkey kidney cell line MARC-145 (12) and in primary culture of porcine pulmonary alveolar macrophages (PAMs). PAMs are the main target cells for PRRSV during acute infection of pigs (31).
PRRSV-infected pigs develop a delayed appearance of neutralizing antibodies (15) and a weak cell-mediated immune response (39). PRRSV inhibits synthesis of type I interferons (IFNs) in infected pigs (1, 5, 17). IFNs could not be detected in the lungs of pigs in which PRRSV actively replicated. PRRSV infection of PAMs and MARC-145 cells in vitro leads to very low levels of IFN-α expression (1, 23). Suppression of innate immunity is believed to be an important factor contributing to the PRRSV modulation of host immune responses.
Type I IFNs, such as IFN-α and -β, acting in concert with IFN-γ, are critical to innate immunity against viruses and play an important role in the modulation of adaptive immunity (34). Activation of IFN signaling leads to induction of antiviral responses. The signaling of type I IFNs is initiated after IFN-α and -β bind to their receptors on the cell surface (8, 32, 33). The receptor binding activates Janus kinase (JAK) and Tyk2 to phosphorylate the signal transducers and activators of transcription (STATs) STAT1 and STAT2. Phosphorylated STAT1 and STAT2 form heterotrimers with interferon regulatory factor 9 (IRF9) and translocate into the nucleus to induce expression of IFN-stimulated genes (ISGs), which result in the establishment of an antiviral state (8, 32, 33).
It was found that PRRSV suppresses IFN-β production in MARC-145 cells by interfering with the RIG-I signaling pathway (17) and that PRRSV NSP1β inhibits interferon production (3, 6, 13). Overexpression of PRRSV NSP1β in HEK293T cells interferes with the nuclear translocation of STAT1-green fluorescent protein (GFP), as observed under fluorescence microscopy (6). However, whether PRRSV can inhibit type I IFN signaling and induction of IFN-stimulated genes, especially in primary PAMs, is not known.
To further define the mechanisms of PRRSV-induced inhibition of innate immunity, we examined the effects of PRRSV infection on type I IFN signaling. In the present study, we found that PRRSV inhibited type I IFN signaling and downstream gene expression. The nuclear translocation of STAT1/STAT2/IRF9 heterotrimers was blocked, while the IFN-induced phosphorylation of STAT1 and STAT2 was not affected. PRRSV NSP1β is the possible viral protein that is responsible for the inhibition of IFN signaling. The interference of IFN signaling was also demonstrated in primary culture of PAMs.
MATERIALS AND METHODS
Cells and viruses.
MARC-145 (12), HEK293, and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). PRRSV VR2385 (19) was used to inoculate MARC-145 cells at a multiplicity of infection (MOI) of 0.5 to 1. A licensed modified live virus (MLV) vaccine strain, Ingelvac PRRS MLV (herein named MLV), was kindly provided by Kay S. Faaberg (National Animal Disease Center, Ames, IA). Virus titers were determined in MARC-145 cells for the median tissue culture infectious dose (TCID50), as described previously (40).
Primary PAMs were prepared from bronchoalveolar lavage fluid of 8-week-old, PRRSV-negative piglets. The preparation and subsequent culture of PAMs in RPMI 1640 culture medium were conducted as previously described (25).
For virus inactivation, supernatant containing virus was treated in a UV cross-linker for two 10-min pulses separated by a 1-min interval. Virus inactivation was confirmed by inoculation of MARC-145 cells and the absence of cytopathic-effect development 72 h postinfection (hpi).
For interferon stimulation, recombinant human IFN-α A/D (Sigma-Aldrich, St. Louis, MO) was added to the cell culture at a final concentration of 1,000 U/ml, unless stated differently in Results and the figure legends. The cells were harvested at 30 min to 1 h or later (depending on the experiments) for further analysis.
Plasmids.
pEGFP-C1-STAT1, for STAT1-enhanced GFP (STAT1-eGFP) expression, was obtained from Addgene (36). The NSP1α sequences were amplified from cDNA of VR2385 and MLV, respectively, with primers 85NSP1F1 and 85NSP1R1, and the NSP1β sequence was amplified with primers 85NSP1F2 and 85NSP1R2 (Table 1), which contain restriction sites for EcoRI or XhoI to facilitate directional cloning. The two fragments were cloned separately into the pCMVTag2B vector. The resulting recombinant plasmids produced FLAG-tagged NSP1α and NSP1β. In each plasmid, cloning was confirmed by restriction enzyme digestion and DNA sequencing. NSP1β was also cloned into a red fluorescent protein (RFP) reporter vector to express NSP1β-RFP.
TABLE 1.
List of primers used for real-time PCR
| Primer namea | Primer sequence (5′ to 3′) |
|---|---|
| ISG15-F1 | CACCGTGTTCATGAATCTGC |
| ISG15-R1 | CTTTATTTCCGGCCCTTGAT |
| ISG56-F1 | CCTCCTTGGGTTCGTCTACA |
| ISG56-R1 | GGCTGATATCTGGGTGCCTA |
| Actin-F1 | ATCGTGCGTGACATTAAG |
| Actin-R1 | ATTGCCAATGGTGATGAC |
| sISG15-F1 | GGTGCAAAGCTTCAGAGACC |
| sISG15-R1 | GTCAGCCAGACCTCATAGGC |
| sIFI56-F1 | TCAGAGGTGAGAAGGCTGGT |
| sIFI56-R1 | GCTTCCTGCAAGTGTCCTTC |
| sRPL32-F1 | CTCTTCCAAGAACCGCAAAG |
| sRPL32-R1 | GCTGAGCCACAACTGGAACT |
| sCCL2-F1 | CACCAGCAGCAAGTGTCCTA |
| sCCL2-R1 | TCCAGGTGGCTTATGGAGTC |
| sCXCL10-F1 | TTCGCTGTACCTGCATCAAG |
| sCXCL10-R1 | CAACATGTGGGCAAGATTGA |
| sMX1-F1 | AGCGCAGTGACACCAGCGAC |
| sMX1-R1 | GCCCGGTTCAGCCTGGGAAC |
| sOAS2-F1 | CACAGCTCAGGGATTTCAGA |
| sOAS2-R1 | TCCAACGACAGGGTTTGTAA |
| sRNaseL-F1 | GCCAGACCTAGTGGCTTCTG |
| sRNaseL-R1 | AGAGGCCCAGAGAGTTGTGA |
| 85NSP1F1 | GCGAATTCTCTGGGATGCTTGATCG |
| 85NSP1R1 | CCGCTCGAGTTACATAGCACACTCAAAGG |
| 85NSP1F2 | GCGAATTCGCTACTGTCTATGACATTG |
| 85NSP1R2 | CCGCTCGAGTTAGCCGTACCACTTGTGAC |
F1 (or F2), forward primer; R1 (or R2), reverse primer. An “s” before a primer name indicates that the primer is for a porcine gene. An “85” before a primer name indicates that the primer is based on sequences of PRRSV VR2385.
