Abstract
The NS1 protein from influenza A viruses contains a four-amino-acid sequence at its carboxyl terminus that is termed the PDZ-binding motif (PBM). The NS1 PBM is predicted to bind to cellular PDZ proteins and functions as a virulence determinant in infected mice. ESEV is the consensus PBM sequence of avian influenza viruses, while RSKV is the consensus sequence of human viruses. Currently circulating highly pathogenic H5N1 influenza viruses encode an NS1 protein with the ESEV PBM. We identified cellular targets of the avian ESEV PBM and identified molecular mechanisms involved in its function. Using glutathione S-transferase (GST) pull-down assays, we found that the ESEV PBM enables NS1 to associate with the PDZ proteins Scribble, Dlg1, MAGI-1, MAGI-2, and MAGI-3. Because Scribble possesses a proapoptotic activity, we investigated the interaction between NS1 and Scribble. The association between NS1 and Scribble is direct and requires the ESEV PBM and two Scribble PDZ domains. We constructed recombinant H3N2 viruses that encode an H6N6 avian virus NS1 protein with either an ESEV or mutant ESEA PBM, allowing an analysis of the ESEV PBM in infections in mammalian cells. The ESEV PBM enhanced viral replication up to 4-fold. In infected cells, NS1 with the ESEV PBM relocalized Scribble into cytoplasmic puncta concentrated in perinuclear regions and also protected cells from apoptosis. In addition, the latter effect was eliminated by small interfering RNA (siRNA)-mediated Scribble depletion. This study shows that one function of the avian ESEV PBM is to reduce apoptosis during infection through disruption of Scribble's proapoptotic function.
The influenza A virus NS1 protein is a multifunctional protein that counteracts multiple antiviral mechanisms of the innate immune system (reviewed in references 14 and 24). NS1 inhibits PKR, RNase L, and TRIM25/RIG-I (10, 15, 30, 31, 36). NS1 also inhibits posttranscriptional processing of cellular pre-mRNAs by binding and inhibiting two cellular proteins: the 30-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF30) and the poly(A)-binding protein II (PABII) (6, 33). The NS1 block to pre-mRNA processing prevents the production of alpha/beta interferon and other cellular mRNAs involved in antiviral action. Additionally, NS1 activates phosphatidylinositol 3-kinase (PI3K) by binding to its p85β regulatory subunit, thereby enhancing viral replication (13).
Genetic and structural studies have identified two functional domains in the ∼230-amino-acid-residue NS1 protein (14). Residues 1 to 73 comprise an RNA-binding domain (RBD) that binds to double-stranded RNA (27). Resides 74 to ∼210 comprise the effector domain (ED), which contains binding sites for a number of cellular proteins involved in innate immunity, including PKR, CPSF30, and PABII (3). A large-scale sequencing study of avian influenza virus isolates identified a four-amino-acid sequence at the carboxyl terminus of NS1 that was termed the PDZ-binding motif (PBM) (35). PBMs confer binding to a class of cellular proteins that contain a characteristic structure known as the PDZ domain (20, 41). In general, PDZ domain-containing proteins act as scaffolds to assemble large protein complexes and function in cell signaling and cellular polarity (34).
The NS1 PBM from avian influenza viruses has the consensus sequence ESEV (78% of viral isolates [35]), while that of human viruses is RSKV (85% of viral isolates [35]). Because X-S/T-X-VCOOH is a consensus sequence for a class I PBM (34, 45), both the avian and human virus consensus PBMs are predicted to bind to PDZ proteins. The sequence differences between these PBMs likely confer differences in specificity and avidity to PDZ protein targets. Depending upon its sequence, each PBM is therefore likely to target a distinct set of cellular PDZ proteins. Thus, each NS1 PBM sequence may have its own signature of PDZ proteins that are targeted during infection. A previous study with recombinant NS1 proteins and synthetic peptides encompassing single PDZ domains demonstrated that the PBMs from avian and human influenza viruses possess distinct binding properties in vitro (35). Both the avian ESEV and human RSKV PBMs have been shown to function as virulence determinants in infected mice (21). In infected mice, the ESEV PBM displayed a more severe phenotype than the RSKV PBM and was associated with severe alveolitis, hemorrhage, and spread of virus in the lungs of infected mice. A recent study confirmed that the avian ESEV PBM possesses a more severe virulence phenotype in mice than the human RSKV PBM (40).
In this study, we sought to identify cellular PDZ proteins that bind to the ESEV PBM from avian influenza viruses. Using in vitro binding assays, we found that the ESEV PBM enables NS1 to bind specifically to the PDZ proteins Scribble, Dlg1, MAGI-1, MAGI-2, and MAGI-3. Because Scribble has recently been shown to posses a proapoptotic function (48), we focused our analysis on the interaction between NS1 and Scribble. Using recombinant proteins, we found that the ESEV PBM confers direct and highly specific binding of NS1 to Scribble. To investigate the role of the ESEV PBM during influenza virus infections, we constructed recombinant H3N2 viruses which express a H6N6 NS1 protein containing either the ESEV or mutant ESEA PBM. We found that the recombinant virus with the ESEV PBM replicated at up to 4-fold-higher levels than the recombinant virus with the mutant ESEA PBM. We also found that infection of cells with a virus expressing an NS1 protein with the ESEV PBM resulted in the sequestration of Scribble into cytoplasmic puncta in infected cells and reduced apoptosis. Our results indicate that the ESEV PBM functions to protect influenza virus-infected cells from apoptosis through the inactivation of Scribble's proapoptotic function.
MATERIALS AND METHODS
Cells and preparation of cell extracts.
Cultures of 293T, A549, MDCK, and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum and antibiotics. Cell extracts were prepared by lysing cells in EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40, 5 mM dithiothreitol [DTT]) containing protease inhibitors as described previously (19). Where indicated, cell extracts were spun at full speed in a microcentrifuge, the supernatant was removed, and the pelleted material was resuspended in 200 μl loading buffer for SDS gels. Equal volumes of soluble cell extracts and resuspended pelleted material, representing equivalent numbers of cells, were loaded on SDS-polyacrylamide gels.
Plasmids and recombinant protein purification.
