Abstract
Previous studies with Venezuelan equine encephalitis virus and Sindbis virus (SINV) indicate that alphaviruses are capable of suppressing the cellular response to type I and type II interferons (IFNs) by disrupting Jak/STAT signaling; however, the relevance of this signaling inhibition toward pathogenesis has not been investigated. The relative abilities of neurovirulent and nonneurovirulent SINV strains to downregulate Jak/STAT signaling were compared to determine whether the ability to inhibit IFN signaling correlates with virulence potential. The adult mouse neurovirulent strain AR86 was found to rapidly and robustly inhibit tyrosine phosphorylation of STAT1 and STAT2 in response to IFN-γ and/or IFN-β. In contrast, the closely related SINV strains Girdwood and TR339, which do not cause detectable disease in adult mice, were relatively inefficient inhibitors of STAT1/2 activation. Decreased STAT activation in AR86-infected cells was associated with decreased activation of the IFN receptor-associated tyrosine kinases Tyk2, Jak1, and Jak2. To identify the viral factor(s) involved, we infected cells with several panels of AR86/Girdwood chimeric viruses. Surprisingly, we found that a single amino acid determinant, threonine at nsP1 position 538, which is required for AR86 virulence, was also required for efficient disruption of STAT1 activation, and this determinant fully restored STAT1 inhibition when it was introduced into the avirulent Girdwood background. These data indicate that a key virulence determinant plays a critical role in downregulating the response to type I and type II IFNs, which suggests that the ability of alphaviruses to inhibit Jak/STAT signaling relates to their in vivo virulence potential.
Members of the genus Alphavirus in the family Togaviridae include a variety of human pathogens with a nearly global distribution. These viruses are transmitted through mosquito vectors and are considered a threat due to their potential to cause large-scale epidemics. The Old World alphaviruses, such as chikungunya virus (CHIKV) and Ross River virus (RRV), have been linked to explosive epidemics of infectious arthritis, while the New World alphaviruses Venezuelan equine encephalitis virus (VEEV) and eastern equine encephalitis virus (EEEV) have caused sporadic outbreaks of potentially fatal encephalitis. Sindbis virus (SINV), the prototype alphavirus, is an Old World alphavirus responsible for cases of self-limited arthralgia in humans. Infection of mice with SINV has also provided an excellent model of alphavirus-induced encephalomyelitis (14, 31, 33, 40, 41).
Upon alphavirus infection, viral RNA is recognized by host pattern recognition receptors, including the cytoplasmic RNA sensors PKR, MDA-5, and/or RIG-I, which can activate IRF-3/7-dependent signaling pathways to induce beta interferon (IFN-β) and IFN-α4 production (23, 30, 45, 52). Infection of mice with SINV results in detectable type I IFN (IFN-α/β) levels in the serum by 12 h postinfection (hpi) (33). IFN-γ (type II IFN), which is secreted by specific immune effector cells, is detectable in the serum at slightly later times (24 h) post-SINV infection (33). The cellular response to secreted IFN-α/β and IFN-γ involves separate but overlapping signaling cascades that result in transcription of IFN-stimulated genes (ISGs), several of which have known antiviral functions. These signaling pathways have been well studied (for detailed reviews, see references 46 and 53). In brief, IFN-α/β and IFN-γ bind distinct, ubiquitously expressed cell surface receptors, the IFN-α/β receptor (IFNAR) and the IFN-γ receptor (IFNGR), respectively. Ligation of the IFNAR results in dimerization of the receptor subunits, IFNAR1 and IFNAR2, which allows apposition and autophosphorylation of Jak1/Tyk2 kinases that constitutively associate with each subunit. Once activated, these Jaks phosphorylate the receptor subunits, allowing the recruitment and phosphorylation of STAT1 and STAT2, which dimerize and associate with IRF-9 to form the ISGF3 complex, which binds IFN-stimulated response elements (ISREs) to drive transcription. In contrast, the activated type II IFN receptor complex is composed of IFNGR1/2 subunits and Jak1/Jak2 kinases that activate STAT1, which predominantly forms homodimers to drive expression of IFN-γ-stimulated genes containing IFN-γ-activated sequence (GAS) elements in their promoters. The presence of both ISREs and GAS elements within a single ISG promoter partially explains the overlap between the IFN-γ and IFN-α/β responses. Thus, STAT1 is a central component of the type I and type II IFN responses.
The type I IFN system is known to play a critical role in the control of most virus families, including alphaviruses (for an excellent review, see reference 49). Compared to immunocompetent adult mice that control most SINV strains, mice with targeted deletions of the IFNAR succumb rapidly to infection (50). The average survival time of SINV-infected mice is reduced further when they are doubly deficient in both the IFNAR and the IFNGR (51), suggesting that both type I and type II IFNs are important in limiting SINV replication and pathogenesis in adult mice. The IFN-γ response also has direct antiviral activity, but it is thought to play an important role at later times post-SINV infection, when CD8+ and CD4+ T-cell-derived IFN-γ acts together with antiviral antibody to mediate noncytolytic clearance of SINV from central nervous system (CNS) neurons (for a detailed review, see reference 24). Since STAT1 is central to both type I and type II IFN responses, nonfatal SINV infection results in 100% lethality in STAT1−/− mice (9, 51), although some STAT1-independent IFN-α/β and IFN-γ responses with anti-SINV activities have been described (22).
