Abstract
The V protein of the paramyxovirus subfamily Paramyxovirinae is an important virulence factor that can interfere with host innate immunity by inactivating the cytosolic pathogen recognition receptor MDA5. This interference is a result of a protein-protein interaction between the highly conserved carboxyl-terminal domain of the V protein and the helicase domain of MDA5. The V protein C-terminal domain (CTD) is an evolutionarily conserved 49- to 68-amino-acid region that coordinates two zinc atoms per protein chain. Site-directed mutagenesis of conserved residues in the V protein CTD has revealed both universal and virus-specific requirements for zinc coordination in MDA5 engagement and has also identified other conserved residues as critical for MDA5 interaction and interference. Mutation of these residues produces V proteins that are specifically defective for MDA5 interference and not impaired in targeting STAT1 for proteasomal degradation via the VDC ubiquitin ligase complex. Results demonstrate that mutation of conserved charged residues in the V proteins of Nipah virus, measles virus, and mumps virus also abolishes MDA5 interaction. These findings clearly define molecular determinants for MDA5 inhibition by the paramyxovirus V proteins.
Immune responses to virus infections are initiated by cellular recognition of specific pathogen-associated molecular patterns (PAMPs), such as viral nucleic acids, by cellular pattern recognition receptor (PRR) proteins. Mammalian PRRs for viral nucleic acid responses include a subset of the transmembrane Toll-like receptors (1, 12, 31), as well as a number of intracellular proteins that sense cytosolic nucleic acids of foreign origin. One group of these intracellular PRRs includes proteins encoded by the retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (49). These proteins, collectively referred to as RIG-I-like receptors (RLRs), are characterized by the presence of tandem caspase activation and recruitment domain (CARD) motifs at their N termini (17) coupled to a C-terminal DECH box RNA helicase domain (20, 56, 57). A third related protein, LGP2, has significant sequence similarity with RIG-I and MDA5 in the helicase region but lacks the N-terminal CARD.
Interaction with nonself RNAs enables the RLRs to transmit a signal through the CARD-containing mitochondrial adaptor protein IPS-1 (also named Cardif, MAVS, and VISA) (21, 30, 48, 54). Available evidence suggests that IPS-1 acts as a scaffold that facilitates a serine kinase-based signaling cascade that leads to the activation of immediate responding transcription factors, including NF-κB and interferon (IFN) regulatory factor 3 (IRF3). These transcription factors accumulate in the nucleus and bind to the enhancer of the IFN-β gene promoter, resulting in transcription of the IFN-β mRNA. Secreted IFN-β can then bind to type I IFN receptors, leading to downstream signaling via the JAK-STAT signal transduction pathway. IFN-β binding to the receptor results in tyrosine phosphorylation of STAT1 and STAT2, which heterodimerize and complex with a DNA binding subunit, IRF9, to form the trimeric complex known as the interferon-stimulated gene factor 3 (ISGF3) (13, 22). ISGF3 rapidly translocates to the nucleus and binds to IFN-stimulated response element (ISRE) sequences of IFN-stimulated gene (ISG) promoters to induce their transcription (26, 43). The induced ISG products produce a cellular antiviral state that provides a broadly effective barrier that protects the cell against virus infections.
The importance of the IFN system in mediating antiviral defense is emphasized by the fact that many viruses have evolved mechanisms to evade or antagonize this innate immune response (18, 25, 47). This phenomenon has been well documented for the Paramyxoviridae family of enveloped nonsegmented negative-strand RNA viruses, which have evolved mechanisms to escape or prevent both IFN production and IFN-responsive signal transduction. In many cases, the paramyxovirus IFN evasion activities are mediated by the virus-encoded V protein. The paramyxovirus V protein is produced from the polycistronic P gene (50) and is identifiable by a C-terminal domain (CTD) that is highly conserved among diverse virus species. The V protein has been demonstrated experimentally and crystallographically to coordinate two zinc atoms per protein chain (29, 39). The crystal structure of the parainfluenza virus 5 (PIV5) V protein was determined in complex with cellular DDB1 (DNA damage-binding protein 1) and revealed that the CTD exists as a unique zinc finger fold with a first “finger” formed between an invariant histidine (H171) and the first conserved cysteine (C190, also referred to here as C1) (see Fig. 1B, below). A second, shorter loop is formed between the second (C2) and third (C3) conserved cysteines, C194 and C206. The remaining cysteines, C208, C211, C215, and C218 (C4 to C7) loop back to support coordination at both zinc atoms (see Fig. 1B). The entire conserved domain shares no structural or sequence homology with cellular proteins but is readily identifiable in paramyxovirus proteomes (27).
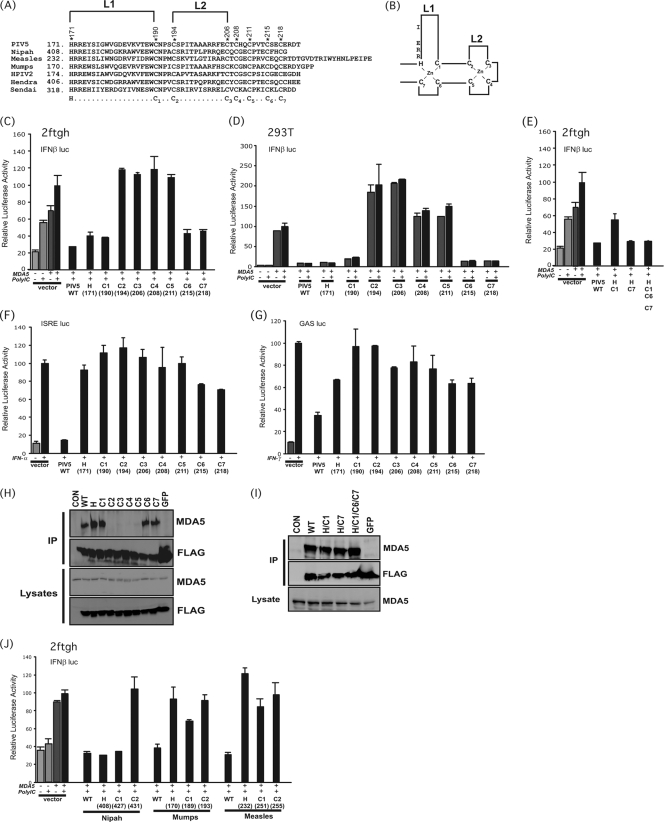
FIG. 1.