Confocal fluorescence microscopy.
MARC-145 cells were seeded directly onto Lab-Tek chamber slides, cultured overnight, and transfected with STAT1-eGFP plasmid. At 4 h after transfection, the cells were infected with PRRSV VR2385 at an MOI of 1. IFN-α was added to the cells at a final concentration of 1,000 U/ml at 16 to 24 hpi. One hour after IFN-α treatment, the cells were fixed with 2% paraformaldehyde and mounted onto slides by using SlowFade Gold antifade reagent containing 4′6′-diamidino-2-phenylinodole (DAPI) (Invitrogen, Carlsbad, CA). The STAT1-eGFP distribution in the cells was visualized by confocal fluorescence microscopy.
To determine STAT1-eGFP nuclear translocation in HeLa cells after IFN treatment, the cells were cotransfected with STAT1-eGFP and NSP1β-RFP plasmids. At 24 h after transfection, the cells were treated with IFN-α at 300 U/ml for 1 h and fixed for confocal microscopy as described above.
Western blot analysis.
Cells were lysed in Laemmli sample buffer. The samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis as described previously (41). Briefly, the cell lysates were resolved in a 12% polyacrylamide gel. The separated proteins were then transferred onto a nitrocellulose membrane and probed with rabbit anti-STAT1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Specific reaction products were detected using goat anti-rabbit IgG conjugated with horseradish peroxidase (Sigma) and revealed using a chemiluminescence substrate. The chemiluminescence signal was recorded digitally by a ChemiDoc XRS imaging system (Bio-Rad Laboratories, Hercules, CA). β-Tubulin was detected on the same blot membrane to normalize protein loading. Digital signal acquisition and analysis were conducted using the Quantity One program, version 4.6 (Bio-Rad). The expression of other proteins was detected through corresponding antibodies against STAT2, histone H1, phospho-STAT2 (Tyr690, herein named STAT2-Y690) (Santa Cruz Biotechnology), and phospho-STAT1 (Tyr701, herein named STAT1-Y701) (Millipore, Billerica, MA). Antibodies against STAT1-Y701 and STAT2-Y690 were used to detect IFN-activated phosphorylation of STAT1 and STAT2 after IFN-α stimulation. Convalescent antiserum from a pig inoculated with PRRSV VR2385 was used to detect PRRSV proteins in the lysates of the virus-infected cells.
Subcellular fractionation.
The nuclear fraction was extracted from MARC-145 cells by using a CelLytic NuClear extraction kit (Sigma). The cells were inoculated with VR2385 at an MOI of 1 and, at 16 to 24 hpi, treated with IFN-α at 1,000 U/ml for 1 h. Cell collection, lysis, and subcellular fractionation were done following the manufacturer's instructions. The nuclear and cytoplasmic fractions were subjected to Western blotting. Antibodies against β-tubulin and histone H1 were used to assess the fractionation. Separation of cytoplasmic and nuclear fractions of HEK293 cells was done similarly.
IP.
MARC-145 cells were infected with VR2385 and treated with IFN-α as described above. The cells were lysed with lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with a protease inhibitor cocktail (Sigma). The lysate was clarified by centrifugation at 14,000 × g for 5 min at 4°C. Antibodies against STAT1 or STAT2 (Santa Cruz) were added to the supernatant. Immunoprecipitation (IP) with protein G agarose (KPL Inc., Gaithersburg, MD) was done following the manufacturer's instructions. The IP samples with antibody against STAT1 were subjected to Western blotting with STAT2-Y690 antibody. The IP samples with antibody against STAT2 were subjected to Western blotting with STAT1-Y701 antibody.
RNA isolation and real-time RT-PCR.
Total RNA was isolated from HEK293 and MARC-145 cells and PAMs with TRIzol reagent (Invitrogen) by following the manufacturer's instructions. Real-time PCR primers used in this study are listed in Table 1. Reverse transcription (RT) of RNA and real-time quantitative PCR were conducted as previously described (25, 26). Transcripts of ribosomal protein L32 (RPL32) or β-actin were also amplified from the samples of PAMs and MARC-145 or HEK293 cells, respectively, and used to normalize the total amount of input RNA. Relative transcript levels were quantified by the 2−ΔΔCT (threshold cycle) method (16) and are shown as relative fold change in comparison with the level for the mock-treated control.
Statistical analysis.
The significant differences in cellular RNA level between the groups of IFN-treated cells in the presence or absence of virus infection were assessed by Student's t test. A two-tailed P value of less than 0.05 was considered significant.
RESULTS
PRRSV interferes with IFN-α induction of ISG expression.
Type I IFN signaling leads to elevated expression of a variety of cellular genes, including ISG15 and ISG56 (8, 32, 33). To examine the effect of PRRSV replication on type I IFN signaling, we inoculated MARC-145 cells with PRRSV VR2385 and treated the cells with IFN-α at 16 to 24 hpi. Quantitative RT-PCR was conducted to assess the transcript levels of ISG15 and ISG56 1 h after IFN-α treatment. The transcript levels of ISG15 and ISG56 in IFN-treated cells increased 5.5- and 43.2-fold, respectively, in comparison with the levels in mock-treated cells (Fig. 1 A). In PRRSV-infected MARC-145 cells after IFN stimulation, the transcript levels of ISG15 and ISG56 were, respectively, 3- and 2.6-fold lower than those in the mock-infected cells (Fig. 1A). The expression levels of the two genes in PRRSV-infected cells without IFN-α treatment were similar to those in mock-inoculated cells, indicating that PRRSV infection did not affect the basal expression level of these genes.
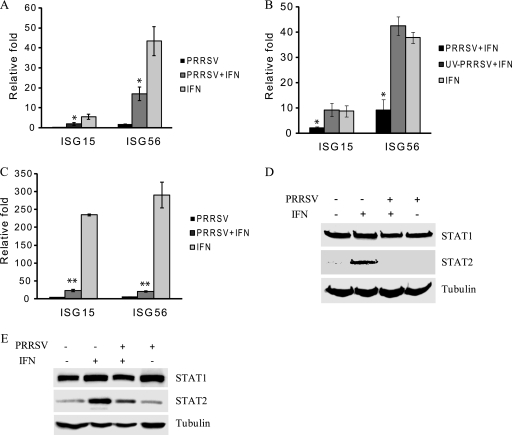
FIG. 1.