Plasmids containing NS1 genes from an H6N6 avian influenza virus isolate (A/Blue-winged teal/MN/993/1980) and an H3N2 human influenza virus isolate (A/Memphis/14/1998) were kindly provided by Clayton Naeve (35). All plasmid variants of the H6N6 NS1 protein were from the same avian virus isolate (A/Blue-winged teal/MN/993/1980); all plasmid variants of H3N2 were from the same human virus isolate (A/Memphis/14/1998). These NS1 genes were used as PCR templates to generate Escherichia coli and mammalian expression plasmids. For expression in E. coli, GST-NS1 plasmids were constructed that contained the NS1 ED fused to glutathione S-transferase (pGEX-2T/NS1ED). Both the H6N6 and H3N2 NS1 EDs contained residues 73 to 230 fused to GST. GST fusion proteins were expressed and purified from E. coli as described previously (19). Mammalian expression plasmids for PDZ proteins were the following: pcDNA3/HA-MAGI-2 (44), pcDNA/MAGI-3-V5 (44), pcDNA/Flag-Scribble (32), GW1/HA-Dlg1-I2 (9), GW1/HA-Dlg1-I3 (9), GW1/HA-MAGI-1b (12), GW1/HA-MAGI-1c (12), GW1/HA-MUPP1 (26), and pRK5/Myc-PATJ (25). To construct mammalian expression vectors for fragments of Scribble, PCR was utilized to insert fragments of Scribble into pCMV-Tag3B (Stratagene). To create a Scribble fragment predicted to lose the ability to bind to the NS1 PBM (39), four alanine substitutions were introduced into residues 872 to 876 in the β2 domain of the second Scribble PDZ domain; this protein was termed 1F2R-4A (L872A G873A F874A I876A) (see Fig. 2). An additional single alanine substitution in the second Scribble PDZ domain was generated, and the protein was termed 1FR-1A (F874A). To construct E. coli expression vectors for Scribble, PCR was utilized to insert the 1st and 2nd Scribble PDZ domains into the histidine-tagged vector pET46 Ek/LIC (Novagen). Expression and purification of His-tagged Scribble proteins were carried out according to a commercial protocol (Qiagen).
Influenza A viruses and infections.
Recombinant influenza viruses were generated in which an H6N6 NS1 gene (A/Blue-winged teal/MN/993/1980) was inserted into the background of the A/Udorn/72 virus strain using the reverse genetic system described by Takeda and colleagues (42). Viruses with a wild-type ESEV or mutant ESEA PBM were plaque purified twice, and stocks were grown in 10-day-old embryonated chicken eggs. Titers of viral stocks were determined on MDCK cells. Reverse transcription-PCR (RT-PCR) assays were performed with viral stocks to verify identities of the wild-type and mutant H6N6 NS1 genes. For virus growth experiments, confluent MDCK cells were infected at a multiplicity of infection (MOI) of 0.01, samples were collected at the indicated time points, and virus titers were determined by plaque assays on MDCK cells.
GST-NS1 binding and kinase assays.
Cultures of 293T cells were transfected with PDZ protein expression plasmids using calcium phosphate precipitation. Cell extracts were prepared 48 h posttransfection. GST-NS1 bound to glutathione-Sepharose beads was incubated with precleared cell extracts at 4°C for 1 h with gentle mixing. After binding, Sepharose bead complexes were washed three times with EBC buffer, and Sepharose beads were resuspended in SDS-PAGE loading buffer. The samples were loaded onto SDS-polyacrylamide gels which were either stained with Coomassie blue or analyzed in immunoblots. For kinase assays, GST-NS1-loaded glutathione-Sepharose beads were incubated with DNase and RNase and then incubated with precleared 293T extracts; the glutathione-Sepharose bead complexes were washed with EBC plus 5 mM DTT plus 0.03% SDS after the binding step. Bead complexes were washed in TKB buffer (50 mM Tris [pH 7.4], 10 mM MgCl2) and then resuspended in 25 μl kinase reaction mix (TKB plus 2.5 mM MnCl2+5 mM DTT plus 5 μM ATP) plus 1 μl of γ-labeled 32P-ATP (5 μCi). After 1 h of incubation at room temperature, bead complexes were washed three times with TKB and loaded onto an SDS-polyacrylamide gel. The gel was dried, and kinase reaction products were examined by autoradiography.
Immunoblots and immunofluorescence.
Antisera used were the following: goat anti-Scribble (Santa Cruz Biotechnology), rabbit anti-NS1 (Abcam), rabbit anti-PARP-1 (Cell Signaling), and mouse anti-FLAG (M2; Sigma). Immunoblotting was performed as described previously (16). A549 cells were grown on glass coverslips in 24-well culture dishes until confluent. The cells were infected with wild-type or ESEA mutant virus at an MOI of 1 in serum-free DMEM containing tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin (2 μg/ml) and incubated at 37°C with 5% CO2. At 10 h postinfection, cells were washed in ice-cold phosphate-buffered saline (PBS) and fixed in 4% formaldehyde for 30 min at 4°C. Cells were permeabilized in 0.5% Triton X-100 in PBS for 20 min at room temperature and blocked with 5% nonfat dry milk in TBST (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween) overnight at 4°C. Coverslips were incubated with primary antibody dilutions of anti-NS1 and anti-hScrib in 2.5% nonfat dry milk in TBST overnight at 4°C, followed by incubation with appropriate secondary antibodies conjugated to green and red fluorochromes (Invitrogen) for 1 h at room temperature. Cells were fixed a second time in 4% formaldehyde and quenched with 1 mg/ml NaBH4 in PEM [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 5 mM EGTA, and 2 mM MgCl2]. Nuclei were counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS. Coverslips were mounted on slides using ProLong Gold antifade reagent (Invitrogen). Deconvolution microscopy was conducted as described previously (17). A Z series of focal planes was captured and deconvolved using the Applied Precision DeltaVision restoration microscopy system with the softWoRx software program (Applied Precision). A single focal plane from each Z series was then further processed in Adobe Photoshop.
TUNEL assays and siRNA depletions.
A549 cells were grown on glass coverslips in 24 well culture dishes until confluent. The cells were infected with wild type or ESEA mutant virus at a MOI of 5 in serum-free DMEM containing TPCK trypsin (2 μg/ml) and incubated at 37°C with 5% CO2. Cells were fixed at 12 and 24 h postinfection. Nuclear DNA fragmentation was identified by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay. Fluorescein-conjugated dUTP was used to label free 3-hydroxyl ends using an in situ cell death detection kit (Roche). Nuclei were counterstained with DAPI. TUNEL-positive cells and nuclei were observed with a fluorescence microscope (Olympus IX70 with a Microfire [Optronics] camera). Two random fields of at least 250 cells from each coverslip were counted. Small interfering RNAs (siRNAs) targeting Scribble were those used previously (43) and were purchased from Ambion; Scribble target sequences were 5′-CAGGATGAAGTCATTGGAACA-3′. For depletions, HeLa cells were grown on glass coverslips in a 24-well culture dish until 25% confluent. Cells were then transfected with negative-control siRNAs (Qiagen) or Scribble-targeted siRNAs by Oligofectamine (Invitrogen) according to the manufacturer's directions. At 48 h posttransfection, cells were infected at an MOI of 5 and processed for TUNEL assays as described for A549 cells.
RESULTS
Identification of cellular PDZ proteins that specifically bind in vitro to the NS1 protein with the avian ESEV PBM.