Role of zinc-coordinating residues in MDA5 interference by the PIV5 V protein. (A) Sequence alignment of paramyxovirus V protein C-terminal domains. The invariant histidine and seven cysteine residues implicated in zinc coordination are highlighted with asterisks above, and the seven cysteine residues are also labeled C1 to C7 in the consensus format below. The larger and smaller fingers are depicted as L1 and L2, respectively. (B) Schematic diagram of the PIV5 V protein zinc finger structure. L1 and L2 refer to the larger and smaller finger loops, respectively. Relative positions of key amino acids investigated in this study are illustrated. (C) MDA5 signaling interference by FLAG-tagged PIV5 V protein variants in human fibrosarcoma 2fTGH cells. Cells were transfected with the FLAG-tagged PIV5 V protein expression plasmids, an expression vector for MDA5, the −110 IFN-β luciferase (IFN-β luc) reporter gene, and a control Renilla luciferase vector. Cells were left unstimulated (-) or were stimulated (+) by transfection with poly(I:C) (5 μg/ml). Cells were lysed 6 h later and processed for luciferase assay. Bars illustrate average values (n = 3); standard deviations are indicated. (D) Same experiment as in panel C, but using human HEK293T cells. (E) Analysis of double mutants (H/C1 and H/C7) or quadruple mutant (H/C1/C6/C7) in MDA5 signaling interference assays in 2fTGH cells. The experiment was performed as described for panel C. (F) Effects of PIV5 V protein mutants on IFN-α signaling. The experiment was similar to that shown in panel C, but a 5× ISRE-luciferase reporter gene was used and stimulation (+) was with 1,000 U/ml of human IFN-α. (G) Effects of PIV5 V protein mutants on IFN-γ signaling. The experiment was similar to that shown in panle F, but a 4× GAS-luciferase reporter gene was used and stimulation (+) was with 5 ng of IFN-γ/ml. (H) Coimmunoprecipitation of MDA5 with PIV5 V protein point mutants. Independent plates of HEK293T cells were transfected to express either the V proteins or MDA5. Cell extracts were prepared and mixed in equal ratios for IP with FLAG M2 affinity resin, and the eluates were evaluated by MDA5 and FLAG immunoblotting. (I) Same experiment as shown in panel H, but analyzing the activity of double and quadruple mutants. (J) Effects of histidine and cysteine mutations on MDA5 signaling interference by diverse V proteins. Analysis of signaling interference was analyzed as described for panel C, but using the WT and mutant V proteins indicated from Nipah virus, mumps virus, and measles virus.
Paramyxovirus V protein-mediated evasion of IFN signaling occurs by direct interference with the IFN-responsive STAT proteins (18, 41). It has been observed that individual viruses within the family use distinct mechanisms to inhibit STAT proteins in a genus-specific fashion. For example, the rubulaviruses PIV5, human PIV type 2 (HPIV2), and mumps virus use their V protein to prevent IFN signaling by targeting STAT proteins for ubiquitin modification and proteasome-mediated degradation (11, 14, 23, 36-38, 53, 55, 58). Biochemical studies of STAT targeting by PIV5 and mumps virus V proteins indicate that they coordinate the assembly of multicomponent E3 ubiquitin ligase complexes from a number of cellular proteins. These V protein-dependent targeting complexes have been termed VDC to reflect the component protein constituents: V protein, the β-propeller adapter DDB1, and the cullin family member Cul4A (3, 28, 51). In addition, both genetic and biochemical evidence has demonstrated fundamental roles of STAT1 and STAT2 as both targets and cofactors for the E3 complexes (3, 28, 51). For example, cells lacking STAT2 are defective for STAT1 targeting by PIV5 V protein, and both STATs are found in association with affinity-purified V protein (38, 51).
In contrast to the Rubulavirus-encoded ubiquitin ligase activities, other strategies have been elucidated for V-mediated STAT interference. The Henipavirus V proteins bind to both STAT1 and STAT2 and efficiently inhibit IFN signaling by sequestering STAT1 and STAT2 in high-molecular-weight cytoplasmic complexes, preventing their IFN-induced activation by tyrosine phosphorylation (45, 46). The V protein from measles virus, a Morbillivirus, also binds to STAT1 and STAT2, effectively preventing their IFN-induced nuclear import (6, 34, 42). Neither Henipavirus nor Morbillivirus V proteins are capable of interacting with DDB1, Cul4A, or other known ubiquitin ligase machinery and do not target STATs to the proteasome. For these viruses, discrete regions of the V protein have been described that are necessary and sufficient for STAT association and IFN signaling interference. STAT1 is a primary target for Nipah and Hendra virus V protein, which is contacted by a short peptide in the N terminus (44). Measles virus also contacts STAT1 via a short N-terminal region that includes a tyrosine-based sequence motif but uniquely targets STAT2 via nonconserved residues in the CTD (6, 42).
In addition to the diverse mechanisms of IFN signaling interference, it has been established that paramyxovirus V proteins can also interfere with MDA5, a cellular RLR that can trigger IFN-β gene expression, as well as the related cellular RNA helicase protein, LGP2 (2, 35). The V protein specifically interacts with the MDA5 helicase domain through a minimal V protein-binding region that is shared with LGP2 but does not interact with the RLR prototype, RIGI (35). The highly conserved V protein CTD is necessary and sufficient for MDA5 and LGP2 associations, a feature that is common to all Paramyxovirus V proteins tested (2, 7, 8, 35). Although the mechanistic details remain poorly characterized, the V protein has also been shown to interfere with MDA5 oligomerization (8) and enzymatic activity (35).
Despite its high selectivity and importance for virus-host interactions, little is known about the molecular determinants of the V protein-MDA5 interface. By using the PIV5 V protein as a model, specific conserved amino acids that are required for V protein interference with MDA5 signal transduction were identified. Results indicate both general and virus-specific requirements for conserved V protein residues in MDA5 interaction and RLR signaling interference.
MATERIALS AND METHODS
Cell culture.
Human HEK293T and 2fTGH cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% Cosmic calf serum (HyClone) and 1% penicillin-streptomycin (Gibco-BRL) as described previously (38).