Like most viruses, alphaviruses employ strategies to interfere with the host type I IFN response. Most work on these viruses has focused on virus-mediated nonspecific shutoff of cellular transcription and translation (2, 3, 17, 19-21). Recent studies with both SINV and VEEV suggested that both Old World and New World alphaviruses are also able to disrupt signaling events required for the cellular response to IFN-α/β and IFN-γ, namely, by inhibiting the accumulation of tyrosine-phosphorylated STAT1 and STAT2 (57, 63). This activity appeared to be mediated by the viral nonstructural proteins and was independent of shutoff of host macromolecular synthesis (57). However, the importance and relevance of upstream Jak/STAT signaling inhibition in the face of potent and generalized shutoff of ISG transcription have not been demonstrated. In fact, the transcriptional induction of several ISGs did not correlate with the degree to which STAT1/2 activation was inhibited in mouse neuron cultures infected with VEEV replicon particles (VRP) (63).
To further define the potential role that viral inhibition of Jak/STAT signaling plays in alphavirus pathogenesis, we evaluated whether viruses with differing virulence profiles exhibited the ability to inhibit Jak/STAT signaling. We focused our analysis on two SINV isolates, S.A.AR86 (called AR86 hereafter) and a closely related strain, GirdwoodS.A. (called Girdwood hereafter). In adult mice, a virus derived from the infectious clone of AR86 (S300) causes lethal disease following intracranial inoculation (27, 28, 58, 60), while the closely related Girdwood virus (clone G100) is avirulent, even though both S300 and G100 replicate to similar levels within the CNS of mice at early times postinfection (60). The difference in virulence is mediated by four genetic determinants within AR86, which result in a gain of full virulence when introduced into the avirulent G100 background. Here we report that the adult mouse virulence potential of SINV strains S300, G100, and TR339 correlates directly with the relative abilities of these viruses to disrupt the activation of Jak proteins and/or STAT1/2 in response to both IFN-β and IFN-γ. Strikingly, we found that a single virulence determinant unique to AR86, a threonine at nsP1 position 538, is both necessary and sufficient for rapid and efficient inhibition of STAT1 activation. The previously demonstrated importance of this determinant for adult mouse neurovirulence and for avoiding clearance from the CNS (27, 60) suggests that inhibition of Jak/STAT signaling contributes to alphavirus pathogenesis, perhaps through downregulation of the response to type I IFNs and/or through suppression of noncytolytic SINV clearance from CNS neurons, which involves IFN-γ signaling (5-8).
MATERIALS AND METHODS
Cell culture and reagents.
Vero-81 cells (designated Vero cells; ATCC CCL-81) and BHK-21 cells were grown at 37°C under 5% CO2. Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM)-F-12 medium (Gibco) supplemented with 10% fetal bovine serum (HyClone), l-glutamine (0.29 mg/ml; Gibco), nonessential amino acids (1×; Gibco), penicillin-streptomycin (1×; Gibco), and sodium bicarbonate (final concentration of 1.2 g/liter; Gibco). BHK-21 cells were maintained in α-minimal essential medium (α-MEM) (Gibco) containing 10% donor calf serum (DCS; HyClone) and 10% tryptose phosphate broth (Sigma) and supplemented with l-glutamine and penicillin-streptomycin as described above. For virus and replicon production in BHK-21 cells, fetal bovine serum (10%; Lonza) was used in place of DCS. Recombinant human IFN-β (Calbiochem) was resuspended in sterile DMEM containing 10% fetal bovine serum, and aliquots were stored at −80°C. The biologic activity of each IFN-β preparation was determined by a type I IFN bioassay on A549 cells, where protection from encephalomyocarditis virus (EMCV)-induced cytopathic effect was scored relative to that of the NIH human IFN-β standard (Gb23-902-531), as previously described (54). Recombinant human IFN-γ was used at the activity reported by the manufacturer (R&D Systems).
Virus and replicon production.
The infectious clone-derived viruses used are listed in Table 1, which annotates the mutations that each clone carries. Viruses were generated from plasmid templates named with a “p” prefix (i.e., S300 virus was generated from plasmid pS300). S300, G100, S350, G106, S363, S340, S343, and S344 were all generated as previously described (60). Both pS300 and pS55, which differ only in the linearization site used, encode wild-type AR86; thus, S300 and the previously designated S55 strain (27, 28) are synonymous. Similarly, S51 is synonymous with S340 (27, 60). Primer-directed mutagenesis of pG100 was used to construct clones pG119, pG121, and pG120 as described previously (60). pTR339, which carries the consensus A.R.339 sequence, was generated previously by replacing cell culture-adaptive mutations in the E2 gene and is representative of the original virulent A.R.339 natural isolate (34, 42).
TABLE 1.