PRRSV inhibits expression of IFN-stimulated genes in MARC-145 cells. (A) Reduction of ISG15 and ISG56 transcripts, as detected by real-time RT-PCR. Cells were inoculated with PRRSV VR2385, incubated for 24 h, and then treated with IFN-α for 1 h. (B) UV-inactivated PRRSV has no effect on expression of ISG15 and ISG56, as detected by real-time RT-PCR. Cells were inoculated with PRRSV VR2385 or UV-inactivated VR2385, incubated for 24 h, and then treated with IFN-α for 1 h. (C) Reduction of ISG15 and ISG56 transcripts in PRRSV-infected cells 15 h after IFN treatment. Cells were inoculated with PRRSV VR2385, incubated for 24 h, and then treated with IFN-α for 15 h. Significant differences between the two groups for each transcript are denoted by a single asterisk and a double asterisk, which indicate P values of <0.05 and <0.01, respectively. Error bars represent standard deviations. (D) Inhibition of IFN-induced STAT2 protein expression by Western blotting. Cells were infected with PRRSV VR2385 or mock infected for 24 h and then treated with IFN-α for 8 h. The same blot was incubated with β-tubulin antibody as a protein loading control. (E) Western blotting of STAT1 and STAT2 from the cells 24 h after IFN-α treatment in the presence or absence of PRRSV infection.
To test whether viral replication is needed for the interference effect, we inactivated the PRRSV virions by UV illumination and verified the viral inactivation by the lack of viral replication 72 h after inoculation of the cells. When the UV-inactivated virus was used to inoculate the cells, the expression of the ISGs after IFN-α treatment was similar to that in the mock-inoculated cells. No difference in the transcript levels of the two ISGs was detected between the cells receiving mock inoculum and those receiving UV-inactivated PRRSV after the IFN-α treatment (Fig. 1B). This result indicates that active PRRSV replication was needed for the transcript reduction of the two ISGs after IFN stimulation.
When incubation of the cells was extended to 15 h after IFN-α treatment, the transcript levels of ISG15 and ISG56 increased 234- and 290-fold, respectively, in comparison with the levels in mock-treated cells (Fig. 1C). PRRSV-infected cells had significantly lower levels of expression of ISG15 and ISG56 (10- and 14-fold, respectively) after IFN treatment than did mock-infected cells. The transcript levels of STAT1 and STAT2 were also assessed, and no significant difference was noticed between cells with and without the IFN treatment (data not shown). Together, the data demonstrate that PRRSV infection of MARC-145 cells interferes with IFN stimulation of ISG15 and ISG56 expression.
Type I IFN signaling leads to an elevation in expression of a variety of proteins encoded by IFN-responsive genes, including STAT2 (8, 32, 33). MARC-145 cells, in the presence or absence of PRRSV infection, were treated with IFN-α for 8 h and harvested for Western blotting. After IFN-α stimulation, the STAT2 protein level in the IFN-treated cells increased considerably (Fig. 1D), which is consistent with STAT2 function as an ISG. However, STAT2 protein in VR2385-infected cells after IFN-α treatment remained at a low basal level similar to that for mock-treated MARC-145 cells. The levels of STAT1 in all samples were similar, possibly because of the high basal level of STAT1 protein in the cytoplasm. When the IFN-treated cells were incubated for 24 h, the STAT2 protein level in mock-infected cells was much higher than that in the VR2385-infected cells (Fig. 1E). The STAT2 protein level in VR2385-infected cells was similar to that in mock-infected cells in the absence of IFN-α treatment. The STAT1 level in mock-infected cells was slightly increased at 24 h after IFN-α stimulation. A slight increase was also noticed in VR2385-infected cells without addition of IFN (Fig. 1E), which might be caused by sample loading, shown by a higher intensity of the β-tubulin band (Fig. 1E, last lane). These results indicate that PRRSV infection interfered with IFN-α signaling and, thus, resulted in the reduction of downstream gene expression.
An immunofluorescence assay with PRRSV N-specific monoclonal antibody was conducted to examine the percentage of PRRSV-infected cells at 24 hpi. Positive staining was observed in over 95% of cells by confocal microscopy (data not shown). The high percentage of PRRSV-infected cells indicated that the interference of IFN-induced STAT2 protein expression was due to PRRSV infection.
PRRSV does not alter the IFN-induced phosphorylation of STAT1 and STAT2.
STAT1 and STAT2 are key players in the IFN-α-activated JAK/STAT signaling pathway (8, 32, 33). Phosphorylation of STAT1 and STAT2 is an early step in the pathway after type I IFNs bind to their receptors. To determine if PRRSV interferes with the IFN-induced activation of these two proteins, we tested the phosphorylation status of STAT1 and STAT2 in MARC-145 cells 1 h after IFN-α treatment. The levels of phosphorylated STAT1 at tyrosine 701 (STAT1-Y701) and STAT2 at tyrosine 690 (STAT2-Y690) were greatly increased after IFN-α treatment (Fig. 2 A). No difference was observed between the IFN-treated cells with and without PRRSV infection. In mock-treated cells, the phosphorylation of these two proteins was below detection level. This result showed that PRRSV replication did not change STAT1 and STAT2 phosphorylation status after IFN-α stimulation in comparison with that for mock-infected cells. To make sure that the PRRSV proteins were similarly expressed in the PRRSV-infected cells with and without IFN-α treatment, the cell lysates were subjected to Western blotting with convalescent pig antiserum. Both lanes showed similar band patterns and intensities of the PRRSV proteins (Fig. 2A).
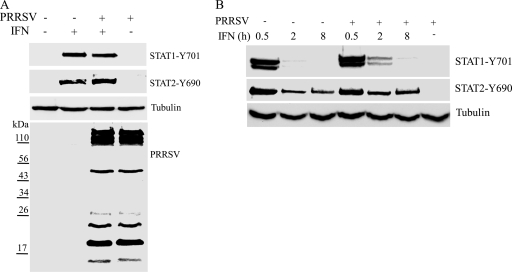
FIG. 2.
Phosphorylation status of STAT1 and STAT2 in PRRSV-infected MARC-145 cells after IFN-α stimulation. (A) Western blotting with antibodies against STAT1-Y701 and STAT2-Y690. Cells were infected with PRRSV VR2385 or mock infected and, at 24 hpi, treated with IFN-α for 1 h. The same blot was incubated with β-tubulin antibody as a protein loading control. Convalescent pig antiserum was used to blot the membrane to show the PRRSV proteins in the lysates of PRRSV-infected cells. Positions of prestained molecular mass markers are shown on the left. (B) Western blotting with antibodies against STAT1-Y701 and STAT2-Y690 in VR2385-infected or mock-infected cells 0.5, 2, and 8 h after IFN-α treatment.