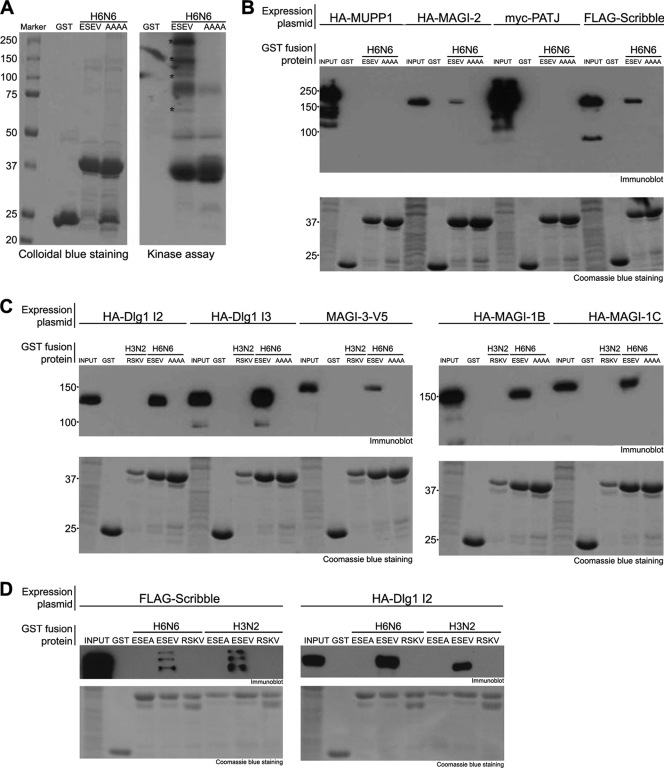
To identify cellular proteins that are targets of the avian NS1 PBM, we carried out in vitro binding assays with recombinant NS1 proteins and 293T cell extracts. We generated a GST fusion of the effector domain (ED) plus PBM of an H6N6 avian NS1 protein; this protein was termed GST-avNS1. As a specificity control, we constructed a mutant of the GST-avNS1 protein in which all of the ESEV residues of the PBM were altered to alanine; this fusion protein was termed GST-avNS1-AAAA. We anticipated that the background of cellular proteins that bind to both GST fusion proteins would hinder the detection of proteins whose binding is specific to the NS1 PBM. We reasoned, however, that cellular protein kinase activities may be associated with proteins that bind to the NS1 PBM, and in vitro kinase assays might reveal substrates that were specifically associated with the GST-avNS1 protein. We used this strategy previously to identify cyclin T1/CDK9 as a protein kinase that binds specifically to the activation domains of the HIV-1 and HIV-2 Tat proteins (18, 19).
GST-avNS1 and GST-avNS1-AAAA were expressed in and purified from E. coli by binding to glutathione-Sepharose beads. Bead complexes were incubated with extracts prepared from 293T cells, washed, and then incubated under protein kinase reaction conditions. 32P-ATP-labeled products of the in vitro reactions were examined on an SDS-polyacrylamide gel (Fig. 1A). A number of high-molecular-mass substrates (>60 kDa) were observed in reactions with the GST-avNS1 but not GST-avNS1-AAAA protein (indicated by asterisks in Fig. 1A). These data suggest that the NS1 PBM mediates specific in vitro associations with several relatively large cellular proteins. The nature of the kinase(s) responsible for phosphorylation of the cellular proteins substrates in Fig. 1A is unknown.
FIG. 1.
GST-NS1 in vitro binding assays. (A) The indicated GST fusion proteins (NS1 ED + PBM) attached to glutathione-Sepharose beads were incubated with 293T cell extracts. Bead complexes were washed and then incubated with [γ-32P]ATP under kinase reaction conditions. Products of kinase reactions were analyzed on a 9% SDS-polyacrylamide gel. A Colloidal blue stain (showing GST-NS1 proteins used in the assay) and autoradiograph of the dried gel are shown. Major substrates of the kinase reaction with the GST-avNS1 protein are indicated by asterisks. (B and C) Cultures of 293T cells were transfected with expression plasmids for the indicated epitope-tagged PDZ proteins. Cell extracts were prepared and incubated with the indicated GST proteins (NS1 ED + PBM) attached to glutathione-Sepharose beads. Proteins bound to GST fusion proteins were examined in immunoblots using the appropriate antisera to detect epitope tags. The amounts of input GST-NS1 proteins used in binding assays are shown at the bottom (Coomassie-stained gel). (D) Cultures of 293T cells were transfected with expression plasmids for epitope-tagged Scribble or Dlg1. Cell extracts were prepared and incubated with the indicated GST proteins containing the H6N6 or H3N2 ED plus the indicated PBM. Scribble or Dlg1 bound to GST fusion proteins was examined in immunoblots. The amounts of input GST-NS1 proteins used in binding assays are shown at the bottom (Coomassie-stained gel).
These binding data led us to screen PDZ proteins with molecular masses of >60 kDa for the ability to specifically associate in vitro with the GST-avNS1 protein. Expression plasmids for widely expressed epitope-tagged PDZ proteins were transfected into 293T cells, cell extracts were prepared, and binding reactions were carried out with GST-avNS1 proteins (Fig. 1B and C). The MUPP1 and PATJ proteins did not detectably associate with either NS1 fusion protein. However, MAGI-1, MAGI-2, MAGI-3, Scribble, and Dlg1 (I2 and I3 isoforms) bound to the GST-avNS1 but not to the GST-avNS1-AAAA protein. The I2 and I3 isoforms of Dlg1 are generated by alternative splicing and contain a different small insertion element outside the three Dlg1 PDZ domains (29). These data suggest that the avian NS1 protein binds in vitro to MAGI-1, MAGI-2, MAGI-3, Scribble, and both Dlg1 isoforms and that this association requires the NS1 PBM. Additionally, a GST fusion containing the ED plus PBM of a human H3N2 virus NS1 protein with an RSKV PBM did not associate in vitro with Dlg1, MAGI-1, MAGI-2, or MAGI-3 (Fig. 1C).
To further characterize the binding properties of the ESEV and RSKV PBM sequences, we examined GST fusions of ED plus PBM in which the PBM sequences were swapped between the EDs of the H6N6 and H3N2 NS1 proteins (Fig. 1D). The H6N6 and H3N2 ED fusion proteins containing the ESEV PBM bound Scribble and Dlg1 in vitro, while the ED fusion protein with the RSKV PBM did not. The H6N6 ED plus RSKV PBM likewise failed to bind to MAGI-1, -2, and -3 (data not shown). These data show that the ESEV PBM enables the NS1 protein to bind specifically to Scribble and Dlg1, as well as MAGI-1, -2, and -3.
Our binding data shown in Fig. 1 differ from findings of a previous study that examined interactions between GST-NS1 fusion proteins and filter arrays containing single PDZ domains (35). In binding assays with the filter arrays, a GST-NS1 protein with the ESEV PBM bound to single PDZ domains of MUPP1 and Scribble but not MAGI-3 and Dlg1. However, our characterization of the binding of GST-NS1 fusion proteins to both Scribble and Dlg1 found that two PDZ binding domains in each protein are required for specific binding to NS1 with the ESEV PBM (see Fig. 2 below; also data not shown).