Plasmids and transfection and reporter gene assays.
PIV5 V, Nipah V, mumps virus V, and measles virus V cloned into a mammalian expression plasmid (pEF FLAG) downstream of an in-frame N-terminal FLAG epitope tag were used as templates to make the different point mutants. The point mutations were made using Stratagene's QuikChange mutagenesis XLII kit. All plasmid constructs were verified by DNA sequencing.
High-efficiency transient transfection of HEK293T cells was carried out on 60 to 80% confluent cultures in 100-mm dishes by using standard calcium phosphate procedures (4). The 2fTGH cells were transfected using polyethyleneimine transfection reagents (Polysciences Inc.) according to the manufacturer's instructions.
Luciferase reporter gene assays were performed in HEK293T or 2fTGH cells as described previously (53). Briefly, the cells were transfected with reporter gene, Renilla luciferase, and either empty vector or the cDNA expression plasmids. For IFN-γ response, the reporter gene contained four copies of the m67-SIE (GAS) linked to a TATA box and the firefly luciferase open reading frame (ORF). The IFN-α/β-responsive reporter gene contains five copies of the ISG54 ISRE upstream of the TATA box and the firefly luciferase ORF. After 24 h, transfection medium was replaced with fresh medium or medium supplemented with 1,000 U/ml IFN-α or 5 ng/ml IFN-γ. At 14 h post-IFN treatment, the cells were harvested and assayed for firefly and Renilla luciferase activities (dual luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of the Renilla luciferase.
For MDA5 signaling assays, cells were transfected with the −110 IFN-β luciferase reporter gene along with a Renilla luciferase control. After 24 h of transfection, the cells were stimulated with 5 μg/ml of poly(I:C) (Amersham Pharmacia) by transfection with Lipofectamine or Lipofectamine 2000 (Invitrogen). The cells were harvested after 6 h and assayed for firefly and Renilla luciferase activities. Relative luciferase activity was measured using Dual Luciferase reporter assay system (Promega). Data are plotted as average values (n = 3), with error bars representing standard deviation.
Cell extracts, immunoblotting, and immunoprecipitation.
For preparation of cell extracts, samples were washed once with ice-cold phosphate-buffered saline and subsequently lysed with whole-cell extract buffer (WCEB) as described previously (38, 51). For mixed lysate experiments, the lysates expressing V protein and MDA5 were mixed in a ratio of 1:1 and were used for immunoprecipitation (IP). For detection of endogenous MDA5, cells transfected with the V protein expression vectors were treated with IFN-α for 16 h prior to lysis and immunoprecipitation.
For immunoblotting, proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, probed with antibodies, and visualized by chemiluminescence (NEN Life Sciences). For immunoprecipitation, lysates were prepared in WCEB and precleared with Sepharose beads. Typically, 5% of the clarified lysate was analyzed prior to IP and the remainder is subject to IP. Antibody-protein complexes were purified with FLAG M2 affinity resin (Sigma) and washed with WCEB. After elution with SDS, proteins were separated by SDS-PAGE and processed for immunoblotting. The antibodies used were anti-STAT1 (C-20; Santa Cruz Biotechnology), anti-STAT2 (C-24; Santa Cruz Biotechnology), anti-FLAG (Sigma), anti-DDB1 (Invitrogen), or an anti-MDA5 monoclonal antibody generated by the Monoclonal Antibody Facility at the Lurie Cancer Center, Northwestern University, Chicago, IL (D. Pollpeter and C. M. Horvath, unpublished data).
Indirect immunofluorescence.
For immunofluorescence, 2fTGH cells were grown on Permanox chamber slides (Nalgene Nunc) and transfected and stained as described previously (45). Images were obtained using a Leica TCSSP confocal microscope, and representative fields with both transfected and untransfected cells in the same field are illustrated whenever possible for direct comparison. Due to the catalytic nature of VDC activity, we regularly observe that STAT1 staining is reduced to background levels by PIV5 V protein, even in cells with nearly undetectable V protein accumulation levels.
RT-PCR analysis.
HEK293T cells were transfected with V protein expression plasmids along with MDA5 or MDA5 K335A expression plasmids. After 24 h the cells were transfected with poly(I:C) or left untreated. Total RNA was extracted using TRIzol reagent (Invitrogen). Samples were treated with DNase I (Invitrogen), and 1 μg of RNA was subjected to the reverse transcriptase Superscript III (Invitrogen) for cDNA production. Quantitative real-time PCR was carried out using the MX3005P real-time PCR system (Stratagene) with SYBR green detection. IFN-β mRNA abundance was measured by real-time reverse transcription-PCR (RT-PCR) and are depicted normalized to the level of glyceraldehyde-3-phosphate dehydrogenase mRNA. Error bars represent standard deviations of duplicate samples.
Antiviral assays.
Antiviral responses were measured in a cytopathic effect protection assay using vesicular stomatitis virus (VSV, Indiana strain) as a reporter virus. HEK293T cells were transfected for expression of V proteins along with MDA5 or MDA5 K335A. Supernatant was harvested 30 h later, diluted, and added to 2fTGH cells. Following 8 h of incubation with the supernatant, 2fTGH cells were infected with VSV (Indiana strain) at 6 × 103 PFU/well in serum-free medium for 1 h. Medium was changed and cells incubated for 20 h before staining with 1% methylene blue in 50% ethanol.
RESULTS
Cysteine requirements for MDA5 interaction.
The CTD of the paramyxovirus V protein is sufficient to mediate association with MDA5 (2, 7, 8, 35) and is highly conserved among the diverse virus species (Fig. 1 A). To identify specific V protein residues required for MDA5 interaction, conserved amino acids were targeted for alanine substitution. Using the PIV5 V protein as a model system, two sets of conserved residues were targeted. Initial experiments focused on the invariant histidine and cysteine residues that are required for zinc coordination and are central to V protein structure and function (Fig. 1A and B) (3, 27, 28). Subsequent experiments targeted conserved noncysteine residues (Fig. 2A). All mutant proteins were tagged with the FLAG peptide epitope at their amino terminus to enable uniform detection and immunoaffinity purification. No obvious differences in expression or accumulation levels were observed for any of the V protein mutants.
