Abstract
Vaccines designed to elicit AIDS virus-specific CD8+ T cells should engender broad responses. Emerging data indicate that alternate reading frames (ARFs) of both human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) encode CD8+ T cell epitopes, termed cryptic epitopes. Here, we show that SIV-specific CD8+ T cells from SIV-infected rhesus macaques target 14 epitopes in eight ARFs during SIV infection. Animals recognized up to five epitopes, totaling nearly one-quarter of the anti-SIV responses. The epitopes were targeted by high-frequency responses as early as 2 weeks postinfection and in the chronic phase. Hence, previously overlooked ARF-encoded epitopes could be important components of AIDS vaccines.
CD8+ T cells control AIDS virus replication (5, 9, 17, 21); however, their role in prophylactic AIDS vaccines is topic for debate. CD8+ T cells recognize infected cells by the presence of virus-derived peptides bound to major histocompatibility complex class I (MHC-I) molecules on the cell surface. The nine defined human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) proteins have long been thought to be the sole sources of virus-derived, MHC-I bound epitopes because researchers assume the classical viral protein annotations to represent the totality of the viral translation products despite increasing evidence to the contrary. Our laboratory and others have shown that MHC-I-bound epitopes can be derived from translation of viral alternate reading frames (ARFs), termed cryptic epitopes (2, 4, 6, 10, 15, 16). Collectively, these data indicate that cryptic CD8+ T cell responses might be more common, and more important, than previously appreciated.
Rhesus macaques infected with a molecularly cloned strain of SIV offer several important advantages for studying specific CD8+ T cell responses (22). Since the exact sequence of the inoculum is known, it is possible to track precisely the CD8+ T cell responses against all possible viral ARF translations. We used a gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay to screen SIVmac239-infected rhesus macaques in both the acute and chronic stages of infection for T cell responses against an overlapping peptide set (15-mers, overlapping by 11) spanning the entire potential ARF-encoded proteome in the “sense” direction. Altogether, we defined eight novel MHC-I epitope-containing translation products putatively ranging in length from 32 to 71 amino acids, each containing from one to five epitopes. We found that, in some animals, the cryptic epitope-directed response can be a dominant component of the total antiviral response, comprising nearly a quarter of the total response. Together, our data indicate that translation and immune recognition of viral ARFs are common features of AIDS virus infection.
Search for ARF-specific responses.
To search for cryptic epitope-specific CD8+ T cell responses, we synthesized peptides (15-mers, overlapping by 11) spanning all of the potential translation products of SIVmac239 (Fig. 1, gray boxes). We then used the IFN-γ ELISPOT assay to test SIV-infected (and uninfected) macaques during the acute and chronic phases of infection for responses against these peptides. The use of overlapping peptide sets, as opposed to those algorithmically predicted to bind specific MHC molecules, has the distinct advantage of not requiring a priori knowledge of which peptides either will be translated or will bind specific MHC molecules. The unfortunate trade-off, however, is that these peptides likely are less sensitive for detecting responses, particularly low-frequency responses, due to their requirement for processing prior to MHC-I binding and presentation (25). We used overlapping peptides to examine the repertoire of ARF-specific CD8+ T cells in SIV-infected macaques, given the large number of MHC-I mRNAs expressed by the rhesus macaque MHC genomic region (18) and the limited number of algorithms for predicting macaque MHC-I-peptide binding (20).
FIG. 1.
A peptide set, 15-mers overlapping by 11, spanning all potential “sense” alternate reading frames (ARFs; shown in gray), was synthesized. Classically defined, functional proteins are shown in black.
Detection of ARF-specific responses.