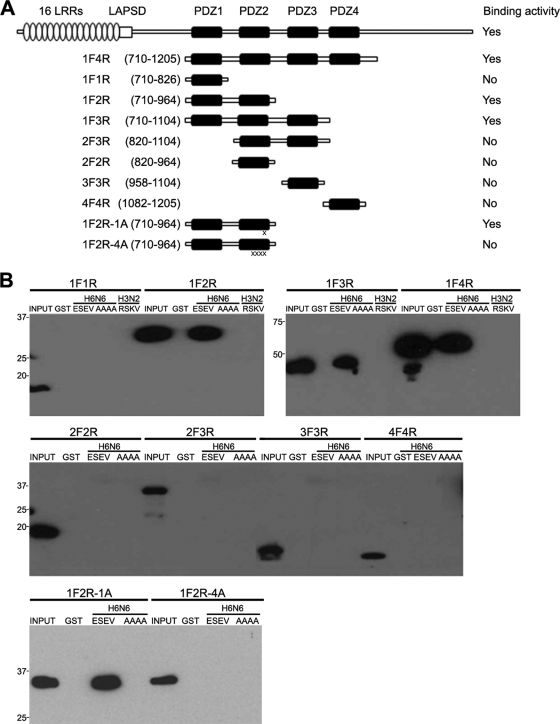
FIG. 2.
Requirements of Scribble for binding in vitro to the avian influenza virus NS1 protein. (A) The domain structure of Scribble is shown: 16 leucine-rich repeats (LRRs), a LAPS domain (LAPSD), and four PDZ domains. Portions of Scribble expressed from Myc-tagged vectors are shown; the vectors 1F2R-1A and 1F2R-4A contain one and four alanine substitutions in PDZ domain two. The binding activities of Scribble proteins for avian influenza virus NS1 are summarized. (B) The indicated Myc-tagged Scribble vectors were transfected into 293T cells, extracts were prepared, and binding assays were performed with indicated GST proteins. Products of binding assays were evaluated in immunoblots.
Characterization of interaction between avian influenza virus NS1 and Scribble.
Scribble is an evolutionary conserved protein that is involved in cellular polarity (2) and also possesses a proapoptotic function (48). Scribble has previously been found to be a target of viral proteins encoded by papillomavirus, human T-cell leukemia virus type 1 (HTLV-1), and the flavivirus tick-borne encephalitis virus (TBEV). Scribble is targeted directly by high-risk human papillomavirus E6 proteins for degradation through proteasome-mediated proteolysis (32). The Tax protein of HTLV-1 associates with Scribble and sequesters it into cytoplasmic puncta, possibly to enhance the NFAT pathway (1). Scribble is targeted directly by the TBEV NS5 protein, and this interaction inhibits interferon-mediated JAK-STAT signaling (46). Given these well-documented functional interactions of viral proteins with Scribble, our subsequent experiments focused on this PDZ protein.
We next determined which of the PDZ domains in Scribble are involved in the binding to the avian NS1 protein. Scribble contains 16 leucine-rich regions at its amino terminus, followed by a LAPSD domain and four PDZ domains (Fig. 2A). We constructed a series of Myc-tagged vectors that express fragments of Scribble, transfected these vectors into 293T cells, and assessed their ability to bind in vitro to the GST-avNS1 and GST-avNS1-AAAA proteins (Fig. 2B). Deletion of the 16 leucine-rich repeat (LRR) and LAPS domains (1F4R) did not affect binding to the GST-avNS1 protein, indicating that these domains are not involved in binding to NS1. Fragments of Scribble containing only a single PDZ domain did not bind to GST-avNS1. However, fragments containing the first three (1F3R) or the first two (1F2R) PDZ domains bound to the GST-avNS1 but not GST-avNS1-AAAA protein. A fragment containing the second and third Scribble PDZ domains (2F3R) was unable to bind to either the avNS1 or avNS1-AAAA fusion protein. Taken together, these data indicate that the first two PDZ domains of Scribble are necessary and sufficient for a specific interaction in vitro with the avian NS1 protein.
A GST fusion of the ED of an H3N2 NS1 protein with an RSKV PBM sequence failed to bind to a Scribble fragment containing all four PDZ domains or just the first two PDZ domains (Fig. 2B). To further define the interaction between the first two Scribble PDZ domains and the avian NS1 fusion protein, we constructed Myc-tagged vectors containing a single or four alanine substitutions in the second Scribble PDZ domain. The analysis of these proteins indicated that the single alanine substitution (1F2R-1A) did not impair binding to the GST-avNS1 protein. However, four contiguous alanine substitutions (1F2R-4A) in the second Scribble PDZ domain (residues 872 to 876) abrogated this interaction, as expected from previous studies on determinants within PDZ domains that specify binding to ligands (39).
Direct binding between the avian influenza virus NS1 protein and Scribble.
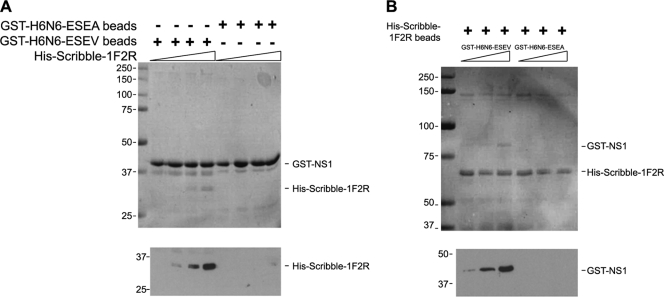
The data presented thus far indicate that the avian NS1 protein requires an intact PBM for an association with Scribble in GST pull-down assays. These data do not, however, demonstrate a direct interaction between NS1 and Scribble, since the association between these two proteins may involve a bridging protein(s). To examine if NS1 and Scribble interact directly, we performed in vitro binding assays with purified recombinant proteins. In one set of binding experiments, the His-tagged Scribble fragment 1F2R (see Fig. 3A) was expressed and purified from E. coli, while GST-avNS1 proteins, the wild type and the ESEA mutant, were expressed in E. coli and bound to glutathione-Sepharose beads. Purified His-Scribble-1F2R was then incubated with the GST-avNS1 bead complex and washed, and the amount of His-Scribble-1F2R that bound to the GST-avNS1 bead complex was evaluated in an SDS-polyacrylamide gel (Fig. 3A). A Coomassie blue stain of the gel indicated that Scribble bound to the GST-avNS1 but not the GST-avNS1-ESEA fusion protein. An immunoblot of the binding reactions with a His antiserum confirmed specific binding of Scribble-1F2R to the GST-avNS1 bead complexes.