SINV infectious clones used for this study
| Virus | Background strain and mutations |
|---|---|
| S300 | Wild-type S.A.AR86 |
| G100 | Wild-type GirdwoodS.A. |
| TR339 | Wild-type A.R.339 |
| G106 | AR86 nt 43 to 6411 in Girdwood background |
| S350 | Girdwood nt 43 to 6411 in AR86 background |
| S363 | AR86 with nsP1 position 538 mutation (Thr→Ile), 18-aa insertion of G100 nsP3 aa 386 to 403 (un-Δ), Cys→opal mutation at AR86 nsP3 position 537, and Ser→Leu mutation at E2 243 |
| S340 | AR86 with nsP1 position 538 mutation (Thr→Ile) |
| S343 | AR86 with 18-aa insertion of G100 nsP3 aa 386 to 403 |
| S344 | AR86 with Cys→opal mutation at nsP3 position 537 |
| G119 | Girdwood with nsP1 position 538 mutation (Ile→Thr) |
| G120 | Girdwood with opal→Cys mutation at nsP3 position 555 |
| G121 | Girdwood with deletion of nsP3 aa 386 to 403 |
Infectious SINV clones contained the viral cDNA inserted between an SP6 promoter and a unique restriction site, either PmeI or XbaI, used for linearization. In brief, linearized cDNA templates were used for SP6 in vitro transcription reactions (Ambion). Capped, polyadenylated transcripts were then electroporated into BHK-21 cells, and after 24 h, supernatants were harvested and clarified at 3,000 rpm for 15 min and the virus was concentrated through 20% sucrose at 24,000 rpm for more than 4 h at 4°C. Virus pellets were resuspended in virus diluent (1× Dulbecco's phosphate-buffered saline [DPBS; Gibco] supplemented with 1% DCS, 0.122 mg/ml CaCl2, and 0.10 mg/ml MgCl), aliquoted, and stored at −80°C. All concentrated full-length virus stocks were titrated by plaque assay on BHK-21 cells.
For replicon particle packaging, a tripartite helper system has been described previously (26). The replicon RNA genomes used in this study carried the green fluorescent protein (GFP) gene in place of the viral structural genes and were thus capable of GFP expression from the subgenomic 26S RNA promoter but were propagation defective. Construction of the AR86-based pREP89 (26) and Girdwood-based pRgird (29) replicon clones was described previously, and the GFP gene was inserted 3′ of the 26S promoter 5′-untranslated region (5′-UTR) by use of the ClaI site, as described previously for insertion of other foreign genes (29). Helper transcripts for the capsid helper construct pCAP86 (26) and the glycoprotein helper pGIRDGLY (29) were generated as previously described. Replicon particles were packaged by coelectroporating BHK-21 cells with transcripts of replicon genomes and helper RNAs, and particles were harvested and concentrated as described above. The resulting replicon particles, REP89-gfp (AR86-based) and Rgird-gfp (Girdwood-based), contained equivalent coats and were titrated on Vero cells by counting GFP-positive cells. The packaging of VRP, which carry the wild-type (V3000) nonstructural genes but contain adaptive mutations in the E2 glycoprotein gene (pV3014 coat) to maximize tissue culture infectivity, was described previously (57).
Metabolic labeling.
To assess relative amounts of de novo protein synthesis, Vero cells were infected with S300 or G100 at a multiplicity of infection (MOI) of 20 PFU per cell or with diluent alone (mock) for the indicated times. Prior to each indicated harvest time, cells were starved for 2 h with DMEM deficient in cysteine-methionine (Gibco) and then labeled for 1 h with medium containing 35S-labeled cysteine-methionine (33 μCi; Amersham Pro-Mix). Cells were then rinsed in ice-cold 1× PBS and lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Igepal CA-630 [Sigma], Complete Mini protease inhibitors [Roche]). Lysates were clarified by centrifugation, and equal volumes were resolved by 8% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The gel was fixed in buffer containing 10% acetic acid and 40% methanol and then was dried and exposed to a phosphorimager screen, which was scanned using an FX personal molecular imager (Bio-Rad). To quantify relative de novo host protein synthesis, a host protein that resolved at the expected molecular weight of actin was quantified using Quantity One software (Bio-Rad), and each value was normalized to the average signal of this band for three mock-infected samples.
Virus infections and IFN treatment.
For all experiments, Vero cells were infected for 1 h at 37°C with inocula containing the indicated viruses or replicon particles prepared in virus diluent. After virus binding, warm medium was added to cells without removing the inocula. At the indicated hpi, cell supernatants were replaced with medium containing IFN-β or IFN-γ prepared immediately prior to stimulation. Cells were treated for 20 min with IFN and washed once or twice with ice-cold 1× PBS, and extracts were prepared as described below.
Immunoblot analysis.