To further confirm that PRRSV replication has no effect on the IFN-α-activated phosphorylation of these two proteins, we harvested cells at 0.5, 2, and 8 h after IFN-α addition. The levels of STAT1-Y701 and STAT2-Y690 were highest 0.5 h after IFN addition (Fig. 2B). The level of STAT1-Y701 in cells 8 h after IFN-α addition decreased substantially to below detection level. At all time points, the levels of STAT1-Y701 and STAT2-Y690 in the PRRSV-infected MARC-145 cells were similar to those in mock-infected cells after IFN-α treatment (Fig. 2B). In PRRSV-infected and mock-treated cells, STAT1-Y701 and STAT2-Y690 were below detection level. Therefore, we conclude that PRRSV infection does not affect the IFN-α-induced phosphorylation of STAT1 and STAT2.
STAT1/STAT2/IRF9 heterotrimer formation is not altered.
The IFN-induced activation of STAT1 and STAT2 results in the formation of STAT1/STAT2 heterodimers that further associate with IRF9 to form the mature ISG factor 3 (ISGF3) (8, 32, 33). Since the IFN-induced phosphorylation of STAT1 and STAT2 was not significantly changed in PRRSV-infected cells, we further analyzed the ISGF3 complex in MARC-145 cells by immunoprecipitation followed by Western blotting. IP with STAT1 antibody and then blotting with antibody against STAT2-Y690 showed the presence of phosphorylated STAT2 in the samples from the IFN-treated cells regardless of PRRSV infection (Fig. 3). Similarly, IP with STAT2 antibody and then blotting with antibody against STAT1-Y701 showed the presence of phosphorylated STAT1 in the two IFN-treated samples. In contrast, no specific signal was detected in samples from cells without IFN stimulation. This result indicated that the ISGF3 heterotrimer formation after IFN treatment in VR2385-infected cells is not significantly affected.
FIG. 3.
IP detection of STAT1/STAT2 heterodimer formation in MARC-145 cells after IFN-α treatment. Cells were infected with PRRSV VR2385 or mock infected and, at 24 hpi, treated with IFN-α for 1 h. WB, Western blot.
PRRSV interferes with ISGF3 nuclear translocation.
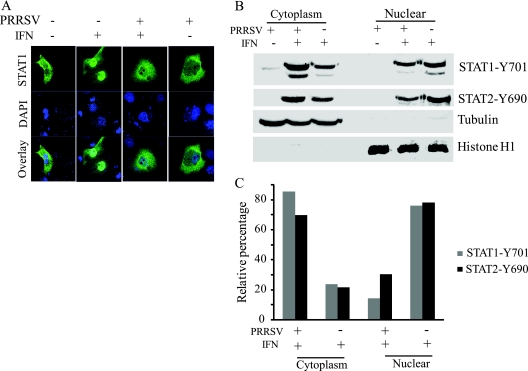
The ISGF3 complex translocates to the nucleus to initiate gene transcription by binding to interferon-stimulated response elements (ISREs). To examine the translocation step of the JAK/STAT signaling pathway in PRRSV-infected cells, we transfected MARC-145 cells with pEGFP-C1-STAT1 and subsequently infected the cells with VR2385. At 24 hpi, the cells were treated with IFN-α for 1 h, fixed, and mounted for confocal microscopy. In the IFN-treated cells without PRRSV infection, the major portion of STAT1-eGFP was translocated to the nucleus (Fig. 4 A). However, the majority of the STAT1-eGFP protein remained in the cytoplasm of PRRSV-infected cells after IFN-α treatment. This result indicated that VR2385 inhibits STAT1 nuclear translocation.
FIG. 4.
Blockage of nuclear translocation of ISGF3 heterotrimers in PRRSV-infected MARC-145 cells. (A) PRRSV inhibits nuclear translocation of STAT1-eGFP, as observed by confocal microscopy. Cells were transiently transfected with STAT1-eGFP plasmid and inoculated with VR2385 4 h later. Cells were treated with IFN-α at 24 hpi and fixed 1 h later. (B) Phosphorylated STAT1 and STAT2 in nuclear and cytoplasmic fractions. Subcellular fractionation of the cells 1 h after IFN treatment and Western blotting was determined. The same blot was incubated with antibodies against β-tubulin and histone H1 as controls for loading and fractionation. (C) Densitometry analysis of the digital image shown in panel B. The band intensity of each fraction is shown as the relative percentage of the sum density of corresponding cytoplasmic and nuclear fractions from the same treatment. Normalization for cytoplasmic and nuclear fractions was done with tubulin and histone H1, respectively.
To confirm this observation, we conducted nuclear and cytoplasmic fractionation of the cells after IFN treatment. Antibodies against STAT1-Y701 and STAT2-Y690 were used to detect the presence of the phosphorylated proteins in the two fractions. After IFN-α treatment of mock-infected cells, more STAT1-Y701 and STAT2-Y690 were found in the nuclear than in the cytoplasmic fraction (Fig. 4B), as expected. In contrast, in PRRSV-infected cells after IFN stimulation, more STAT1-Y701 and STAT2-Y690 were detected in the cytoplasmic than in the nuclear fraction. The absence of β-tubulin in the nuclear fraction and histone H1 in the cytoplasmic fraction verified a successful subcellular fractionation. Densitometry analysis of the digital images of the blotting results showed that 76% of STAT1-Y701 and 78% of STAT2-Y690 were detected in the nuclear fraction of mock-infected cells, while only 14% and 30% of the two proteins, respectively, were detected in the nuclear fraction of VR2385-infected cells (Fig. 4C). The remaining portions of the phosphorylated proteins remained in the cytoplasmic fractions. The fractionation result was consistent with the observation from confocal microscopy and indicated that PRRSV infection strongly blocks nuclear translocation of the ISGF3 (STAT1/STAT2/IRF9) complex.
NSP1β inhibits the IFN-induced expression of ISGs by blocking nuclear translocation of STAT1.
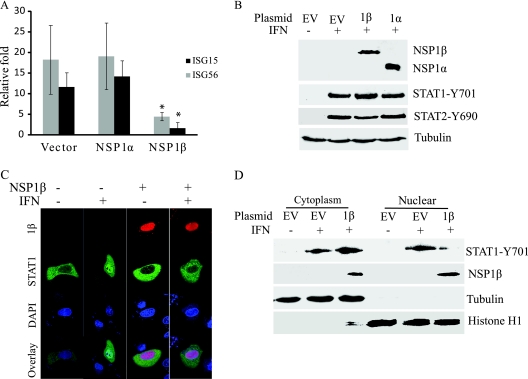
Since VR2385 inhibits type I IFN signaling, we wished to determine which PRRSV protein is responsible for the effect. NSP1α and NSP1β were selected for the analysis, as recent studies indicate their roles in the IFN pathway (3, 6). To determine if NSP1α or NSP1β of VR2385 can inhibit IFN signaling, we cloned NSP1α and NSP1β into a pCMVTag2B vector, separately, and transfected HEK293 cells. The expression of ISG15 and ISG56 in HEK293 cells 12 h after IFN-α stimulation was determined by real-time PCR. Results showed that the cells with NSP1β expression had 4- and 7-fold-lower ISG15 and ISG56 transcript levels, respectively, than cells transfected with an empty vector (Fig. 5 A). The levels of ISG15 and ISG56 transcripts in the cells with NSP1α expression were similar to those in the cells transfected with an empty vector. These results indicate that NSP1β inhibited ISG expression in the cells.