FIG. 3.
Direct binding of Scribble to avian influenza virus NS1. (A) The GST-avNS1 and GST-avNS1-ESEA proteins bound to glutathione-Sepharose beads were incubated with purified His-Scribble-1F2R (see Fig. 4A) and washed, and proteins eluted with 2× SDS sample buffer. Eluates were analyzed on a 10% SDS-polyacrylamide gel. A Coomassie blue stain of the gel is shown. Immunoblotting with anti-His antiserum was performed on eluates to detect His-Scribble. (B) His-Scribble-1F2R was bound to Ni beads and incubated with purified GST-NS1 and GST-NS1-ESVA proteins. After binding, bead complexes were washed, and proteins were then eluted with 2× SDS sample buffer. Eluates were analyzed on a 10% SDS-polyacrylamide gel. A Coomassie blue stain of the gel is shown. Immunoblotting with anti-GST antiserum was performed on eluates to detect GST-NS1 proteins.
In reciprocal binding experiments, GST-avNS1 proteins were expressed and purified from E. coli, while His-Scribble-1F2R was expressed in E. coli and purified by binding to Ni beads. Purified GST-avNS1 proteins were incubated with His-Scribble-1F2R bead complexes and washed, and binding of NS1 to Scribble was evaluated in an SDS-polyacrylamide gel. A Coomassie blue stain of the gel indicated that the GST-avNS1 protein but not a GST-avNS1-ESEA protein bound to the His-Scribble-1FR2 bead complex. An immunoblot of the binding reactions with a GST antiserum confirmed specific binding of GST-avNS1 to the His-Scribble-1F2R bead complex (Fig. 3B). These data demonstrate a direct protein-protein interaction between the avian NS1 protein and Scribble, and this is dependent upon the NS1 PBM.
Generation of recombinant H3N2 influenza viruses that express an H6N6 NS1 protein with an ESEV PBM or a mutant ESEA PBM.
To enable us to carry out biochemical and immunofluorescence analyses to examine the function of the NS1 ESEV PBM during influenza virus infections in mammalian tissue culture cells, we generated two recombinant H3N2 influenza viruses (Udorn strain) that encode an H6N6 NS1 protein with either the wild-type ESEV or mutant ESEA PBM. These viruses were termed the ESEV and ESEA viruses. The practical advantage of these chimeric viruses is that experiments can be carried out under BSL2 containment conditions, allowing a detailed biochemical analysis of viruses that differ by only one residue in the avian ESEV consensus PBM. The ESEA mutation overlaps with the NEP (NS2) coding sequence and alters a phenylalanine at residue 73 to a leucine. Given that a number of alterations in the PBM which alter the NEP coding sequence do not affect viral replication in vitro (21, 40) and our analysis of the phenotypes of the ESEV and ESEA viruses (see below), it is unlikely that that this substitution affects the replication phenotype of these recombinant influenza viruses.
To compare in vitro replication of the ESEV and ESEA viruses, we infected MDCK cells at an MOI of 0.01 and assayed viral yield at 24 and 48 h postinfection (Fig. 4). Replication of the ESEV virus was 2.6-fold and 4-fold higher than that of the ESEA virus at 24 and 48 h postinfection, respectively. We observed a similar enhancement of replication by the ESEV PBM in independent experiments with these two recombinant viruses (data not shown). We observed similar enhanced replication of the ESEV virus relative to the ESEA virus in A549, HeLa, and Calu-3 cells (data not shown). These data suggest that the ESEV PBM can enhance viral replication in mammalian tissue culture cells.
FIG. 4.
ESEV PBM enhances viral replication in MDCK cells. Duplicate cultures of confluent MDCK cells were infected at an MOI of 0.01 with recombinant H3N2 viruses encoding the H6N6 NS1 protein with ESEV or mutant ESEA PBM. Viral replication at 24 and 48 h postinfection was measured by plaque assays in MDCK cells, with triplicate culture dishes used to quantify plaques for each time point of the duplicate infections. Viral yields at 24 h postinfection (p.i.) were 3.6 × 106 and 1.4 × 106 for the ESEV and ESEA viruses, respectively. Viral yields at 48 h p.i. were 8.1 × 106 and 2.0 × 106 for the ESEV and ESEA viruses, respectively. Asterisk indicates P value of 0.005 according to Student's t test; error bars are standard deviations.
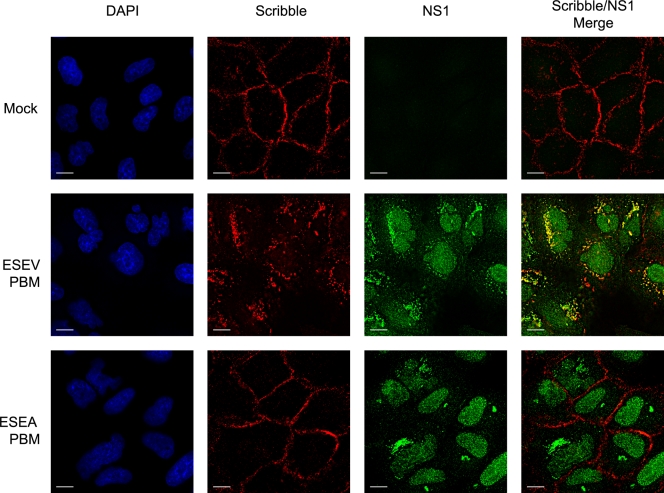
Colocalization of Scribble and NS1 with ESEV PBM in cytoplasmic puncta during infection.
To examine the effect of the PBM on cellular localization of Scribble, A549 cells were infected with the ESEV or ESEA virus and an immunofluorescence analysis was performed at 10 h postinfection (Fig. 5). In mock-infected cells, Scribble was concentrated at the plasma membrane. While infection with the ESEA virus did not affect the localization of Scribble, infection with the ESEV virus resulted in a striking redistribution of Scribble into cytoplasmic puncta that were concentrated in perinuclear regions of infected cells. There was extensive colocalization of Scribble with NS1 in the perinuclear puncta, as indicated by the yellow signal in the merged images in Fig. 5.
FIG. 5.
The avian NS1 protein colocalizes with Scribble in cytoplasmic puncta. A549 cells were infected at an MOI of 1 with the indicated influenza viruses, and cells were fixed at 10 h postinfection for immunofluorescence analysis with NS1 and Scribble antisera. DAPI stains were performed to visualize cell nuclei. Scale bar, 10 μm.