For direct immunoblotting, Vero cells were lysed in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Igepal CA-630, 0.1% SDS, 0.5% deoxycholate, Complete Mini protease inhibitor cocktail tablets, phosphatase inhibitor cocktail [Sigma]) on ice for more than 5 min, after which cells were scraped, lysates were clarified for 10 min at 4°C, and total protein content was quantified using a Coomassie Plus protein assay (Thermo). Equal amounts of total protein from the samples were denatured for 5 min at 95°C in SDS sample buffer, resolved by SDS-polyacrylamide gel electrophoresis (8%), and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Membranes were blocked for 1 h in 5% nonfat dry milk in PBS-T (137 mM NaCl, 2.7 mM KCl, 0.88 mM KH2PO4, pH 7.5, 0.1% Tween 20), exposed to primary antibody overnight at 4°C, washed five times in PBS-T, incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature, developed with ECL-Plus detection reagent (Amersham), and exposed to film. Dilutions of the following primary antibodies were prepared as recommended by the manufacturer: STAT1 (total), phopho-STAT1 (Tyr701), Tyk2 (total), phospho-Tyk2 (Tyr1054/1055), Jak2 (total), and phospho-Jak2 (Tyr1007/1008) antibodies were all purchased from Cell Signaling Technology; STAT2 (total) and phospho-STAT2 (Tyr689 of mouse STAT2) antibodies were purchased from Upstate; Jak1 (total) antibody was from BD Transduction Laboratories; and phospho-Jak1 (Tyr1022/1023) antibody was from Biosource. Actin antibody was purchased from Santa Cruz. Goat anti-VEEV nsP2 was kindly provided by Alphavax, Inc., and monospecific anti-SINV nsP2 rabbit polyclonal serum was a generous gift from Charles Rice. HRP-conjugated IgG secondary antibodies were purchased from Amersham (anti-rabbit and anti-mouse) and Sigma (anti-goat). For detection of immunoprecipitated protein, light-chain-specific HRP-conjugated IgG (anti-rabbit and anti-mouse) secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
Where indicated, membranes were stripped for 30 min at 50°C in stripping buffer (62.5 mM Tris-HCl, pH 6.7, 2% SDS, 100 mM β-mercaptoethanol), washed at least four times with PBS-T, and then blocked and reprobed as described above.
To ensure the linearity of immunoblotting parameters, 2-fold dilutions of extracts from mock-, S300-, and G100-infected cells treated with IFN-β at 8 h postinfection were resolved by SDS-PAGE and transferred to PVDF membranes as described above. The membranes were probed for p-STAT1, total STAT1, and total Jak1, and then densitometry analysis was performed with ImageJ software (http://rsb.info.nih.gov/ij/). At the antigen quantities loaded in these experiments (25 to 50 μg total protein), detection of p-STAT1 in virally infected lysates was within the linear range, but when the membranes were stripped and reprobed to assess total STAT1 levels, the resulting signal demonstrated saturation at this antigen concentration. However, a dilution series of mock-, S300-, and G100-infected lysates verified that there was no difference between total STAT1 levels at lower antigen concentrations that fell within the linear range. Finally, where indicated, quantitation of total Jak1 levels from input lysates was performed under linear conditions (<30 μg total protein; 1:500 dilution of anti-Jak1 primary antibody).
Immunoprecipitations.
To analyze activation of Jaks through tyrosine phosphorylation, Vero cells were seeded in 6-well plates, infected and treated with IFN-β as described above, and incubated on ice with 200 μl of phosphorylation lysis buffer (47) (0.5% Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 50 mM HEPES [pH 7.4], 200 μM sodium orthovanadate, 10 mM sodium pyrophosphate, 100 mM NaF, Complete protease inhibitor tablets [Roche]) per well for at least 10 min. Wells were scraped at 4°C, lysates were harvested, pooled (2 wells per group), and clarified, and total protein content was quantified as described above. Equivalent amounts of lysate (0.5 to 0.8 mg total protein) were precleared with protein G agarose (Sigma) plus preimmune normal serum for 2 h at 4°C and then exposed to the indicated total Jak antibody overnight at 4°C. Pull-down assays were performed at 4°C for 2 h with protein G agarose, and the immunoprecipitates (IPs) were washed five times with phosphorylation lysis buffer prior to dissociation with 2× SDS sample buffer. IPs were subjected to immunoblotting with the indicated antibodies as described above, with the exception that Tyk2 (total) antibody from BD Transduction Laboratories was used for immunoprecipitation and Tyk2 (total) antibody from Cell Signaling Technology was used for immunoblotting. As a negative control, lysates from a mock-infected and IFN-treated group were subjected to immunoprecipitation with isotype-matched controls: normal mouse IgG2a, IgG2b (eBiosciences), and normal rabbit IgG (Sigma). A portion of the input lysates (5 to 7% of that used for immunoprecipitation) was subjected to direct immunoblot analysis to assess the levels of total Jak, phospho-STAT1, and SINV nsP2 as described above.
IFA.
To assess the percentage of infected cells in all STAT1 activation assays, parallel cultures in each experiment were stained at 10 h postinfection in an indirect immunofluorescence assay (IFA). Infected cells were washed twice with ice-cold 1× PBS, fixed in ice-cold methanol for 10 min at −20°C, air dried, and stored at 4°C until being stained. Cells were incubated for 15 min in PBS-glycine (100 mM [pH 7.2]) and then blocked for 1 h at room temperature in blocking buffer (10% normal goat serum [Sigma], 3% bovine serum albumin [BSA], 0.05% Tween 20 prepared in 1× PBS). Cells were then stained with anti-SINV hyperimmune serum for 1 h at room temperature, washed three times in IFA wash buffer (3% BSA, 0.05% Tween 20 prepared in 1× PBS), and then stained with Alexa Fluor 488-goat anti-mouse IgG (Invitrogen) and DAPI (4′,6-diamidino-2-phenylindole [Roche]; 10 μg/ml). The percentage of infected cells was calculated by acquiring fluorescent images and counting the number of positively stained cells (green) relative to the total number of DAPI-positive nuclei. Since threonine at nsP1 position 538 is associated with delayed expression of 26S RNA (28), cells infected with viruses encoding this determinant (e.g., S300) gave lower fluorescence signals and their calculated infectivities were most likely underestimated.