We examined whether recognition of ARF-encoded CD8+ T cell epitopes was a common feature of AIDS virus infection by first screening chronic-phase T cell responses in SIV-infected macaques. Pools of 8 to 10 overlapping peptides were used for ELISPOT analysis. We detected T cell responses against peptides representing eight distinct translation products. These immunogenic peptides are encoded by ARFs of Pol, Tat/Vpr, Env, and Env/Rev and putatively range in size from 32 to 71 amino acids. Not all of the predicted translation products contain translation initiation codons. In such cases, alternate start codons, such as CUG (encoding leucine) (14), or ribosomal frameshifts (3, 11, 19), might account for translation.
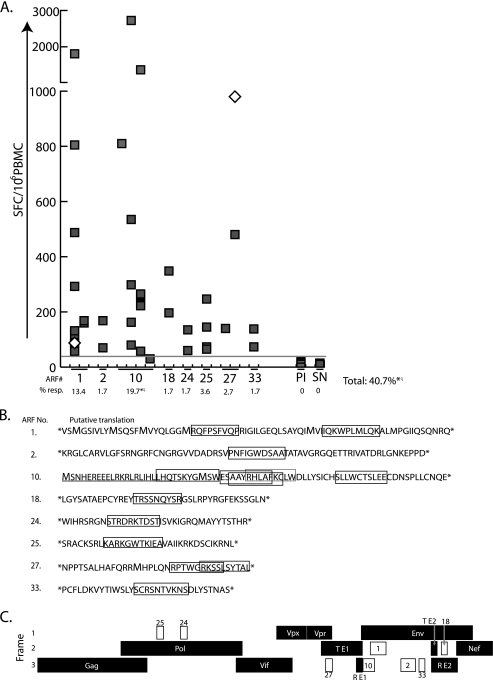
To begin to identify the actual determinants recognized by these CD8+ T cell responses, we next performed ELISPOT assays to break down the responses to individual 15-mer peptides. The magnitude of the positive responses varied widely, from 55 to >2,500 spot-forming cells (SFC) per 106 lymphocytes (Fig. 2A). An additional response, against ARF10, was not significant by our analysis, but we were able to determine its existence through the follow-up experiments outlined below. In most cases, the responses broke down to either a single peptide or two overlapping peptides, with both 15-mers containing the likely epitope determinant. However, in three cases, the pool responses broke down to more than one distinct epitope. ARF1 encodes two epitopes, ARF27 encodes two, and ARF10 encodes five. All responses at the limit of statistical significance were verified by in vitro CD8+ T cell growth cultures (data not shown). Additionally, responses against two epitopes (15-mers from ARF1, as well as ARF27) were detected in animals vaccinated with SIVmac239 D-Nef (Fig. 2A, empty diamonds) prior to pathogenic SIV challenge, demonstrating that an effective, live attenuated vaccine induces cryptic epitope-specific responses.
FIG. 2.
Magnitude of cryptic epitope-specific T cell responses and locations of the epitopes. (A) An IFN-γ ELISPOT assay was used to measure the magnitude of cryptic epitope-specific responses. Each square represents the results for one animal, and empty diamonds represent the results for animals vaccinated with live-attenuated SIVmac239 D-Nef, assayed prior to pathogenic SIV challenge. The individual ARFs are indicated on the x axis, with each column representing an individual 15-mer from that ARF. In cases where the positive response was against two overlapping peptides, we show the 15-mer with the largest response based on the reasoning that the optimal epitope is most efficiently liberated from this peptide. The horizontal gray line represents the limit of statistical significance. PI, preinfection samples from at least one animal responding to each cryptic epitope; SN, SIV-naïve animals (n = 20). The percentage of animals (out of 112 tested) responding to any 15-mer within a given ARF is shown below each ARF. Responses were considered positive if the response exceeded 50 SFC per 106 cells and were determined using a one-tailed t test and an alpha of 0.05, where the null hypothesis (H0) is as follows: background level ≥ treatment level. An asterisk indicates that these percentages (from ARF10 and the total) include the results for five additional animals that responded to the RW9 epitope in ARF10, as part of a previous study (16), but that were not tested for responses against the other ARFs due to lack of samples. Hence, these percentages are calculated as the fraction of 117 animals showing responses (rather than for 112 animals for the other ARFs). The percentages marked with ζ also include one animal that made a real, but statistically nonsignificant, response to the SE10 epitope in ARF10. (B) Putative ARF-encoded translation products and the immunogenic epitopes from SIVmac239. Translation products were given arbitrary identification numbers. Predicted amino acid sequences between stop codons are shown, except for that for ARF10, which is translated by extension from the first exon of Rev (underlined) (15). The newly mapped epitopes are boxed in black, and the previously published RW9 epitope (in ARF10) (16) is boxed in gray. Potential translation initiation codons are shown in an enlarged font. (C) The ARFs showing positive responses are represented by white boxes, with their ARF numbers corresponding to those provided in panel B. RE1 and RE2, Rev exons 1 and 2, respectively. TE1 and TE2, Tat exons 1 and 2, respectively.