The relocalization of Scribble into perinuclear puncta by an NS1 protein with the ESEV PBM during infection may be similar to the relocalization of several PDZ proteins by the adenovirus E4-ORF1 protein. In the case of E4-ORF1, the PDZ proteins MUPP1, PATJ, MAGI-1, and ZO-2 become aberrantly sequestered in complexes in the cytoplasm and partition into an insoluble cell fraction when cell lysates are prepared (22). To determine if Scribble might partition into similar complexes during influenza virus infections in 293T and A549 cells, we prepared cell extracts at 8 h postinfection and examined the soluble and insoluble fractions in immunoblots (Fig. 6). We also examined Dlg1 in these experiments, since this PDZ protein also binds specifically to NS1 having a wild type PBM in GST pull-down assays (Fig. 1). Scribble and Dlg1 were present in the soluble fraction in extracts from both mock-infected cells and cells infected with the virus encoding the ESEA mutant NS1 protein. In contrast, both proteins were predominantly present in the insoluble fraction of cells infected with the virus that expresses the NS1 protein with the ESEV PBM. This result is consistent with the immunofluorescence analysis in Fig. 5 and indicates that the cytoplasmic puncta contain insoluble Scribble bound to wild-type NS1.
FIG. 6.
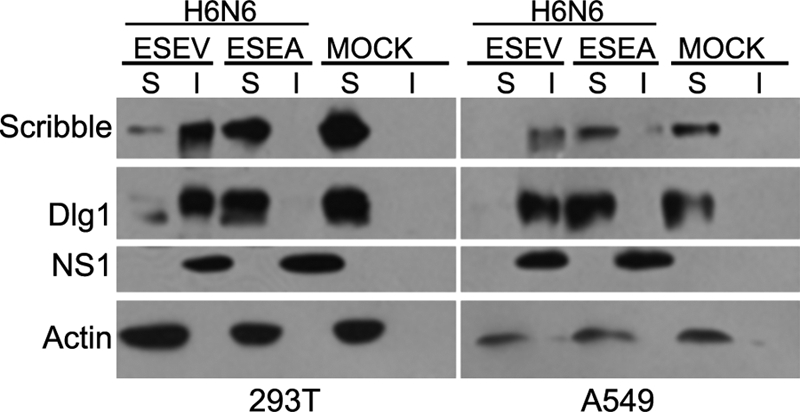
Avian NS1 sequesters Scribble in insoluble complexes. Cultures of 293T or A549 cells were infected with the indicated influenza viruses at an MOI of ∼2, and whole-cell extracts were prepared at 8 h postinfection. Cell lysates were fractionated into soluble and insoluble fractions. Amounts of soluble (S) and insoluble (I) cell fractions that represented equal numbers of cells were loaded on an 8% SDS-polyacrylamide gel. The indicated proteins were examined by immunoblotting.
The NS1 ESEV PBM reduces apoptosis during infection through functional inactivation of Scribble.
The relocalization of Scribble into cytoplasmic puncta and insoluble complexes by the ESEV but not ESEA virus suggests that the NS1 ESEV PBM may inactivate Scribble. Because Scribble has been shown to possess a proapoptotic function (48), we compared apoptosis in infections of the ESEV and ESEA viruses. Cultures of A549 cells were infected at an MOI of 5, and the TUNEL assay was used to quantify the percentages of apoptotic cells at 12 and 24 h postinfection in four independent experiments (Fig. 7A). Infections with the ESEA virus resulted in a statistically significantly greater number of apoptotic cells at both 12 and 24 h postinfection than was found for infections with the ESEV virus. At 24 h postinfection, there were about 2-fold more apoptotic cells in infections with the ESEA virus. We observed similar results in TUNEL assays in infections of A549 cells with these two viruses (data not shown). We also used an independent assay for apoptosis to confirm our results with the TUNEL assay. This was done by immunoblot analysis to examine cleavage of poly(ADP-ribose) polymerase 1 (PARP-1) at 6 h postinfection with the ESEV and ESEA viruses (Fig. 7B). Approximately 31% of the full-length PARP-1 was cleaved in infection with the ESEA virus, while approximately 19% was cleaved in infection with the ESEV virus. The results of both TUNEL and PARP-1 cleavage assays indicate that the NS1 ESEV PBM can function to protect cells from apoptosis.
FIG. 7.
ESEV PBM of NS1 reduces apoptosis during infection. (A) HeLa cells were infected with the indicated influenza viruses at an MOI of 5, and cells were fixed at 12 and 24 h postinfection. The percentages of apoptotic cells in two fields of at least 250 cells were quantified by a TUNEL assay. Error bars indicate standard deviations from four independent experiments (*, P < 0.05 by Student's t test). (). Cultures of HeLa cells were infected with the ESEV or ESEA virus at an MOI of 5, and cell extracts were prepared at 6 h postinfection. Cleavage of PARP-1 was evaluated in an immunoblot, and β-actin was used as a loading control. Quantitation of the immunoblot indicated that approximately 19% or 31% of full-length PARP-1 was cleaved in infection with the ESEV or ESEA virus, respectively. (C) HeLa cells were transfected with the indicated siRNAs and, 48 h later, infected with the indicated viruses at an MOI of 5. The percentages of apoptotic cells in random fields of at least 250 cells were quantified by a TUNEL assay. Error bars indicate standard deviations from four independent experiments (**, P < 0.05 by Student's t test). An immunoblot verified that the siRNAs were effective in Scribble depletions.
If the lower level of apoptosis seen in the ESEV NS1 infections is largely the result of functional inactivation of Scribble's proapoptotic function, then infections in cells depleted of Scribble by siRNAs should reduce the level of apoptosis in the mutant ESEA NS1 infections to that in the wild-type ESEV NS1 infection. To test this prediction, we used siRNAs that have previously been shown to be effective for Scribble depletions (43). HeLa cells were transfected with siRNAs against Scribble or control siRNAs, and at 48 h posttransfection, cells were infected with influenza viruses. TUNEL assays were used to quantify apoptotic cells at 24 h postinfection in four independent experiments (Fig. 7C). In cells transfected with control siRNAs, a statistically significantly greater number of apoptotic cells were observed with the ESEA NS1 virus than with the wild-type ESEV NS1 virus, similar to results in A549 cells. In contrast, in cells transfected with siRNAs against Scribble, apoptotic rates were similar in infections of either virus, demonstrating that the increased apoptosis in cells infected with the mutant ESEA virus is largely mediated by Scribble. An immunoblot analysis verified that Scribble was effectively depleted by the siRNAs (Fig. 7C). These data indicate that the reduced level of apoptosis seen with the wild-type ESEV NS1 protein results from inactivation of Scribble's proapoptotic function, likely through its sequestration into cytoplasmic puncta (see Fig. 5).