RESULTS
Differential regulation of STAT1 phosphorylation by strains of SINV.
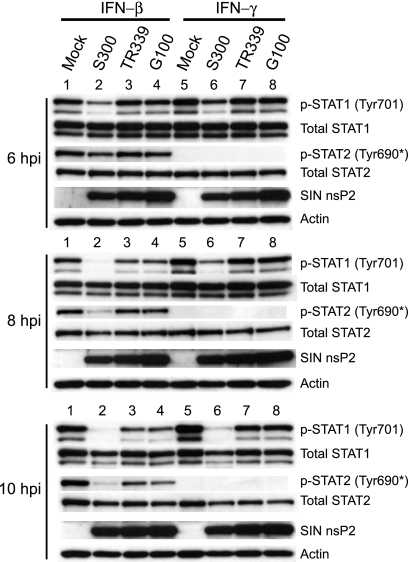
The alphaviruses VEEV and SINV have previously been reported to antagonize IFN-α/β and IFN-γ responses by limiting the accumulation of tyrosine-phosphorylated STAT1/2 after stimulation with these cytokines (57, 63). To determine whether SINV strains that have different mouse neurovirulence profiles differ in the ability to disrupt type I and type II IFN-mediated STAT activation, we assessed STAT phosphorylation in cells infected with SINV strain AR86 (infectious clone S300), which is nearly 100% lethal in adult mice (27, 58), and two SINV strains that do not cause disease in adult mice—TR339 and Girdwood (clone G100). Since various alphaviruses induce quantitatively different amounts of type I IFN both in vivo and in vitro (10, 18, 33, 50), it was important to normalize the amount of IFN present in each culture. For this reason, Vero cells were infected, since these cells are highly permissive for alphavirus infection, are unable to synthesize endogenous type I IFN due to a defective genetic locus (13, 43), and are therefore widely used to compare virus-mediated IFN antagonism specific to the IFN signaling pathway (4, 32, 48, 57, 63). After 6, 8, or 10 h of infection with each virus, Vero cells were stimulated with recombinant IFN-β or IFN-γ to assess STAT1 activation, as indicated by phosphorylation at tyrosine 701, which is required for STAT1 to form transcriptionally active dimers (38) (Fig. 1). Strikingly, infection with the neurovirulent S300 strain resulted in a more rapid and more complete disruption of STAT1/2 activation in response to IFN-β and of STAT1 activation in response to IFN-γ than those for infection with nonneurovirulent G100 or TR339 virus (Fig. 1). Repeated experiments demonstrated that by 8 h postinfection, S300 inhibited IFN-β- and IFN-γ-mediated STAT1 activation similarly at a range of doses (50 to 2,000 U per ml; data not shown). Total STAT levels were not affected by infection with any virus at any time point, indicating that the loss of STAT1 phosphorylation did not result from a decrease in STAT1 expression or from specific STAT1 degradation (Fig. 1). Moreover, differential STAT1 phosphorylation between SINV-infected cultures cannot be explained by differences in infectivity, since G100 and TR339 each infected >95% of cells, as indicated by indirect immunofluorescence staining of parallel cultures at 10 hpi, using anti-SINV hyperimmune serum (data not shown). These data corroborate those previously reported by Yin et al. (63) and indicate that SINV TR339 and G100 partially inhibit the activation of STAT1/2 in response to IFN-α/β. Thus, it appears that several SINV strains are capable of inhibiting Jak/STAT signaling, but importantly, a strain with enhanced virulence exhibits more potent activity.
FIG. 1.
SINV neurovirulent strain AR86 efficiently inhibits STAT1/2 activation in response to type I and type II IFNs. Vero cells, which respond to but do not secrete type I IFNs, were infected at an MOI of 10 PFU/cell with SINV strains AR86 (S300), TR339, and Girdwood (G100). At 6, 8, or 10 hpi, the cells were treated with IFN-β or IFN-γ (1,000 U/ml) for 20 min, after which whole-cell lysates were prepared. Equivalent protein from the samples was then subjected to immunoblotting using the indicated antibodies. Four independent experiments yielded results consistent with the data shown. *, the phospho-specific STAT2 antibody used (Upstate) is directed against mouse STAT2 phosphorylated at tyrosine 689 but cross-reacts with the corresponding position of human (primate) STAT2 (pY-690).