FIG. 5.
PRRSV NSP1β protein inhibits IFN signaling. (A) NSP1β inhibits expression of ISG15 and ISG56 in HEK293 cells. Cells were transiently transfected with NSP1α and NSP1β plasmids or an empty vector and, 48 h after transfection, treated with IFN-α at 300 U/ml. The cells were harvested 12 h after IFN treatment. Significant differences in ISG15 and ISG56 transcript levels between the two groups of NSP1β and empty vector are denoted by an asterisk, which indicates a P value of <0.05. (B) NSP1α and NSP1β have no effect on IFN-induced phosphorylation of STAT1 in HEK293 cells. Cells were harvested for STAT1-Y701 detection 1 h after IFN treatment. (C) NSP1β inhibits nuclear translocation of STAT1-eGFP in HeLa cells, as observed by confocal microscopy. Cells were transiently transfected with STAT1-eGFP and NSP1β-RFP plasmids. At 24 h after transfection, the cells were treated with IFN-α at 300 U/ml for 1 h. (D) NSP1β inhibits STAT1 nuclear translocation. HEK293 cells were transiently transfected with NSP1β plasmid or an empty vector and, 48 h after transfection, treated with IFN-α at 300 U/ml for 1 h. Subcellular fractionation of the cells and Western blotting were conducted to detect phosphorylated STAT1 in nuclear and cytoplasmic fractions. The same blot was incubated with antibodies against NSP1β, β-tubulin, and histone H1 as controls for loading and fractionation. EV, empty vector; 1α, NSP1α; 1β, NSP1β.
To determine the mechanism of the NSP1β inhibition of IFN signaling, we analyzed the phosphorylation of STAT1 and STAT2. After IFN-α stimulation, the HEK293 cells with NSP1α or NSP1β expression had levels of STAT1-Y701 and STAT2-Y690 similar to those in cells transfected with an empty vector (Fig. 5B), which indicated that neither protein has an effect on IFN-activated phosphorylation of STAT1.
Since NSP1β does not affect the IFN-stimulated phosphorylation of STAT1, we speculated that it might interfere with STAT1 nuclear translocation, as with VR2385 in MARC-145 cells. To test this speculation, we transfected HeLa cells with STAT1-eGFP and NSP1β-RFP plasmids. HeLa cells were used in this experiment, as they attach to cover glass better than HEK293 cells. At 24 h after transfection, the cells were treated with IFN-α for 1 h and observed under confocal microscopy. In cells expressing both STAT1-eGFP and NSP1β-RFP, the majority of STAT1 remained in the cytoplasm (Fig. 5C), indicating that NSP1β inhibits STAT1 nuclear translocation.
To confirm this observation, we conducted subcellular fractionation of HEK293 cells to determine the distribution of phosphorylated STAT1. In HEK293 cells with NSP1β expression, the majority of STAT1-Y701 remained in the cytoplasm 1 h after IFN stimulation, while cells transfected with the empty vector had the majority of STAT1-Y701 in the nucleus (Fig. 5D). NSP1β was detected in both cytoplasmic and nuclear fractions. These results indicate that NSP1β blocks the IFN-stimulated nuclear translocation of ISGF3.
In PAMs, VR2385 interferes with IFN-α signaling similarly to that observed in MARC-145 cells.
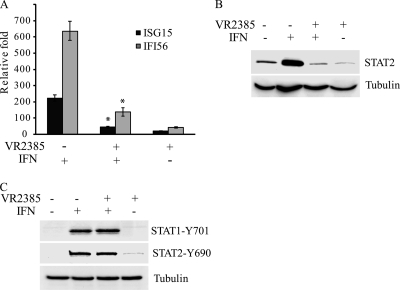
Since PAMs are the major targets for PRRSV infection in pigs, PRRSV infection of PAMs was conducted for greater physiological relevance to the viral infection in its natural host. We tested whether VR2385 has a negative effect on IFN signaling in PAMs, as observed in MARC-145 cells. Primary PAMs were infected with VR2385 at an MOI of 0.05 for 15 h and then treated with IFN-α for 8 h. Quantitative RT-PCR showed that the IFN treatment of mock-infected cells increased transcript levels of ISG15 and IFI56 (the porcine gene equivalent to ISG56) 223- and 637-fold, respectively (Fig. 6 A). Upon IFN-α stimulation, the VR2385-infected PAMs had significantly lower levels (5.1- and 4.6-fold, respectively) of ISG15 and IFI56 transcripts than did mock-infected cells. We concluded that PRRSV VR2385 interferes with the expression of IFN-induced genes in PAMs after IFN-α stimulation.
FIG. 6.
VR2385 interferes with IFN signaling in PAMs. (A) Real-time RT-PCR detection of ISG15 and IFI56 from PAMs 8 h after IFN-α treatment in the presence or absence of VR2385 infection. Significant differences between the two IFN-treated groups are denoted by an asterisk, which indicates a P value of <0.05. (B) Western blotting of STAT2 from PAMs 8 h after IFN-α treatment in the presence or absence of VR2385 infection. (C) Western blotting with the antibodies against phosphorylated STAT1 and STAT2 from PAMs 1 h after IFN-α treatment in the presence or absence of VR2385 infection.
The STAT2 protein level in the PAMs after PRRSV infection and IFN treatment was also assessed. Similarly to the results with MARC-145 cells, the STAT2 protein level in mock-infected PAMs after IFN-α stimulation increased significantly (Fig. 6B). The STAT2 protein in VR2385-infected PAMs after IFN treatment remained at the basal level, similar to that of VR2385-infected cells without addition of IFN. This result indicated that VR2385 blocks the IFN-induced STAT2 elevation in PAMs. VR2385 infection had no detectable effect on the basal level of STAT2 in PAMs without external IFN.
To assess the activation of the JAK/STAT signaling pathway in PAMs after IFN treatment, we detected phosphorylated STAT1 and STAT2. The levels of STAT1-Y701 and STAT2-Y690 in PAMs with VR2385 infection were similar to those in mock-infected PAMs (Fig. 6C). This result suggests that VR2385 does not alter the phosphorylation of either STAT1 or STAT2 in PAMs receiving IFN treatment.
Effect of the low-virulence PRRSV vaccine strain on type I IFN signaling.