DISCUSSION
We have shown in this study that the avian influenza virus ESEV PBM allows the NS1 protein to bind directly to Scribble. This interaction appears to result in relocalization of Scribble into cytoplasmic puncta in infected cells, and this is associated with inhibition of Scribble's proapoptotic function. It is noteworthy that the RSKV PBM, the consensus sequence from human influenza viruses, does not mediate an association with Scribble (Fig. 1C and D). The ESEV PBM contributes to virulence in mice (21, 40), and it was previously postulated that this increased virulence is the consequence of NS1 binding to a set of PDZ proteins and the perturbation of their functions (21, 35). Consequently, it is reasonable to postulate that the binding of NS1 to Scribble via the ESEV PBM contributes to virus virulence. We have also shown that the ESEV PBM can enhance viral replication in vitro (Fig. 4). We note that introduction of the ESEA mutation in the NS1 PBM also alters phenylalanine to leucine at residue 73 in the overlapping NEP coding sequence. Given that a number of alterations in the PBM which alter the NEP coding sequence did not affect viral replication in vitro in previous studies (21, 40) and our observation that siRNA depletion of Scribble eliminates the PBM protection of apoptosis (Fig. 7C), it is unlikely that the altered NEP protein is responsible for the reduced replication of the virus that expresses the mutant ESEA PBM.
Scribble is also targeted during infections by several viruses that utilize replication strategies that differ markedly from that of influenza virus. The HTLV-1 Tax protein associates with Scribble and sequesters it into cytoplasmic puncta, similar to the sequestration by the influenza virus NS1 protein (1). The papillomavirus E6 protein binds Scribble and directs its degradation by proteasome-mediated proteolysis (28). The NS5 protein from tick-borne encephalitis virus binds directly to Scribble and inhibits the JAK/STAT pathway and interferon response by a mechanism that remains to be determined (46). These findings suggest that Scribble is involved in several cellular processes that are critical for replication of many different types of viruses.
It is not known how, or if, inhibition of the proapoptotic function of Scribble by the ESEV PBM enhances virulence. The influenza virus PB1-F2 protein induces apoptosis (5), and the antiapoptotic function of the ESEV PBM could fine-tune the extent of apoptosis during influenza virus infection. Uncontrolled apoptosis may not be optimal for influenza virus replication, and the antiapoptotic function of the NS1 PBM may limit the extent or timing of apoptosis in infected cells. Recent evidence has demonstrated that the influenza virus M2 protein functions to inhibit autophagy and promote apoptosis (11), and it has been suggested that a fine balance exists between influenza virus regulation of autophagy and apoptosis, with extensive cross talk between these two systems through the action of multiple viral proteins (37). The enhancement of viral replication by the ESEV PBM relative to the mutant ESEA PBM (Fig. 4) may be the result of a lower level of apoptosis which allows cells to remain viable longer and thereby supports greater levels of viral replication.
It is likely that the binding and inactivation of Scribble do not constitute the only mechanism mediated by the ESEV PBM that causes increased virulence in influenza virus infection, but rather, Scribble inactivation may act in concert with additional effects on other PDZ proteins that are targeted by the ESEV PBM. Our GST pull-down assays indicate that the ESEV PBM also specifically targets Dlg1, MAGI-1, MAGI-2, and MAGI-3 (Fig. 1). Perturbations of Dlg1, MAGI-1, MAGI-2, and MAGI-3 by the ESEV PBM may contribute to virulence. Scribble and Dlg1 function in common pathways to control anterior-posterior polarity (2), and it is possible that a disruption of this polarity by the ESEV PBM contributes to virulence. The MAGI proteins participate in the assembly of multiprotein complexes on the inner plasma membrane and are found at tight junctions (8, 38). It is possible that the association of the ESEV PBM with MAGI proteins may disrupt tight junctions in infected epithelial cells and this may be involved in the edema observed in the lungs in H5N1 infections (23, 47). Future studies will be required to delineate which of the multiple PDZ proteins that are targeted by the ESEV PBM play important roles in viral virulence and what molecular mechanism are involved.
We believe that the Scribble-mediated antiapoptotic activity of the ESEV PBM is not likely to act through augmentation of NS1-induced PI3K activation, since disruption of the NS1 PBM did not alter threonine-308 phosphorylation of the PI3K effector PKB in NS1-transfected cells (data not shown). When expressed from plasmid vectors, NS1 proteins with either a wild-type or mutant PBM associated equivalently with the p85β regulatory subunit of PI3K as determined by coimmunoprecipitations (data not shown).
Our immunofluorescence data show extensive colocalization of wild-type NS1 with Scribble in cytoplasmic puncta that are concentrated in perinuclear regions of infected cells (Fig. 5). These puncta may represent vesicular structures, although our preliminary analysis has not detected colocalization of NS1 and Scribble with markers for a variety of cellular vesicles. In agreement with the immunofluorescence data, biochemical fractionation of infected cells after lysis with nonionic detergent showed a quantitative redistribution of Scribble by the wild-type NS1 protein from the soluble to the insoluble fraction (Fig. 6). Taken together, the biochemical and immunofluorescence data demonstrate that expression of the NS1 protein with the ESEV PBM during infection results in a dramatic redistribution of Scribble, which is likely to be responsible for the disruption of Scribble's proapoptotic function.
The interaction between NS1 and Scribble is highly specific. Substitution of alanine for valine in the last residue of the ESEV PBM removes a single methyl group from the 230-residue NS1 protein, and this abolishes a direct interaction in vitro and the ability of NS1 to sequester Scribble in cytoplasmic puncta in influenza virus-infected cells. The NS1 protein with the ESEV PBM binds directly to the two adjacent amino-terminal PDZ domains of Scribble (Fig. 3). Because NS1 is a dimeric protein (4), it is possible that each PBM in the NS1 dimer binds to a single PDZ domain. The adenovirus E4-ORF1 protein is a precedent for this type of interaction, in which an oligomeric viral protein binds cooperatively to two tandem PDZ domains in target proteins (7). A determination of the structure of NS1 bound to Scribble will resolve this issue. The H6N6 NS1 protein with the ESEV PBM used in this study was derived from a virus isolated from a blue-winged teal. The ESEV PBM is likely to target the Scribble protein of avian species, since PDZ domains 1 and 2 of the chicken and human Scribble proteins are 88% and 70% identical. Additionally, the GLGF motifs of PDZ domains 1 and 2 are conserved between the chicken and human proteins. The GLGF motif is a critical determinant of specific interactions between PDZ domains and their ligands (34).
In summary, our finding that the ESEV PBM targets Scribble to inhibit apoptosis is the first identification of a cellular target and function of the NS1 PBM, and we anticipate that in the future, additional PDZ targets and functions will be identified for the various PBM motifs found in different influenza A virus isolates.
Acknowledgments
This work was supported by grants 5U54AI057156-05 and R21AI083396 (to A.P.R.), R01CA058541 (to R.T.J.), and RO1AI1772 (to R.M.K.) from the National Institutes of Health.