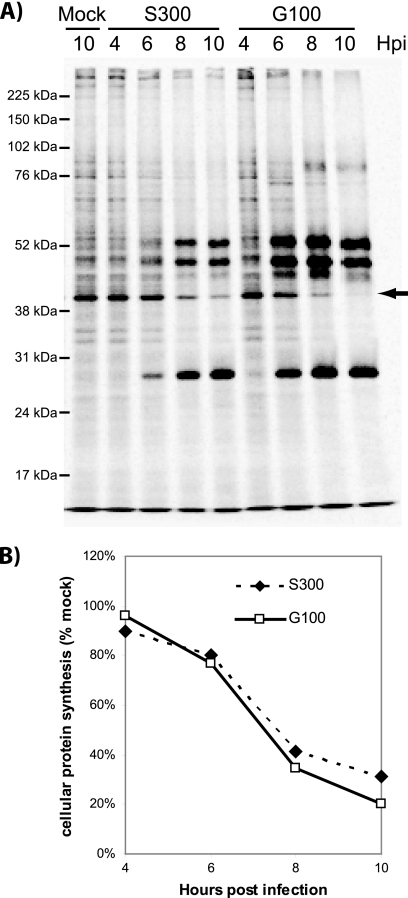
S300 and G100 initiate shutoff of de novo protein synthesis with similar kinetics.
Activation of STAT1 by IFN occurs in the absence of de novo gene expression (15, 16, 44), and we previously demonstrated that the mechanism by which VRP disable Jak/STAT activation does not involve nonspecific shutoff of host transcription/translation, which occurs within several hours after VRP infection (57, 63). However, since maximal STAT1/2 inhibition occurs slightly later during S300 infection than during infection with VRP (8 versus 6 hpi, respectively), it is possible that shutoff of host gene expression could impact STAT1/2 activation at these later times due to turnover of IFN receptor complex components. Therefore, to rule out the possibility that enhanced STAT1 inhibition reflects an ability of S300 to shut off host protein synthesis more efficiently than a nonvirulent SINV, we metabolically labeled Vero cells at various times after infection with S300 and G100. As shown in Fig. 2 A, S300 and G100 both inhibited de novo protein synthesis, with similar kinetics. In fact, when a host protein that migrated at the expected molecular weight of actin was quantified, G100 showed a slightly more rapid and complete shutoff (Fig. 2B), indicating that nonspecific inhibition of gene expression is very unlikely to explain the enhanced ability of S300 to downregulate STAT1/2 tyrosine phosphorylation. This analysis, however, does not rule out the possibility that S300 specifically inhibits expression of a factor required for STAT1 activation but otherwise downregulates global gene expression similarly to G100.
FIG. 2.
S300 and G100 initiate shutoff of de novo protein synthesis with similar kinetics. To measure de novo protein synthesis, Vero cells were infected with diluent (mock), S300, or G100 (MOI = 20 PFU/cell) and then labeled with medium containing 33 μCi 35S-labeled cysteine-methionine per ml for 1 h prior to harvest at the indicated times postinfection (hpi). Prior to each labeling period, cells were starved for 2 h with medium deficient in cysteine-methionine. Extracts of the radiolabeled cells were prepared and resolved by SDS-polyacrylamide gel electrophoresis. (A) The gel was dried, fixed, and exposed to phosphorimaging (Bio-Rad) as described in Materials and Methods. A host protein band that resolved at the expected molecular size of actin (arrow) was quantified using Quantity One software (Bio-Rad). (B) Densities of this band were normalized to those of mock-infected samples and plotted to compare de novo host protein synthesis in S300- versus G100-infected cells. These data are representative of two independent experiments.
Reduced STAT1 phosphorylation in S300-infected cells correlates with defects at the level of or upstream of Jak activation.
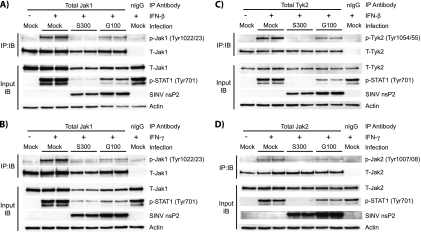
IFN-mediated activation of STAT proteins requires upstream activation of the receptor-associated kinases Jak1/Tyk2 (IFN-α/β) or Jak1/Jak2 (IFN-γ) (11, 55, 56, 61). Therefore, we assessed whether S300 infection interfered with activation of these kinases following IFN treatment. Vero cells were infected with either S300 or G100 and stimulated with either IFN-β or IFN-γ after 8 h of infection, a time that gave nearly maximal STAT1/2 inhibition by S300. After stimulation, total Jak protein was immunoprecipitated and subjected to immunoblotting using phosphotyrosine-specific Jak antibodies. As shown in Fig. 3, STAT1 inhibition (measured by immunoblotting of input lysates) was directly correlated with reduced activation of Tyk2/Jak1 in response to IFN-β (Fig. 3A and C) and with reduced Jak1/Jak2 phosphorylation in response to IFN-γ (Fig. 3B and D). Moreover, though G100 infection resulted in some inhibition of Tyk2/Jak1 and Jak1/Jak2 phosphorylation compared to that in mock-infected cells, inhibition of tyrosine phosphorylation of both STAT1 and Jak proteins was notably greater in S300- than in G100-infected cells. Total Tyk2 and Jak2 protein levels were similar between S300- and G100-infected cells, as determined by direct immunoblotting of input lysates (Fig. 3C and D, input IB panels) and by stripping and reprobing of blots of immunoprecipitated total Jak protein (Fig. 3C and D, IP:IB panels) with antibodies that interact with both the phosphorylated and unphosphorylated forms. Quantitation of total Jak1 protein from input lysates over multiple experiments indicated that total Jak1 levels in S300-infected cells were reduced by as much as 50% relative to levels measured after G100 infection (Fig. 3A and B, input IB panels). However, reduced levels of immunoprecipitated total Jak1 alone cannot explain the reduction in tyrosine-phosphorylated Jak1, since p-Jak1 levels in S300-infected cells were 17% (standard deviation [SD] = 6%) of those measured in G100-infected cells (average over 5 experiments) (Fig. 3A, IP:IB panel). Further experiments are required to evaluate whether the moderate reduction in total Jak1 level within virally infected cells is critical for S300-mediated Jak/STAT signaling inhibition. Taken together, these data are consistent with the hypothesis that S300 infection antagonizes IFN signaling upstream of or at the level of IFN receptor-associated Jak activation.