Next, we mapped the minimal determinants of the CD8+ T cell responses against the proteins encoded by ARFs. To do this, we stimulated lymphocytes from the responding animal with autologous, transformed B-lymphoblastoid cell lines (BLCL) pulsed with 15-mer peptides. Cell lines were tested for reactivity to 9-, 10-, and 11-mer peptides by IFN-γ and tumor necrosis factor alpha (TNF-α) production using an intracellular cytokine-staining (ICS) assay. The putative proteins and the minimal mapped peptides are depicted in Fig. 2B, and their locations in the SIVmac239 genome are depicted in Fig. 2C (all mapped epitopes and their overlapping protein and short names are summarized in Fig. S1 in the supplemental material). We were not able to stimulate cell lines for responses targeting two of the epitopes (ST15 from ARF2 and LW11 from ARF10), because BLCL from the responding animals could not be expanded. In these cases, time point-matched peripheral blood mononuclear cell (PBMC) samples were used in a second ELISPOT assay to determine the optimal antigenic epitope. We expanded a T cell line against the SE10 epitope in ARF10 despite the response not being significant by statistical analyses. This response had the appearance of a positive response; i.e., there were consistent spot numbers in replicate wells with no background. It is possible that other responses were missed due to their being below statistical significance. Importantly, we verified all responses (either from PBMCs or with T cell lines or both) using newly synthesized peptides from alternate vendors to ensure that responses were not directed against low-level contaminants in the original peptide preparations (7).
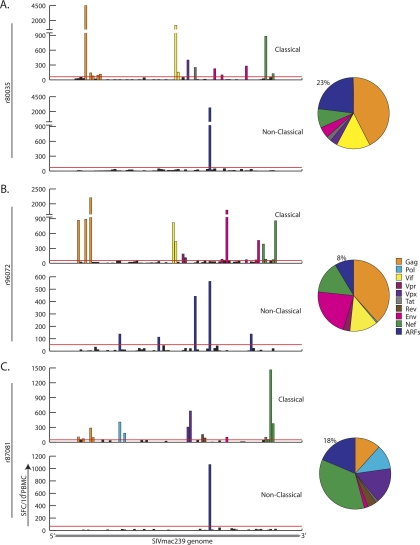
We next measured the total CD8+ T cell response in three chronically SIV-infected macaques. We used our ARF peptide set and pools of overlapping peptides (each pool containing 10 peptides) representing all of the known SIV proteins. Specifically, we tested animals that made high-frequency responses against cryptic epitopes (animals r80035 and r87081) or that made responses against several cryptic epitopes (animal r96072). Hence, these three animals represent the potential (rather than the typical) magnitude and breadth of cryptic epitope-specific responses. Animal r80035 (Mamu-A*01+) made a high-frequency response against a pool containing an immunodominant Gag epitope (CM9) (1) (Fig. 3A, top panel), as well as high-frequency responses against the Vif and Nef proteins. Strikingly, the protein targeted with the second most frequent response was ARF10, encoded by an ARF of Env (Fig. 3A, bottom panel), comprising 23% of the total SIV-specific response (Fig. 3A, pie chart). The ARF10-specific response was directed against two overlapping 10-mer epitopes, EF10 and AC10, and the data presented represent the summation of those responses. Animal r96072 is Mamu-A*02+ and Mamu-B*17+ and made strong responses against Gag, Vif, Env, and Nef (Fig. 3B, upper panel). This animal also made five distinct cryptic epitope-specific responses—against the EF10 epitope in ARF10, the TR9 epitope in ARF18, the ST10 epitope in ARF24, the KA11 epitope in ARF25, and the RL10 epitope in ARF27 (Fig. 3B, lower panel). Together, these responses comprised 8% of the total SIV-specific response (Fig. 3B, pie chart). Animal r87081 does not express MHC-I molecules known to present dominant SIV-derived peptides. This animal made a high-frequency response against a pool of Nef-derived peptides (Fig. 3C, upper panel) as well as a high-frequency response against the EF10 epitope in ARF10 (Fig. 3C, lower panel). Indeed, the EF10 response comprised 18% of the total SIV-specific response in this animal (Fig. 3C, pie chart).