We thank Clayton Naeve (St. Jude Children's Research Hospital) for NS1 plasmids and Robert Lamb (Northwestern University) for plasmids used for influenza virus reverse genetics. We thank Bob Couch and Diane Nino for help in generating viral stocks.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Arpin-Andre, C., and J. M. Mesnard. 2007. The PDZ domain-binding motif of the human T cell leukemia virus type 1 tax protein induces mislocalization of the tumor suppressor hScrib in T cells. J. Biol. Chem. 282:33132-33141. [DOI] [PubMed] [Google Scholar]
- 2.Assemat, E., E. Bazellieres, E. Pallesi-Pocachard, A. Le Bivic, and D. Massey-Harroche. 2008. Polarity complex proteins. Biochim. Biophys. Acta 1778:614-630. [DOI] [PubMed] [Google Scholar]
- 3.Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. [DOI] [PubMed] [Google Scholar]
- 4.Bornholdt, Z. A., and B. V. Prasad. 2008. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, S. H., R. S. Weiss, K. K. Frese, B. V. Prasad, and R. T. Javier. 2008. Functionally distinct monomers and trimers produced by a viral oncoprotein. Oncogene 27:1412-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin, J. L., K. Yoshiura, P. J. Dempsey, G. Bogatcheva, L. Jeyakumar, K. S. Meise, R. S. Pearsall, D. Threadgill, and R. J. Coffey. 2005. Identification of MAGI-3 as a transforming growth factor-alpha tail binding protein. Exp. Cell Res. 303:457-470. [DOI] [PubMed] [Google Scholar]
- 9.Frese, K. K., I. J. Latorre, S. H. Chung, G. Caruana, A. Bernstein, S. N. Jones, L. A. Donehower, M. J. Justice, C. C. Garner, and R. T. Javier. 2006. Oncogenic function for the Dlg1 mammalian homolog of the Drosophila discs-large tumor suppressor. EMBO J. 25:1406-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack, M. U., R. A. Albrecht, T. Urano, K. S. Inn, I. C. Huang, E. Carnero, M. Farzan, S. Inoue, J. U. Jung, and A. Garcia-Sastre. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gannage, M., D. Dormann, R. Albrecht, J. Dengjel, T. Torossi, P. C. Ramer, M. Lee, T. Strowig, F. Arrey, G. Conenello, M. Pypaert, J. Andersen, A. Garcia-Sastre, and C. Munz. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 103:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale, B. G., R. E. Randall, J. Ortin, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:10-76. [DOI] [PubMed] [Google Scholar]
- 15.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann, C. H., and M. A. Mancini. 2001. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114:1491-1503. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, C. H., and A. P. Rice. 1993. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology 197:601-608. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, A. Y., and M. Sheng. 2002. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277:5699-5702. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, D., M. J. Hossain, D. Hickman, D. R. Perez, and R. A. Lamb. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 105:4381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javier, R. T. 2008. Cell polarity proteins: common targets for tumorigenic human viruses. Oncogene 27:7031-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korteweg, C., and J. Gu. 2008. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 172:1155-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 25.Latorre, I. J., M. H. Roh, K. K. Frese, R. S. Weiss, B. Margolis, and R. T. Javier. 2005. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J. Cell Sci. 118:4283-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896-899. [DOI] [PubMed] [Google Scholar]
- 28.Massimi, P., A. Shai, P. Lambert, and L. Banks. 2008. HPV E6 degradation of p53 and PDZ containing substrates in an E6AP null background. Oncogene. 27:1800-1804. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin, M., R. Hale, D. Ellston, S. Gaudet, R. A. Lue, and A. Viel. 2002. The distribution and function of alternatively spliced insertions in hDlg. J. Biol. Chem. 277:6406-6412. [DOI] [PubMed] [Google Scholar]
- 30.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min, J. Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236-243. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 34.Nourry, C., S. G. Grant, and J. P. Borg. 2003. PDZ domain proteins: plug and play! Sci. STKE 2003:RE7. [DOI] [PubMed] [Google Scholar]
- 35.Obenauer, J. C., J. Denson, P. K. Mehta, X. Su, S. Mukatira, D. B. Finkelstein, X. Xu, J. Wang, J. Ma, Y. Fan, K. M. Rakestraw, R. G. Webster, E. Hoffmann, S. Krauss, J. Zheng, Z. Zhang, and C. W. Naeve. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576-1580. [DOI] [PubMed] [Google Scholar]
- 36.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 37.Rossman, J. S., and R. A. Lamb. 2009. Autophagy, apoptosis, and the influenza virus M2 protein. Cell Host Microbe 6:299-300. [DOI] [PubMed] [Google Scholar]
- 38.Sakurai, A., S. Fukuhara, A. Yamagishi, K. Sako, Y. Kamioka, M. Masuda, Y. Nakaoka, and N. Mochizuki. 2006. MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion. Mol. Biol. Cell 17:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skelton, N. J., M. F. Koehler, K. Zobel, W. L. Wong, S. Yeh, M. T. Pisabarro, J. P. Yin, L. A. Lasky, and S. S. Sidhu. 2003. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 278:7645-7654. [DOI] [PubMed] [Google Scholar]
- 40.Soubies, S. M., C. Volmer, G. Croville, J. Loupias, B. Peralta, P. Costes, C. Lacroux, J. L. Guerin, and R. Volmer. 2010. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J. Virol. 84:6733-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaller, M. R. 2006. Act globally, think locally: systems biology addresses the PDZ domain. ACS Chem. Biol. 1:207-210. [DOI] [PubMed] [Google Scholar]
- 42.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 76:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takizawa, S., K. Nagasaka, S. Nakagawa, T. Yano, K. Nakagawa, T. Yasugi, T. Takeuchi, T. Kanda, J. M. Huibregtse, T. Akiyama, and Y. Taketani. 2006. Human scribble, a novel tumor suppressor identified as a target of high-risk HPV E6 for ubiquitin-mediated degradation, interacts with adenomatous polyposis coli. Genes Cells 11:453-464. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 45.Tonikian, R., Y. Zhang, S. L. Sazinsky, B. Currell, J. H. Yeh, B. Reva, H. A. Held, B. A. Appleton, M. Evangelista, Y. Wu, X. Xin, A. C. Chan, S. Seshagiri, L. A. Lasky, C. Sander, C. Boone, G. D. Bader, and S. S. Sidhu. 2008. A specificity map for the PDZ domain family. PLoS Biol. 6:e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werme, K., M. Wigerius, and M. Johansson. 2008. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol. 10:696-712. [DOI] [PubMed] [Google Scholar]
- 47.Xu, T., J. Qiao, L. Zhao, G. Wang, G. He, K. Li, Y. Tian, M. Gao, J. Wang, H. Wang, and C. Dong. 2006. Acute respiratory distress syndrome induced by avian influenza A (H5N1) virus in mice. Am. J. Respir. Crit. Care Med. 174:1011-1017. [DOI] [PubMed] [Google Scholar]
- 48.Zhan, L., A. Rosenberg, K. C. Bergami, M. Yu, Z. Xuan, A. B. Jaffe, C. Allred, and S. K. Muthuswamy. 2008. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135:865-878. [DOI] [PMC free article] [PubMed] [Google Scholar]