FIG. 3.
Reduced STAT1 phosphorylation in S300-infected cells correlates with defects in Jak activation by type I and type II IFNs. Vero cells were infected (MOI = 10 PFU/cell) with S300, G100, or diluent alone (mock) and then treated for 20 min with 1,000 U IFN-β (A and C) or IFN-γ (B and D) per ml. Cell extracts were then prepared as described in Materials and Methods, and total protein content was quantified. Equal amounts of total protein were then subjected to immunoprecipitation with antibodies directed against total Jak1 (A and B), total Tyk2 (C), or total Jak2 (D), and to assess activation, the immunoprecipitates were resolved by SDS-PAGE and immunoblotted using the indicated phospho-specific Jak antibody (IB:IP panels). These blots were then stripped and reprobed using an antibody recognizing the appropriate total Jak. A portion of the input lysates (5 to 7%) was subjected to direct immunoblot analysis to assess total Jak protein levels (input IB panels). These blots were stripped and reprobed consecutive times with phospho-specific STAT1 (Tyr701) and anti-SINV nsP2 polyclonal serum. The data shown in panel A are representative of 5 independent experiments. Data in panels B to D are representative of at least 2 experiments totaling at least 4 independent samples per infection group.
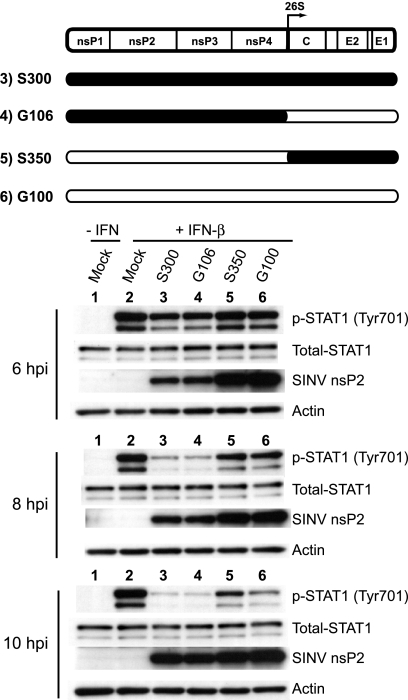
Efficient disruption of STAT1 tyrosine phosphorylation requires determinants within the nonstructural genes of S300.
In order to further define the mechanisms by which SINV strain AR86 (S300) efficiently inhibits STAT1 signaling and IFN receptor complex activation, we first focused on identifying the viral determinants of this activity. Initially, we evaluated infection with S300/G100 chimeric viruses which carry heterologous nonstructural and structural genes. As shown in Fig. 4, phosphorylation of STAT1 at Tyr-701 was again efficiently inhibited in response to IFN-β by 8 h in cells infected with wild-type S300 as well as in cells infected with the chimera carrying the S300 nonstructural genes (G106). Conversely, even though both G100 and the chimera encoding the G100 nonstructural proteins infected >95% of cells (data not shown), neither virus efficiently inhibited STAT1 activation. These data indicate that AR86 (S300) determinants that enable efficient Jak/STAT signaling inhibition are encoded by the viral nonstructural genes. In support of this, STAT1/2 activation by IFN-β was inhibited more efficiently by replicon particles carrying the S300 nonstructural genes than by G100-based replicons (data not shown), again indicating that the enhanced inhibition of STAT1/2 activation by the adult mouse neurovirulent AR86 (S300) virus is mediated by determinants within the viral nonstructural genes.
FIG. 4.
Efficient disruption of STAT1 tyrosine phosphorylation requires determinants encoded by the S300 nonstructural genes. Vero cells were infected for 6, 8, or 10 h with the indicated viruses (MOI = 15 PFU/cell) prior to 20 min of stimulation with 1,000 U IFN-β/ml. Whole-cell extracts were prepared and analyzed as described in the legend to Fig. 1. The schematic of the SINV genome indicates the origin of the nonstructural/structural gene chimeras, where black indicates genes of S300 origin and white indicates genes of G100 origin. Three independent experiments with this chimeric panel yielded results consistent with the data shown.
The presence of threonine at S300 nsP1 position 538 is required for efficient inhibition of STAT1 activation.