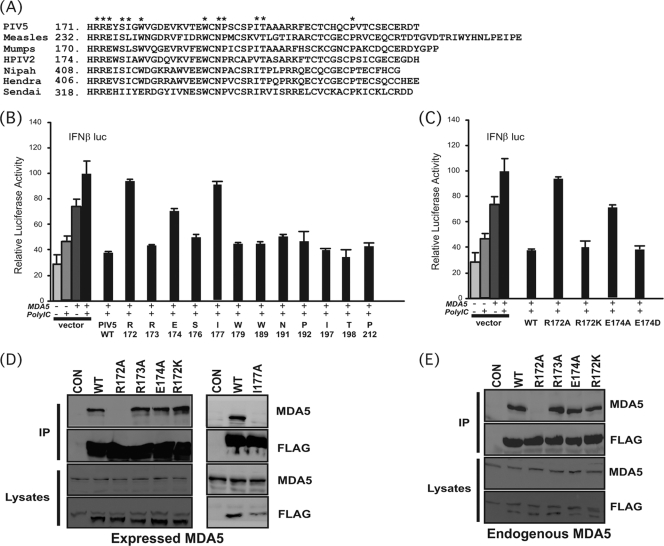
FIG. 2.
Conserved PIV5 amino acids required for MDA5 interference. (A) Sequence alignment of paramyxovirus V protein C-terminal domains. The conserved residues targeted for alanine substitution are illustrated with asterisks. (B) MDA5 signaling interference by PIV5 V proteins. The experiment was similar to that shown in Fig. 1C, but V protein-mediated MDA5 interference was tested by a reporter gene assay in 2fTGH cells. (C) Conservation of charge retains MDA5 interference. The effects of R172K and E174D substitutions were analyzed as described for panel B. (D) Coimmunoprecipitation of V proteins with MDA5. The experiment was similar to that shown in Fig. 1H; WT and mutant V proteins were tested for MDA5 association. (E) Coimmunoprecipitation of endogenous MDA5 with PIV5 V proteins. 2fTGH cells were transfected with V protein expression vectors and then treated with 1,000 U/ml IFN-α for 16 h prior to lysis and immunoprecipitation using FLAG M2 beads followed by immunoblotting with MDA5-specific antibody.
The abilities of the wild-type (WT) and mutated V proteins to interfere with MDA5 signal transduction were tested using an IFN-β promoter-driven luciferase reporter gene assay (32). The V proteins were expressed by transient transfection along with MDA5 and the reporter genes into either human fibrosarcoma 2fTGH cells (Fig. 1C) or HEK293T cells (Fig. 1D), and the cells were stimulated with the MDA5 ligand poly(I:C) for 6 h prior to lysis and luciferase assays. We and others (10, 19) have reported that endogenous responses to transfected poly(I:C) are very low in 293T cells compared to 2fTGH cells (compare Fig. 1C and D). However, MDA5 expression activates the IFN-β reporter gene robustly in both cell lines, and poly(I:C) stimulation can result in enhancement of the response if it is not already saturated by the constitutive signaling activity of expressed MDA5. Expression of the WT PIV5 V protein dramatically reduces the reporter gene activity, consistent with prior observations. The mutant V proteins varied with respect to their ability to interfere with MDA5 signaling. Mutation of histidine 171, or cysteine 190 (C1), 215 (C6), or 218 (C7), resulted in V proteins that retained the ability to disrupt MDA5 signaling, indicating that these amino acids are not required for PIV5-mediated MDA5 interference. In contrast, mutation of cysteine 194 (C2), 206 (C3), 208 (C4), or 211 (C5) resulted in V proteins defective in their ability to disrupt MDA5 signaling, indicating that these residues are essential for MDA5 interference. To control for the remote possibility of alternate cysteine coordination, further mutagenesis was used to create either double mutants with combined H171A/C190A (H/C1) and H171A/C218A (H/C7) changes or a quadruple mutant with combined H171A/C190A/C215A/C218A (H/C1/C6/C7) residues. Despite the multiple changes, the resulting V proteins retained the ability to inhibit MDA5 signaling (Fig. 1E), indicating that the coordination of the first zinc is not required for PIV5 to disrupt MDA5 signaling.
In light of this unexpected dispensability of half of the conserved cysteine residues in MDA5 binding, the V protein mutants were also used in combination with IFN-responsive reporter genes to evaluate their ability to target cellular STAT1 for proteolysis. All the mutant V proteins were defective in their abilities to overcome IFN-α (Fig. 1F) and IFN-γ (Fig. 1G) signaling, consistent with prior findings that the CTD cysteine cluster is important for VDC assembly and STAT1 targeting (3, 28, 52). Together, these findings demonstrate differential requirements for the zinc-coordinating CTD residues in PIV5 V protein in mediating MDA5 interference versus STAT1 degradation.
To mechanistically validate the reporter gene assay results, the ability of the V proteins to interact with MDA5 was tested in a coimmunoprecipitation assay. The FLAG-tagged V proteins and MDA5 were expressed in HEK293T cells, and FLAG immunoprecipitation was carried out to test MDA5 coprecipitation. Only the V proteins that retained the ability to interfere with MDA5 signaling were able to bind MDA5 (Fig. 1H and I). These results confirm the importance of some, but not all, zinc-coordinating residues in PIV5 V protein interference with MDA5.
In order to test if the differential cysteine requirement observed with the PIV5 V protein is a general property of paramyxovirus V proteins, the histidine (H) and the first two cysteine residues (C1 and C2) of V proteins from Nipah virus, mumps virus, and measles virus were mutated to alanine and tested for MDA5 signaling interference (Fig. 1J). For measles or mumps virus, all of the mutations produced proteins defective for MDA5 interference, indicating that for these viruses all cysteine residues are required. However, the Nipah virus V protein behaved similarly to PIV5: the conserved histidine (residue 408) and first conserved cysteine (C1, residue 427) of the Nipah V protein are dispensable for MDA5 interference, but the second cysteine (C2, residue 431) is required.
A conserved CTD arginine is required for MDA5 interference.