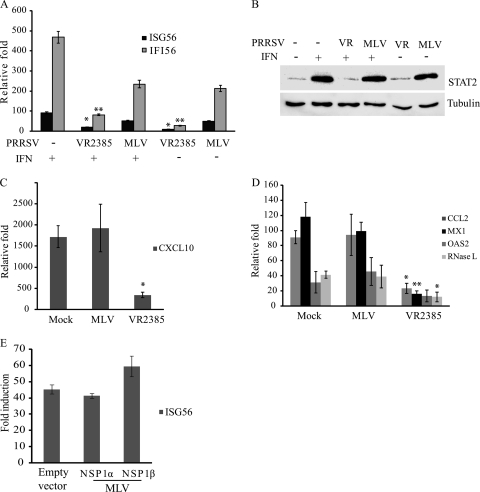
Ingelvac PRRS MLV is a licensed low-virulence vaccine strain. We speculated that MLV had a lower inhibitory effect on IFN signaling than the virulent VR2385 strain. PAMs were infected with MLV and then treated with IFN-α as described above for the VR2385 experiment. The transcript levels of ISG15 and IFI56 in the MLV-infected PAMs receiving IFN treatment increased 50- and 234-fold, respectively (Fig. 7 A), which were slightly lower than, but had no significant difference from, those in mock-infected PAMs after IFN stimulation. Interestingly, the MLV infection of PAMs receiving no external IFN resulted in increases in ISG15 and IFI56 transcripts of 49- and 211-fold, respectively, which were similar to levels in MLV-infected PAMs receiving external IFN stimulation. This result indicates that MLV had no effect on IFN signaling in PAMs.
FIG. 7.
Effect of PRRSV MLV on IFN signaling in PAMs. (A) Real-time RT-PCR detection of ISG15 and IFI56 from PAMs 8 h after IFN-α treatment in the presence or absence of PRRSV infection. Significant differences in the transcript levels between the IFN-treated groups and the nontreated groups are denoted by a single asterisk and a double asterisk, which indicate P values of <0.05 and <0.01, respectively. (B) Western blotting of STAT2 in PAMs 8 h after IFN-α treatment in the presence or absence of PRRSV infection. Samples of mock-treated PAMs were included as controls. VR, VR2385. (C) Real-time RT-PCR detection of CXCL10 transcript in PAMs 8 h after IFN-α treatment in the presence or absence of PRRSV infection. Significant differences between VR2385-infected and mock-infected cells are denoted by an asterisk, which indicates a P value of <0.05. (D) Real-time RT-PCR detection of CCL2, MX1, OAS2, and RNase L transcripts in PAMs 8 h after IFN-α treatment in the presence or absence of PRRSV infection. Significant differences between virus-infected and mock-infected cells are denoted by a single asterisk and a double asterisk, which indicate P values of <0.05 and <0.01, respectively. (E) Real-time RT-PCR detection of ISG56 from HEK293 cells transfected with MLV NSP1α and NSP1β plasmids or empty vector pCMVTag2B. At 48 h after the transfection, the cells were treated with IFN-α at 300 U/ml for 12 h. No significant difference in ISG56 transcript level between the samples was detected.
The viral yields of MLV and VR2385 in the PAMs were also determined by real-time RT-PCR to ensure viral infection of the cells. MLV and VR2385 had viral genomic RNA copy numbers of 6 and 7 log10/ml, respectively, in cell culture supernatant, which suggested that VR2385 had higher viral replication than MLV. This result was consistent with the ISG expression data, which showed high-level ISG15 and IFI56 transcripts in MLV-infected PAMs. The ISG expression might correlate with antiviral responses and result in reduction of MLV replication.
To determine the effect of MLV on IFN-activated STAT2 expression, we detected STAT2 protein levels by Western blotting. Similarly to results for mock-infected cells receiving IFN treatment, the MLV-infected PAMs receiving IFN treatment had an increased level of STAT2 protein (Fig. 7B). A sample of VR2385-infected PAMs was included as a control and had a low STAT2 level. It is interesting to note that the addition of the external IFN to MLV-infected PAMs did not lead to a change in the STAT2 protein level. This result was consistent with the real-time PCR data showing increased transcript levels of ISG15 and IFI56 in the MLV-infected PAMs without the addition of IFN. These results indicated that MLV infection activates IFN signaling in PAMs in the absence of external IFN and that the addition of IFN has no additional effect on the expression of IFN-induced genes in MLV-infected PAMs.
To further examine the difference in IFN signaling between PAMs receiving MLV and those receiving VR2385, we determined transcript levels of IFN-induced chemokine ligand 10 (CXCL10/IP10) (35), chemokine ligand 2 (CCL2, also known as monocyte chemotactic protein 1 [MCP-1]) (30), and three antiviral genes: the myxovirus (influenza virus) resistance 1 (MX1) (11), 2′-5′-oligoadenylate synthetase 2 (OAS2) (42), and RNase L (4) genes. Results showed that, after IFN stimulation, PAMs with VR2385 infection had a significant 5-fold decrease in CXCL10 expression compared to the level in mock-infected cells, while cells with MLV infection had a level of CXCL10 similar to that in mock-infected cells (Fig. 7C). The transcript levels of CCL2, MX1, OAS2, and RNase L in PAMs in the presence of VR2385 infection were 3.9-, 7.3-, 2.4-, and 3.4-fold lower, respectively, than those in mock-infected cells (Fig. 7D). MLV-infected PAMs had levels of these transcripts similar to those in mock-infected cells. These results are consistent with our data described above, indicating that the virulent strain VR2385 can evade IFN-activated antiviral responses.
As VR2385 NSP1β inhibits IFN signaling, we wondered whether MLV NSP1β has any effect. NSP1α and NSP1β of MLV were cloned into the pCMVTag2B vector, and expression of these two proteins from the recombinant plasmids was confirmed by Western blotting. HEK293 cells were transfected with the recombinant plasmids and treated with IFN-α. The transcript level of ISG56 in the cells expressing either NSP1α or NSP1β of MLV was similar to that in the cells transfected with empty vector (Fig. 7E). This result indicated that MLV NSP1α and NSP1β had no effect on IFN signaling and was consistent with the data described above, showing that MLV infection of PAMs has no effect on IFN signaling.
DISCUSSION
Dual infection of pigs with PRRSV and porcine respiratory coronavirus (PRCV) or swine influenza virus (SIV) causes more severe respiratory disease and growth retardation than PRRSV infection alone (38). Both PRCV and SIV infections can induce a high level of bioactive IFN-α (37). How can PRRSV, which is sensitive to pretreatment with IFNs, replicate in pigs coinfected with PRCV or SIV? Our study provides a clue that PRRSV can interfere with type I IFN signaling. We found that PRRSV interferes with IFN-α signaling in MARC-145 cells and PAMs via blocking nuclear translocation of the ISGF3 heterotrimers.