FIG. 3.
The total SIV-specific T cell response was assayed in three animals. We measured PBMC samples for responses against overlapping peptides representing the known SIV proteome (“classical”) and the potential translations of the ARFs (“nonclassical”). Pools of peptides are represented along the x axis in the order in which they appear in the virus, left to right, 5′ to 3′. Significant responses to different proteins are color coded by protein, while all cryptic epitope responses are shown in dark blue. The total contribution of each viral protein and the summation of the cryptic responses are shown in the pie charts. Animal r80035 (A) was Mamu-A*01+; animal r96072 (B) was Mamu-A*02+ and B*17+; and animal r87081 (C) was Mamu-A*11+, a genotype which does not typically present dominant CD8+ T cell epitopes.
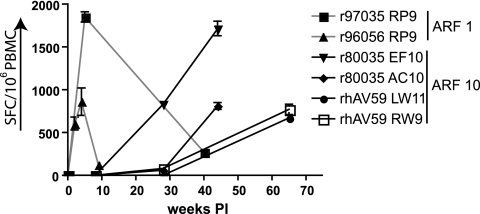
We were able to find both high-frequency responses against single cryptic epitopes and broad responses against several epitopes. We were next interested in the in vivo kinetics of the cryptic epitope-specific responses. The availability of samples limited this search to peptides encoded by ARF1 and ARF10. The responses against the epitopes depicted in Fig. 1 were detected at a variety of time points during SIV infection. To determine when these responses arose, we thawed acute- and chronic-phase PBMC samples from responding animals. Specifically, we thawed samples from two animals responding to the RP9 epitope encoded by ARF1 and from two animals each responding to two different epitopes encoded by ARF10. Surprisingly, high-frequency CD8+ T cells recognized the RP9 epitope, encoded by ARF1, at 2 weeks postinfection (Fig. 4). This response then waned during chronic infection, possibly due to viral escape. In contrast, responses against four of the five epitopes encoded by ARF10 were not detected in the acute phase and were evident later in infection (Fig. 4). The reasons for this are unclear. We showed previously that the RW9 epitope from ARF10 is presented regularly during a single cycle of in vitro SIV infection (15), demonstrating that the in vivo kinetics of the T cell responses are probably not due to the appearance of mutations leading to the translation of ARF10. These data suggest that CD8+ T cells against cryptic epitopes can be high frequency in either the acute or chronic phase of infection.
FIG. 4.
In vivo kinetics of cryptic epitope-specific responses. The magnitude of responses was tested at various time points during SIV infection. Animals responding to peptides encoded by ARF1 and ARF10 were tested in various acute- and chronic-phase time points. Responses against the RP9 epitope in ARF1 (gray lines) were high frequency in the acute phase but diminished rapidly. In contrast, responses against epitopes in ARF10 (black lines) were detected exclusively in the chronic phase, for unknown reasons. PI, postinfection.