Aside from the zinc-coordinating residues, the V protein CTD contains additional conserved amino acids that represent candidates for MDA5 contact residues (Fig. 2A). These invariant residues were replaced with alanine, and the mutated V proteins were tested for MDA5 signaling inhibition (Fig. 2B). Most of the mutant proteins retained the ability to prevent reporter gene activation by MDA5, suggesting that the targeted amino acids may be conserved for reasons unrelated to MDA5 interference. However, substitution of the invariant glutamate at position 174 (E174A) resulted in a partial defect, and substitution of an invariant arginine at position 172 (R172A) or an isoleucine at position 177 (I177A) completely eliminated MDA5 signaling interference.
To further evaluate the importance of the charged residues R172 and E174 in mediating MDA5 interactions, conservative substitutions were engineered to create R172K or E174D mutations. For both positions, conservation of charge resulted in normal activity in MDA5 interference assays (Fig. 2C).
To test if the effect of the V protein mutations was directly related to MDA5 engagement, the V proteins were evaluated for their abilities to interact with MDA5 in two variations of the coimmunoprecipitation assays. In one case, both V and MDA5 proteins were expressed separately in 293T cells and individual lysates were prepared and mixed in a 1:1 ratio for subsequent immunoprecipitation. In the other case, expressed V proteins were tested for interaction with endogenous MDA5. All the proteins retained the ability to interact with MDA5 except R172A and I177A, irrespective of whether the V and MDA5 proteins were provided in trans by mixing lysates from independently expressed proteins (Fig. 2D) or by using the expressed V proteins to copurify endogenous MDA5 (Fig. 2E). The partial defect of the E174A mutant V protein, however, was not clearly recapitulated in the MDA5-binding assay, but the phenotypes associated with the R172A and R172K proteins agreed with the signaling interference assays.
Conserved arginine is universally required for MDA5 interference.
The V proteins from all tested paramyxoviruses interact with MDA5 via their CTD (2, 7, 8, 35), all of which include sequences identical to the R172, R173, and E174 residues of PIV5. The position equivalent to I177 is not completely conserved, as the mumps virus and measles virus V proteins both have a leucine residue at that position. To test the importance of these residues for other viruses, analogous mutations were engineered to create the following: mumps virus R171A, R172A, E173A, and L176A; measles virus R233A, R234A, and E235A; Nipah virus R409A, R410A, E411A, and I414. The WT and variant V proteins were tested for MDA5 signaling interference in the IFN-β reporter gene assay (Fig. 3A to D).
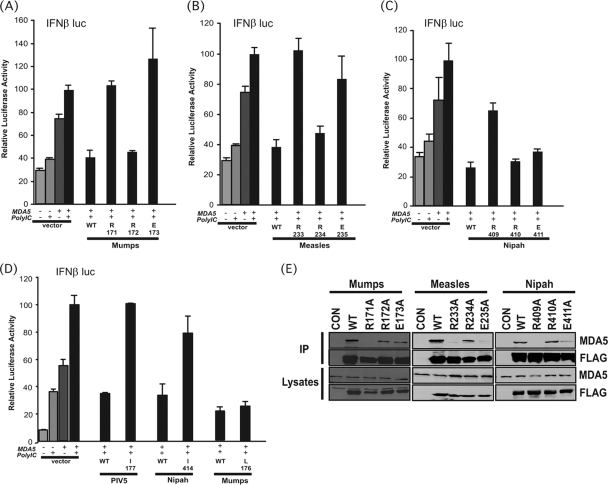
FIG. 3.
General and virus-specific roles of conserved residues for MDA5 interference. (A to C) MDA5 signaling interference by WT and mutant V proteins from mumps virus (A), measles virus (B), and Nipah virus (C) to test the roles of the conserved RRE residues. The experiment conducted as described for that shown in Fig. 1C. (D) Specific roles for the isoleucine/leucine residue in MDA5 interference. As described above, we tested the effects of mutation to the isoleucine of PIV5 and Nipah virus V proteins or the leucine of mumps virus V protein on MDA5 signaling. (E) Coimmunoprecipitation of MDA5 with diverse V protein mutants. As for the experiment shown in Fig. 1H, the variants of mumps virus, measles virus, and Nipah virus were tested for MDA5 associations. CON, control.
For all the V proteins (as for PIV5), mutation of the first conserved arginine prevented MDA5 signaling interference, but mutation of the second arginine had no effect. Mutation of the conserved glutamate disrupted the V proteins of mumps and measles viruses but had little effect on Nipah virus. These results confirm the general importance of the first arginine residue in MDA5 interference and reveal virus-specific variations in the requirement for charged amino acids. Similar to PIV5, mutation of I414 in Nipah virus V protein completely disrupted MDA5 interference, but the corresponding leucine residue of mumps virus was dispensable for V protein activity.
The WT and variant V proteins were also evaluated for MDA5 coimmunoprecipitation (Fig. 3E). In all cases, mutation of the first conserved arginine produced V proteins failed to coprecipitate MDA5, but mutation to the second arginine had no effect on MDA5 interactions. Variable responses for the glutamate substitutions were observed, consistent with a more supporting role for this position in mediating MDA5 interactions. Together, these data indicate both universal and virus-specific contacts for MDA5 interference and further reinforce the general role for the first conserved CTD arginine in MDA5 interference by paramyxovirus V proteins.
MDA5-defective PIV5 V proteins retain STAT interference.