To determine the mechanism of the PRRSV interference of IFN signaling, several experiments were conducted to analyze the JAK/STAT signaling pathway. First, the IFN-induced phosphorylation status of STAT1 and STAT2 was analyzed. We found that PRRSV does not affect the IFN-induced phosphorylation status of STAT1 or STAT2. After IFN treatment, both STAT1 and STAT2 undergo phosphorylation, form heterotrimers with IRF9, translocate to the nucleus, and then undertake dephosphorylation before being redistributed back to the cytoplasm (2, 32). The dephosphorylation rate of STAT2 in MARC-145 cells was lower than that of STAT1, as shown in Fig. 2B.
Second, ISGF3 heterotrimers were assessed by IP and Western blotting. We concluded that PRRSV infection does not alter the ISGF3 heterotrimer formation in MARC-145 cells receiving IFN treatment. As shown in Fig. 3, the band in the lane for PRRSV-infected cells receiving IFN stimulation was weaker than that for mock-infected cells after IFN treatment. This may indicate that there were more proteins in the latter cell lysate than in the former. This assay was designed to show the presence of the ISGF3 complex but not to serve as a quantitative assessment. We were unable to identify IRF9 in the IP pellets perhaps because IRF9 is a 48-kDa protein, which is located very close to the heavy-chain immunoglobulin G band in our Western blot analysis, possibly masking the view of weak bands nearby. However, our data provide further evidence that VR2385 replication interferes with ISGF3 nuclear translocation.
Lastly, ISGF3 heterotrimer nuclear translocation was analyzed. We found that VR2385 infection blocks nuclear translocation of ISGF3 heterotrimers. The nuclear translocation of STAT1-eGFP after IFN treatment was blocked in VR2385-infected cells. The results of our subcellular fractionation assay suggest that interference of ISGF3 nuclear translocation leads to inhibition of IFN-α signaling in PRRSV-infected cells. The data in Fig. 4 showed a small portion of phosphorylated STAT1 and STAT2 present in the nuclei of VR2385-infected cells, which is consistent with the small-scale elevation of ISG15 and ISG56 transcripts in the cells after IFN-α stimulation.
To identify which of the PRRSV proteins is responsible for blocking the ISGF3 nuclear translocation, we cloned NSP1α and NSP1β and expressed them in HeLa and HEK293 cells. Results from confocal microscopy and the subcellular fractionation assay clearly showed that NSP1β blocks STAT1 nuclear translocation, which is consistent with a recent publication (6) showing that overexpression of NSP1β in HEK293T cells blocks STAT1-GFP nuclear translocation. It was also shown in the previous study that NSP1β inhibits IFN-activated phosphorylation of STAT1. In contrast, our results demonstrated that NSP1β does not affect IFN-induced phosphorylation of STAT1 and STAT2, which is consistent with our data that PRRSV infection of MARC-145 cells and PAMs does not affect the IFN-activated phosphorylation of these two proteins. This discrepancy might be caused by the different virus strains and IFN subtypes used but still needs further investigation. The exact mechanism of NSP1β inhibition of STAT1 nuclear translocation is being studied.
After finding that PRRSV infection of MARC-145 cells interfered with IFN-α signaling, we showed that VR2385 inhibited IFN signaling in primary PAMs. PAMs are key sentinel cells in the respiratory system and the primary target cells for PRRSV infection in vivo. Our finding that VR2385 inhibited type I IFN signaling in PAMs has physiological relevance to PRRSV infection of pigs.
We further investigated PRRSV interference of IFN signaling in PAMs. IFN treatment of MLV-infected PAMs does not affect the expression of ISG15 and IFI56 transcripts or STAT2 protein. Possible reasons are that MLV has no effect on IFN signaling or that the cells were less responsive to an external IFN due to priming from MLV-induced endogenous IFN. Another interesting observation was that, in the absence of external IFN addition, the transcript levels of ISG15 and IFI56 and the level of STAT2 protein in the MLV-infected PAMs increased significantly in comparison with those in mock-infected cells. We postulate that (i) MLV has no interference in type I IFN production or (ii) MLV does not inhibit the positive feedback loop for induction of IFN. Our results are more consistent with the latter speculation, since a slight increase in ISG expression was detected in VR2385-infected cells. The MLV-mediated upregulation of IFN-stimulated genes is consistent with the low-virulence nature of this strain. Real-time PCR data further demonstrated that VR2385 inhibited the expression of IFN-inducible genes in PAMs, including CXCL10, CCL2, MX1, OAS2, and RNase L, while MLV-infected PAMs had levels similar to those in mock-infected cells. Reduction of expression of the chemokines and antiviral genes in VR2385-infected cells is consistent with the virulent nature of this strain. The NSP1β proteins of these two PRRSV strains function differently. MLV NSP1β has no effect on IFN signaling, while VR2385 NSP1β inhibits IFN-activated antiviral responses.
The viral interference of type I IFN signaling can be beneficial for viral replication and serves as an important mechanism for the virus to evade the host innate immune response. Severe acute respiratory syndrome (SARS) coronavirus, which belongs to the same order (Nidovirales) as PRRSV, infects macrophages and interferes with the host innate immune response. Both the ORF3b and ORF6 proteins of SARS virus inhibit the synthesis and signaling of type I IFNs (14), while the ORF6 protein alone blocks STAT1 nuclear translocation via sequestration of nuclear import factor karyoperin α2 (KPNA2) (9). We transfected HeLa and HEK293 cells with NSP1β and the FLAG-tagged KPNA1, KPNA2, KPNA3, or KPNA4 plasmid. No change in the nuclear localization of the KPNAs was observed, and no interaction between NSP1β and any of the KPNAs was detected. It seems that PRRSV employs a different mechanism to block the ISGF3 nuclear translocation. It is known that viruses use multiple independent mechanisms to inhibit the IFN response. For example, paramyxovirus V proteins bind to STAT2 and block ISGF3 nuclear accumulation (27, 29), Ebola virus VP24 binds to a nuclear localization signal receptor for phosphorylated STAT1 and blocks STAT1 nuclear accumulation (28), and rotavirus antagonizes the IFN response by inhibiting nuclear translocation of STAT1 and STAT2 (10).
In summary, PRRSV VR2385 inhibits IFN-α signaling in MARC-145 cells and primary PAMs by interfering with ISGF3 nuclear translocation. MLV infection of PAMs can activate IFN signaling without the addition of external IFN. The variable effect on IFN induction might be a factor contributing to the different levels of viral pathogenesis between the two PRRSV strains and have a biological relevance in PRRS vaccine design or improvement.
Acknowledgments
We are grateful to Joseph F. Urban from the Human Nutrition Research Center, USDA, Beltsville, MD, for his gift of the lung lavage fluid from piglets. We thank Kay S. Faaberg from the National Animal Disease Center, Ames, IA, for her gift of Ingelvac PRRS MLV.