Our data suggest that cryptic epitope-specific responses are an important component of the total AIDS virus-specific response. In fact, our data likely represent a substantial underestimate of the total contribution of cryptic epitope-specific responses for three reasons. First, we did not search for epitopes encoded in the antisense direction. Such responses have been detected in HIV-infected humans. Second, we focused almost entirely on frozen samples. Variation in cell viability may have led to missed responses, particularly low-frequency responses. Finally, we used 15-mer peptides for our original screen. These peptides are likely to be significantly less sensitive for detecting many responses than actual optimal epitope peptides.
There is an urgent need for an AIDS vaccine that lowers viral loads and prevents transmission. Recent attempts in humans to elicit broad anti-HIV T cell responses with a vaccine have failed. However, vaccine modalities with similar goals have achieved some success using the rhesus macaque model of HIV infection (12, 13, 23, 24), providing some hope that such a vaccine might succeed. Though it is still unclear which vaccine features determine eventual success, the breadth of the immune response induced (in both the CD8+ and CD4+ T cell compartment) is a good candidate (8, 9). Hence, CD8+ T cells directed against cryptic epitopes may be an important component of the total AIDS virus-specific T cell response. Our data suggest that the inclusion of these novel ARFs in AIDS vaccines could broaden the T cell response and enhance viral control.
Supplementary Material
Acknowledgments
We thank Jonah Sacha and Eva Rakasz for helpful discussions. The peptides spanning the “normal” SIV proteome were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
This work was supported by NIH grants R37 AI052056, R01 A1049120, R01 AI076114, R24 RR015371, R24 RR016038, and R21 PRJ27JP to D.I.W. and grant P51 RR000167 from the National Center for Research Resources, a component of the National Institutes of Health (to the Wisconsin National Primate Research Center, University of Wisconsin---Madison). This research was conducted in part at a facility constructed with support from Research Facilities Improvement grants RR15459-01 and RR020141-01.
Footnotes
Published ahead of print on 25 August 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allen, T. M., B. R. Mothe, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A*01: implications for vaccine design and testing. J. Virol. 75:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal, A., J. Carlson, J. Yan, O. T. Akinsiku, M. Schaefer, S. Sabbaj, A. Bet, D. N. Levy, S. Heath, J. Tang, R. A. Kaslow, B. D. Walker, T. Ndung'u, P. J. Goulder, D. Heckerman, E. Hunter, and P. A. Goepfert. 2010. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J. Exp. Med. 207:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baril, M., D. Dulude, K. Gendron, G. Lemay, and L. Brakier-Gingras. 2003. Efficiency of a programmed −1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M. RNA 9:1246-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, C. T., J. M. Carlson, C. J. Brumme, K. L. Hartman, Z. L. Brumme, L. M. Henry, P. C. Rosato, A. Piechocka-Trocha, M. A. Brockman, P. R. Harrigan, D. Heckerman, D. E. Kaufmann, and C. Brander. 2010. Viral adaptation to immune selection pressure by HLA class I-restricted CTL responses targeting epitopes in HIV frameshift sequences. J. Exp. Med. 207:61-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardinaud, S., A. Moris, M. Fevrier, P. S. Rohrlich, L. Weiss, P. Langlade-Demoyen, F. A. Lemonnier, O. Schwartz, and A. Habel. 2004. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J. Exp. Med. 199:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currier, J. R., L. M. Galley, H. Wenschuh, V. Morafo, S. Ratto-Kim, C. M. Gray, L. Maboko, M. Hoelscher, M. A. Marovich, and J. H. Cox. 2008. Peptide impurities in commercial synthetic peptides and their implications for vaccine trial assessment. Clin. Vaccine Immunol. 15:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, M. E. Feeney, M. M. Addo, M. Lichterfeld, M. P. Lahaie, E. Pae, A. G. Wurcel, T. Roach, M. A. St John, M. Altfeld, F. M. Marincola, C. Moore, S. Mallal, M. Carrington, D. Heckerman, T. M. Allen, J. I. Mullins, B. T. Korber, P. J. Goulder, B. D. Walker, and C. Brander. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7:173-178. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. Capuano III, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrison, K. E., S. Champiat, V. A. York, A. T. Agrawal, E. G. Kallas, J. N. Martin, F. M. Hecht, S. G. Deeks, and D. F. Nixon. 2009. Transcriptional errors in human immunodeficiency virus type 1 generate targets for T-cell responses. Clin. Vaccine Immunol. 16:1369-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giedroc, D. P., and P. V. Cornish. 2009. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 139:193-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen, S. G., C. Vieville, N. Whizin, L. Coyne-Johnson, D. C. Siess, D. D. Drummond, A. W. Legasse, M. K. Axthelm, K. Oswald, C. M. Trubey, M. J. Piatak, J. D. Lifson, J. A. Nelson, M. A. Jarvis, and L. J. Picker. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, J., K. L. O'Brien, D. M. Lynch, N. L. Simmons, A. La Porte, A. M. Riggs, P. Abbink, R. T. Coffey, L. E. Grandpre, M. S. Seaman, G. Landucci, D. N. Forthal, D. C. Montefiori, A. Carville, K. G. Mansfield, M. J. Havenga, M. G. Pau, J. Goudsmit, and D. H. Barouch. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malarkannan, S., T. Horng, P. P. Shih, S. Schwab, and N. Shastri. 1999. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity 10:681-690. [DOI] [PubMed] [Google Scholar]
- 15.Maness, N. J., J. B. Sacha, S. M. Piaskowski, K. L. Weisgrau, E. G. Rakasz, G. E. May, M. B. Buechler, A. D. Walsh, N. A. Wilson, and D. I. Watkins. 2009. Novel translation products from simian immunodeficiency virus SIVmac239 Env-encoding mRNA contain both Rev. and cryptic T-cell epitopes. J. Virol. 83:10280-10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maness, N. J., L. E. Valentine, G. E. May, J. Reed, S. M. Piaskowski, T. Soma, J. Furlott, E. G. Rakasz, T. C. Friedrich, D. A. Price, E. Gostick, A. L. Hughes, J. Sidney, A. Sette, N. A. Wilson, and D. I. Watkins. 2007. AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape. J. Exp. Med. 204:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otting, N., C. M. Heijmans, R. C. Noort, N. G. de Groot, G. G. Doxiadis, J. J. van Rood, D. I. Watkins, and R. E. Bontrop. 2005. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 102:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pande, S., A. Vimaladithan, H. Zhao, and P. J. Farabaugh. 1995. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol. Cell. Biol. 15:298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters, B., H. H. Bui, J. Sidney, Z. Weng, J. T. Loffredo, D. I. Watkins, B. R. Mothe, and A. Sette. 2005. A computational resource for the prediction of peptide binding to Indian rhesus macaque MHC class I molecules. Vaccine 23:5212-5224. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 22.Valentine, L. E., and D. I. Watkins. 2008. Relevance of studying T cell responses in SIV-infected rhesus macaques. Trends Microbiol. 16:605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson, N. A., B. F. Keele, J. S. Reed, S. M. Piaskowski, C. E. MacNair, A. J. Bett, X. Liang, F. Wang, E. Thoryk, G. J. Heidecker, M. P. Citron, L. Huang, J. Lin, S. Vitelli, C. D. Ahn, M. Kaizu, N. J. Maness, M. R. Reynolds, T. C. Friedrich, J. T. Loffredo, E. G. Rakasz, S. Erickson, D. B. Allison, M. J. Piatak, J. D. Lifson, J. W. Shiver, D. R. Casimiro, G. M. Shaw, B. H. Hahn, and D. I. Watkins. 2009. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J. Virol. 83:6508-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, D. H. O'Connor, T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yewdell, J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533-543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.