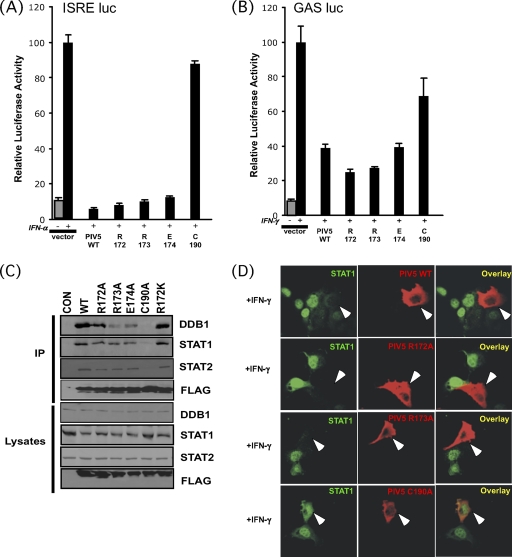
Mutagenesis of conserved amino acids raises a concern that the altered protein's overall structure may be changed sufficiently to create a misfolded variant incapable of binding to MDA5 for nonspecific reasons. To address this possibility for the R172 substitutions, the mutants were evaluated for their ability to interfere with IFN-α/β (Fig. 4A) and IFN-γ (Fig. 4B) signaling by using ISRE or GAS-specific reporter genes. All of the V proteins tested retained the ability to disengage IFN signal transduction, in contrast to the negative-control cysteine mutant C190A (C1). In parallel, the V proteins were assessed for assembly of the VDC ubiquitin ligase complex by FLAG immunoaffinity coprecipitation (3, 28, 51). All of the V proteins were able to coprecipitate the VDC components DDB1, STAT1, and STAT2 (Fig. 4C). In this qualitative assay, a range of signal strength was observed for DDB1 coprecipitation, with WT and R172A substitutions coprecipitating similar amounts of DDB1, while R173A and E174A mutants coprecipitated less DDB1. Similar STAT protein coprecipitation was observed for all proteins, except C190A, which failed to interact with any of the VDC components. Despite the variable level of DDB1 coprecipitation, catalysis of STAT1 destruction was robust for all V proteins. Indirect immunofluorescence demonstrated the loss of STAT1 to background staining levels in cells expressing WT, R172A, or R173A V proteins (Fig. 4D) and the absence of STAT1 targeting by the C190A mutant.
FIG. 4.
Intact STAT1 targeting and IFN signaling interference by MDA5-defective PIV5 V proteins. (A) Effects of PIV5 V protein mutants on IFN-α signaling, as determined using a 5× ISRE-luciferase reporter gene and stimulation (+) with 1,000 U/ml of human IFN-α. (B) Effects of PIV5 V protein mutants on IFN-γ signaling as determined using a 4× GAS-luciferase reporter gene and stimulation (+) with 5 ng of IFN-γ/ml. (C) Assembly of VDC complex by PIV5 V protein mutants. FLAG-purified V proteins were probed with antisera for the VDC component proteins DDB1, STAT1, and STAT2. (D) Degradation of STAT1 by MDA5-defective PIV5 V protein. 2fTGH cells were transiently transfected with expression plasmids for the WT or mutant PIV5 V proteins indicated. Cells were treated with 1,000 U/ml of IFN-γ for 30 min prior to fixation, permeabilization, and sequential staining for a FLAG epitope tag (to detect V protein) and specific antiserum to detect endogenous STAT1. Arrows point to V-expressing cells.
The R172A mutation disrupts antiviral signaling interference.
To verify the defective IFN evasion activity of the R172A mutant V protein in a biologically relevant context, a variation of the standard cytopathic effect (CPE) assay was used to evaluate MDA5-induced antiviral responses. HEK293T cells were transfected with MDA5 along with either a control empty vector or vectors expressing wild-type or mutant V proteins, and conditioned culture medium was collected 30 h later. These IFN-containing supernatants were diluted serially and added to 2fTGH cells for 8 h. Cells were then washed and infected with vesicular stomatitis virus for 20 h prior to staining with methylene blue (Fig. 5A). Medium from cells with no expressed MDA5 did not protect the cells from infection, while medium from cells expressing MDA5 was able to protect from the virus-induced CPE. Expression of WT PIV5 V resulted in greater CPE, due to V-mediated MDA5 interference. Similarly, the R173A mutant V protein interfered with MDA5 activity. In contrast, cells expressing the R172A mutant V protein were unable to interfere with the MDA5-induced antiviral response, and the cells were protected from VSV-induced CPE.
FIG. 5.
R172 and E174 are required for antiviral evasion. (A) HEK293T cells were transfected with empty vector (vec) or expression vectors for WT or mutant PIV5 V proteins, with (+) or without (-) coexpressed MDA5. The supernatant was collected 30 h after transfection, diluted, and transferred to freshly plated 2fTGH cells. After 8 h of incubation, the 2fTGH cells were infected with 6 × 103 PFU of VSV (Indiana strain). Cells were fixed 20 h later and stained with methylene blue in 50% ethanol. (B) Same experiment as shown in panel A, but using the hyperactive MDA5 encoding the K335A mutant to sensitize the assay. (C) Effects of V protein mutations on IFN-β mRNA accumulation. HEK293T cells were transfected with MDA5 or MDA5 K335A mutant expression vectors with or without WT or mutant PIV5 V proteins as indicated. IFN-β mRNA abundance was measured by real-time RT-PCR and is depicted normalized to the level of glyceraldehyde-3-phosphate dehydrogenase mRNA. Error bars represent standard deviations of duplicate samples.
In the CPE assay, the E174A mutant acted like the WT V protein, despite the intermediate MDA5 interference phenotype observed in luciferase assays. To increase the sensitivity of the CPE assay, we took advantage of a hyperactive MDA5 allele which harbors a point mutation, K335A, resulting in approximately 2.5 times the IFN production of WT MDA5 (5). The partial defect of the E174A mutant becomes more apparent in this context, as it fails to completely suppress the IFN production from the hyperactive MDA5 protein (Fig. 5B). In confirmation of these biological CPE assay results, the accumulation of IFN-β mRNA was measured by RT-PCR (Fig. 5C). Consistent with the antiviral assay results, WT and R173A V proteins prevented MDA5-dependent IFN-β mRNA induction, while expression of R172A V protein did not. Again, mutant E174A exhibited an intermediate defect in MDA5 interference.
DISCUSSION
Like other RNA viruses, paramyxoviruses have developed the means to prevent the production of and signaling by the primary innate antiviral cytokines in the type I IFN family. In many cases, both aspects of IFN evasion are ascribed to the virus-encoded V protein (7, 8, 18, 35, 41). The molecular determinants for V-mediated MDA5 engagement and interference were investigated, and the findings indicated requirements for some of the highly conserved residues in the V protein C-terminal domain. Both universal and virus-specific requirements were uncovered for the zinc-coordinating histidine and cysteine residues, and an invariant arginine residue was identified as universally required for MDA5 engagement by all the paramyxovirus V proteins tested.
Cysteine requirements for MDA5 engagement.
The invariant histidine and seven cysteines are defining features of the paramyxovirus V protein CTD, as their identity and spacing are conserved for all family members (24, 50). The PIV5 V protein is the best-studied member of this family and is known to bind to two atoms of zinc per polypeptide chain (39). Structural studies of the PIV5 V protein have demonstrated that the zinc coordination is involved in the formation of a unique zinc finger fold (27). The integrity of this CTD fold is thought to be important for many of the PIV5 V protein biological functions, including those related to host suppression, but is not required for virus replication in cell culture (9, 16).