M. Shen was partially supported by the Shandong Bureau of Education. This project was supported by institutional funds from the University of Maryland.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Albina, E., C. Carrat, and B. Charley. 1998. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 18:485-490. [DOI] [PubMed] [Google Scholar]
- 2.Banninger, G., and N. C. Reich. 2004. STAT2 nuclear trafficking. J. Biol. Chem. 279:39199-39206. [DOI] [PubMed] [Google Scholar]
- 3.Beura, L. K., S. N. Sarkar, B. Kwon, S. Subramaniam, C. Jones, A. K. Pattnaik, and F. A. Osorio. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 84:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisbal, C., C. Martinand, M. Silhol, B. Lebleu, and T. Salehzada. 1995. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 270:13308-13317. [DOI] [PubMed] [Google Scholar]
- 5.Buddaert, W., K. Van Reeth, and M. Pensaert. 1998. In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV). Adv. Exp. Med. Biol. 440:461-467. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z., S. Lawson, Z. Sun, X. Zhou, X. Guan, J. Christopher-Hennings, E. A. Nelson, and Y. Fang. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conzelmann, K. K., N. Visser, P. Van Woensel, and H. J. Thiel. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology 193:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 9.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holloway, G., T. T. Truong, and B. S. Coulson. 2009. Rotavirus antagonizes cellular antiviral responses by inhibiting the nuclear accumulation of STAT1, STAT2, and NF-kappaB. J. Virol. 83:4942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hovnanian, A., D. Rebouillat, M. G. Mattei, E. R. Levy, I. Marie, A. P. Monaco, and A. G. Hovanessian. 1998. The human 2′,5′-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics 52:267-277. [DOI] [PubMed] [Google Scholar]
- 12.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 13.Kim, O., Y. Sun, F. W. Lai, C. Song, and D. Yoo. 2010. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology 402:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labarque, G. G., H. J. Nauwynck, K. Van Reeth, and M. B. Pensaert. 2000. Effect of cellular changes and onset of humoral immunity on the replication of porcine reproductive and respiratory syndrome virus in the lungs of pigs. J. Gen. Virol. 81:1327-1334. [DOI] [PubMed] [Google Scholar]
- 16.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 17.Luo, R., S. Xiao, Y. Jiang, H. Jin, D. Wang, M. Liu, H. Chen, and L. Fang. 2008. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Mol. Immunol. 45:2839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mardassi, H., S. Mounir, and S. Dea. 1995. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Quebec reference strain. Arch. Virol. 140:1405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X. J., P. S. Paul, P. G. Halbur, and M. A. Lum. 1996. Characterization of a high-virulence US isolate of porcine reproductive and respiratory syndrome virus in a continuous cell line, ATCC CRL11171. J. Vet. Diagn. Invest. 8:374-381. [DOI] [PubMed] [Google Scholar]
- 20.Meulenberg, J. J. 2000. PRRSV, the virus. Vet. Res. 31:11-21. [DOI] [PubMed] [Google Scholar]
- 21.Meulenberg, J. J., A. Petersen-Den Besten, E. P. De Kluyver, R. J. Moormann, W. M. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meulenberg, J. J. M., E. J. de Meijer, and R. J. M. Moormann. 1993. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J. Gen. Virol. 74:1697-1701. [DOI] [PubMed] [Google Scholar]
- 23.Miller, L. C., W. W. Laegreid, J. L. Bono, C. G. Chitko-McKown, and J. M. Fox. 2004. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 149:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227:385-392. [DOI] [PubMed] [Google Scholar]
- 25.Patel, D., T. Opriessnig, D. A. Stein, P. G. Halbur, X. J. Meng, P. L. Iversen, and Y. J. Zhang. 2008. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral Res. 77:95-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel, D., D. A. Stein, and Y. J. Zhang. 2009. Morpholino oligomer-mediated protection of porcine pulmonary alveolar macrophages from arterivirus-induced cell death. Antivir. Ther. 14:899-909. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran, A., J. P. Parisien, and C. M. Horvath. 2008. STAT2 is a primary target for measles virus V protein-mediated alpha/beta interferon signaling inhibition. J. Virol. 82:8330-8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rollins, B. J., T. Yoshimura, E. J. Leonard, and J. S. Pober. 1990. Cytokine-activated human endothelial cells synthesize and secrete a monocyte chemoattractant, MCP-1/JE. Am. J. Pathol. 136:1229-1233. [PMC free article] [PubMed] [Google Scholar]
- 31.Rossow, K. D., J. E. Collins, S. M. Goyal, E. A. Nelson, J. C. Hennings, and D. A. Benfield. 1995. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet. Pathol. 32:361-373. [DOI] [PubMed] [Google Scholar]
- 32.Schindler, C., and J. E. Darnell, Jr. 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621-651. [DOI] [PubMed] [Google Scholar]
- 33.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 34.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 35.Taub, D. D., A. R. Lloyd, K. Conlon, J. M. Wang, J. R. Ortaldo, A. Harada, K. Matsushima, D. J. Kelvin, and J. J. Oppenheim. 1993. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J. Exp. Med. 177:1809-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timofeeva, O. A., S. Plisov, A. A. Evseev, S. Peng, M. Jose-Kampfner, H. N. Lovvorn, J. S. Dome, and A. O. Perantoni. 2006. Serine-phosphorylated STAT1 is a prosurvival factor in Wilms' tumor pathogenesis. Oncogene 25:7555-7564. [DOI] [PubMed] [Google Scholar]
- 37.Van Reeth, K., G. Labarque, H. Nauwynck, and M. Pensaert. 1999. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res. Vet. Sci. 67:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Reeth, K., H. Nauwynck, and M. Pensaert. 1996. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet. Microbiol. 48:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, Z., L. Batista, S. Dee, P. Halbur, and M. P. Murtaugh. 2004. The level of virus-specific T-cell and macrophage recruitment in porcine reproductive and respiratory syndrome virus infection in pigs is independent of virus load. J. Virol. 78:5923-5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y. J., D. A. Stein, S. M. Fan, K. Y. Wang, A. D. Kroeker, X. J. Meng, P. L. Iversen, and D. O. Matson. 2006. Suppression of porcine reproductive and respiratory syndrome virus replication by morpholino antisense oligomers. Vet. Microbiol. 117:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y. J., K. Y. Wang, D. A. Stein, D. Patel, R. Watkins, H. M. Moulton, P. L. Iversen, and D. O. Matson. 2007. Inhibition of replication and transcription activator and latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus by morpholino oligomers. Antiviral Res. 73:12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zurcher, T., J. Pavlovic, and P. Staeheli. 1992. Nuclear localization of mouse Mx1 protein is necessary for inhibition of influenza virus. J. Virol. 66:5059-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]