For PIV5, mumps virus, measles virus, and HPIV2, V protein-mediated STAT targeting has been observed to require an intact cysteine cluster, and mutations to individual cysteines are commonly thought to abrogate normal protein interactions and subsequent biological activities (3, 11, 28, 33, 42). For example, recombinant PIV5 V proteins with mutations to any individual cysteine residue fail to interact with DDB1, a central component of the STAT1-targeting VDC ubiquitin ligase complex (28). Our results confirm the importance of each individual PIV5 zinc-coordinating residue for STAT1 destruction and IFN signaling interference (Fig. 1 and 3).
The integrity of the V protein CTD cysteine cluster is also widely assumed to be important for association and interference with MDA5. Indeed, individual alanine substitutions of select cysteine residues of the PIV5 V protein CTD were reported to prevent double-stranded RNA signaling interference, disable the V protein for limiting IFN-β induction by MDA5, and abolish MDA5 binding in a yeast two-hybrid assay system (7, 40). Together, these findings support the contention that the intact CTD cysteine fold is required for MDA5 antagonism by PIV5, and presumably the other V proteins known to target MDA5 (2, 7, 8, 35). In light of these previous findings, it was unexpected that mutation of the conserved histidine and some of the conserved cysteine residues resulted in PIV5 V proteins with unperturbed activity in MDA5 interference assays. In fact, we observed that a quadruple mutant, with an alanine substitution to histidine (H171) as well as cysteines at position 190, 215, and 218 (H/C1/C6/C7), was able to antagonize MDA5 indistinguishably from the wild-type V protein (Fig. 1). These observations distinguish two groups of zinc-coordinating residues for PIV5: the first two (H171 and C190) and last two (C215 and C218) that are involved in the formation of the first finger are dispensable for MDA5 interference, while the central four cysteines (C194, C206, C208, and C211) that are involved in the formation of the smaller second finger are essential for MDA5 interference.
This is the first study to report the effects of mutating all the zinc-coordinating residues of a single V protein on MDA5 signaling for PIV5. We also tested the importance of the histidine and first two cysteine residues of the V proteins from Nipah virus, mumps virus, and measles virus for MDA5 interference to probe the generality of our findings. Nipah virus V protein was similar to PIV5: neither the histidine nor the first cysteine was required for MDA5 interference, but mutation of the second cysteine disrupted V protein activity. In contrast, MDA5 interference by both mumps virus and measles virus V proteins was disrupted by mutation of the histidine or either of the first two cysteines. The reason for this differential cysteine requirement by the different paramyxoviruses is unlikely to be apparent in the absence of detailed structural analysis of V proteins in association with MDA5, but the results clearly indicate that the requirements for zinc coordination are virus specific and cannot be generalized.
Universal role for conserved arginine in MDA5 interference.
In addition to the zinc-coordinating residues, we tested the importance of other conserved amino acids for effects on MDA5 interference. The most dramatic consequences were obtained by mutating the first arginine of the CTD, and a nearby glutamic acid residue, both of which are conserved in all paramyxovirus V proteins and lie at the base of the first CTD finger. A point mutation in the first arginine residue completely abolished MDA5 interference by the PIV5 V protein, and this feature was common to all viruses tested. The disruption of MDA5 interference by the PIV5 R172A mutation is specific and not due to overall deficiencies in the V protein function, as this protein exhibits little defect in its STAT1 targeting activity and can be complemented by lysine substitution. Analogous mutagenesis of this arginine in the V proteins from mumps virus, measles virus, and Nipah virus resulted in defective MDA5 interference. Based on these findings, we conclude that the first conserved arginine residue of the V protein CTD is essential for MDA5 association and signaling interference by diverse paramyxoviruses.
Mutation of the glutamic acid had a partial effect on PIV5 activity and was of variable importance for different viruses. Effects of the analogous glutamic acid mutation ranged from complete disruption of MDA5 interference, observed for mumps virus and measles virus V proteins, to partial disruption for PIV5, to little or no effect in Nipah virus V protein. These findings, coupled with the observation that the E174A mutation is less effective at disrupting a hyperactive MDA5 variant, suggest that this residue plays a supporting role in MDA5 interference. Conservative substitution to aspartic acid in PIV5 (E174D) retained MDA5 targeting, confirming the importance of charge, as observed for R172.
A third residue, isoleucine at position 177, was found to be required for MDA5 interference by PIV5 V protein. Unlike the charged residues, the isoleucine is not completely conserved among V proteins; this position contains a leucine residue in the mumps virus and measles virus V proteins. Results show that the analogous isoleucine of Nipah virus V protein is similarly required for MDA5 interference, but mutation to the leucine at the identical position of mumps virus V protein has no effect. These findings indicate that the isoleucine represents another virus-specific adaptation to MDA5 association. It is an intriguing correlation that the viruses that maintain and require the isoleucine for MDA5 interference (PIV5 and Nipah) also share the dispensability of zinc coordination in the first finger, while those that have a leucine (mumps virus and measles virus) require all cysteines for MDA5 interference.
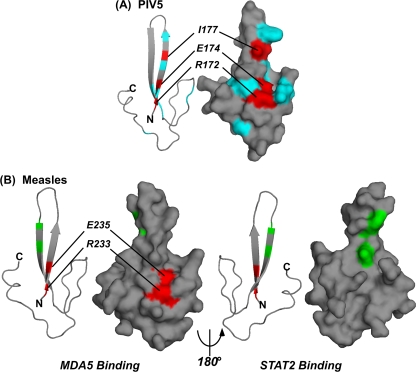
The availability of structural data for the PIV5 V protein enables direct visualization of the positions of the amino acids mutated in this study. The residues required for MDA5 interference are closely juxtaposed in the first zinc finger, indicating a contact surface that is responsible for host evasion by MDA5 interference (Fig. 6A). Modeling of the CTD alone does not adequately explain our observation that mutations to the histidine and cysteine residues that form the base of finger 1 (H, C1, C6, and C7) do not disrupt PIV5-mediated MDA5 interference. It is possible and likely that other regions of the V protein N-terminal domain form stabilizing interactions with the CTD to maintain proper alignment of MDA5-interacting residues. Previous investigation of IFN signaling disruption by the measles virus V protein determined that the CTD is necessary and sufficient to associate with STAT2 and prevent its IFN-induced nuclear import (6, 42). Structural modeling revealed that residues important for STAT2 binding mapped to a single surface of the measles virus CTD, and it was hypothesized that the alternate CTD surface could be available to form multimeric interactions (52) or associate with other cellular targets, such as MDA5 (42). Adding the findings of the present study to the measles virus V protein CTD structural model confirms this theory and indicates that the interaction surfaces for the two cellular targets are indeed distinct (Fig. 6B). Together, these findings suggest that it may be possible to modulate the activity of V proteins with small molecules that selectively attack either the STAT or MDA5 interaction surfaces.
FIG. 6.
Structural models for paramyxovirus V protein CTD contact residues. (A) The PIV5 V protein CTD derived from the crystal structure (PDB accession no. 2B5L), colorized to represent the mutations described here. For simplicity, the zinc atoms are not shown, and their coordinating residues are not colored. Mutations that do not alter MDA5 interference are represented in cyan (R173, S176, W179, W189, N191, P192, I197, T198, and P212). Mutations that impair MDA5 interference are represented in red and labeled. (B) The measles virus V protein sequence was modeled on the simian virus 5 structure (PDB accession no. 2B5L) by use of the Swiss PDB viewer available at http://www.expasy.org/spdbv/ (15). Each pair of images illustrates a ribbon view on the left and a surface view on the right, with front and back faces indicated in a 180° rotation. Coloring corresponds to the residues identified as important for the measles virus V protein association with MDA5 (red) or those important for association with STAT2 (green) (F246R and D248F) (42).
Acknowledgments
We are grateful to Sayantan Bose and Robert A. Lamb for insightful discussions and members of the Horvath lab for critical evaluations of and comments on the manuscript.
This work was supported by NIH grants AI50707 and AI073919.
Footnotes
Published ahead of print on 18 August 2010.
REFERENCES
- 1.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 5.Bamming, D., and C. M. Horvath. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284:9700-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caignard, G., M. Bourai, Y. Jacob, F. Tangy, and P. O. Vidalain. 2009. Inhibition of IFN-alpha/beta signaling by two discrete peptides within measles virus V protein that specifically bind STAT1 and STAT2. Virology 383:112-120. [DOI] [PubMed] [Google Scholar]
- 7.Childs, K., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190-200. [DOI] [PubMed] [Google Scholar]
- 8.Childs, K. S., J. Andrejeva, R. E. Randall, and S. Goodbourn. 2009. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J. Virol. 83:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz, C. D., H. Palosaari, J. P. Parisien, P. Devaux, R. Cattaneo, T. Ouchi, and C. M. Horvath. 2006. Measles virus V protein inhibits p53 family member p73. J. Virol. 80:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, S., K. Eisenacher, A. Kirchhofer, K. Brzozka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169-179. [DOI] [PubMed] [Google Scholar]
- 11.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diebold, S. S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529-1531. [DOI] [PubMed] [Google Scholar]
- 13.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. U. S. A. 87:8555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 17.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 18.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621-4628. [DOI] [PubMed] [Google Scholar]
- 19.Joo, C. H., Y. C. Shin, M. Gack, L. Wu, D. Levy, and J. U. Jung. 2007. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3. J. Virol. 81:8282-8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, D. C., R. V. Gopalkrishnan, Q. Wu, E. Jankowsky, A. M. Pyle, and P. B. Fisher. 2002. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 99:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 22.Kessler, D. S., S. A. Veals, X. Y. Fu, and D. E. Levy. 1990. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 4:1753-1765. [DOI] [PubMed] [Google Scholar]
- 23.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-rich region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 24.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1450-1496. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 25.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 26.Levy, D. E., D. S. Kessler, R. Pine, N. Reich, and J. E. Darnell, Jr. 1988. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 2:383-393. [DOI] [PubMed] [Google Scholar]
- 27.Li, T., X. Chen, K. C. Garbutt, P. Zhou, and N. Zheng. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124:105-117. [DOI] [PubMed] [Google Scholar]
- 28.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 29.Liston, P., and D. J. Briedis. 1994. Measles virus V protein binds zinc. Virology 198:399-404. [DOI] [PubMed] [Google Scholar]
- 30.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 31.Mogensen, T. H., and S. R. Paludan. 2005. Reading the viral signature by Toll-like receptors and other pattern recognition receptors. J. Mol. Med. 83:180-192. [DOI] [PubMed] [Google Scholar]
- 32.Nusinzon, I., and C. M. Horvath. 2006. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol. Cell. Biol. 26:3106-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno, S., N. Ono, M. Takeda, K. Takeuchi, and Y. Yanagi. 2004. Dissection of measles virus V protein in relation to its ability to block alpha/beta interferon signal transduction. J. Gen. Virol. 85:2991-2999. [DOI] [PubMed] [Google Scholar]
- 34.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisien, J. P., D. Bamming, A. Komuro, A. Ramachandran, J. J. Rodriguez, G. Barber, R. D. Wojahn, and C. M. Horvath. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 38.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 40.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 41.Ramachandran, A., and C. M. Horvath. 2009. Paramyxovirus disruption of interferon signal transduction: STATus report. J. Interferon Cytokine Res. 29:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran, A., J. P. Parisien, and C. M. Horvath. 2008. STAT2 is a primary target for measles virus V protein-mediated alpha/beta interferon signaling inhibition. J. Virol. 82:8330-8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reich, N., B. Evans, D. Levy, D. Fahey, E. Knight, Jr., and J. E. Darnell, Jr. 1987. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. U. S. A. 84:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 78:5358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez, J. J., L. F. Wang, and C. M. Horvath. 2003. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 77:11842-11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 49.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17-22. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 52.Ulane, C. M., A. Kentsis, C. D. Cruz, J. P. Parisien, K. L. Schneider, and C. M. Horvath. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 79:10180-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 55.Yokosawa, N., S. Yokota, T. Kubota, and N. Fujii. 2002. C-terminal region of STAT-1alpha is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 76:12683-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 57.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 58.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]