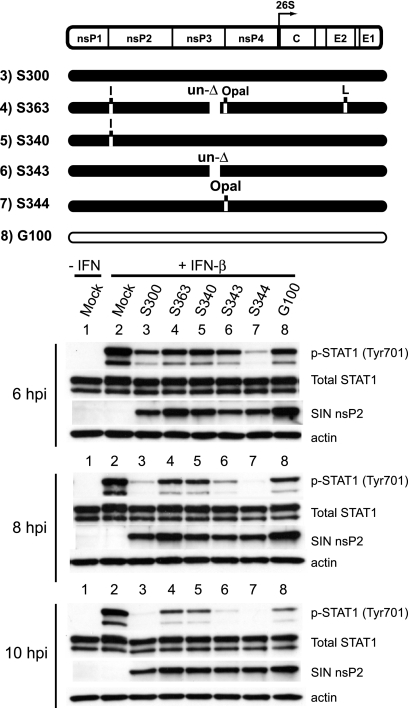
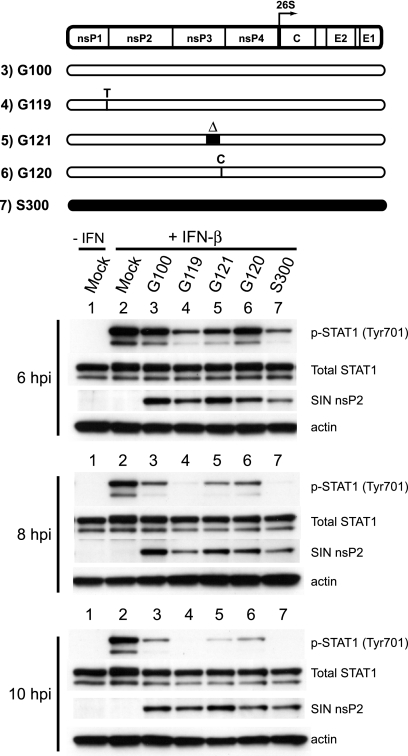
Results from the chimeric virus and replicon studies indicated that determinants within the nonstructural protein coding region mediate the effects on STAT activation. To further define these determinants, we next evaluated whether any of the three previously defined S300 determinants of adult mouse neurovirulence in this region (60) was required for STAT1 inhibition, either individually or in combination. Two of these virulence determinants reside within nsP3—an 18-amino-acid deletion present in the C-terminal region of S300 nsP3 and a cysteine that replaces the G100-carried opal termination codon just upstream of the nsP3/4 cleavage domain. A third virulence determinant, a threonine at nsP1 position 538, is unique to the AR86 strain (S300) among Sindbis viruses. As shown in Fig. 5, introducing the four defined attenuating mutations into S300 (S363) abrogated enhanced STAT1 inhibition, giving results equivalent to those seen during G100 infection. When we evaluated the individual contributions of the three nonstructural virulence determinants, we found that threonine at nsP1 position 538 was required, since S340 (encoding the consensus isoleucine at that position) also failed to inhibit STAT1 activation effectively. Conversely, introduction of either of the G100-carried nsP3 attenuating mutations (S343 and S344) did not result in a major loss of STAT1 inhibition relative to that with S300. We obtained similar results by measuring STAT1 activation in response to IFN-γ (data not shown). While we cannot rule out that other differences between the S300 and G100 nonstructural genes are required for Jak/STAT signaling inhibition, these data clearly demonstrate that threonine at nsP1 position 538 is absolutely required for the enhanced inhibition seen in S300-infected cells.
FIG. 5.
The presence of threonine at S300 nsP1 position 538 is required for efficient inhibition of STAT1 activation. For a loss-of-function analysis, Vero cells were infected with the indicated viruses for 6, 8, or 10 h (MOI = 15 PFU/cell) and then stimulated for 20 min with 1,000 U IFN-β/ml. Whole-cell lysates were harvested and then subjected to Western blotting as described in the legend to Fig. 1. The schematic of the SINV genome denotes the locations of four attenuating mutations of G100 origin (white) that were inserted individually or in combination into the wild-type S300 background (black): isoleucine (I) at nsP1 position 538, within the nsP1/2 cleavage domain, replaces threonine encoded by wild-type S300; an 18-amino-acid insertion (un-Δ) replaces a deletion present in wild-type S300, at nsP3 positions 386 to 403; an opal termination codon replaces the wild-type S300-encoded cysteine at nsP3 position 537; and a leucine (L) replaces a serine encoded by wild-type S300 at position 243 of the E2 glycoprotein. These data are representative of three independent experiments.
Presence of threonine at nsP1 position 538 is sufficient for inhibition of STAT1 activation during S300 infection.
Our loss-of-function analysis demonstrated that threonine at nsP1 position 538, which is unique to the adult mouse neurovirulent AR86 strain, was required for the enhanced STAT1 inhibition by S300. We next evaluated whether replacement of the isoleucine at nsP1 position 538 in G100 with threonine also resulted in enhanced inhibition or whether other determinants present within the nonstructural genes were also required for maximal STAT1 inhibition. Vero cells were infected with the wild-type G100 virus or mutants encoding the AR86 virulence determinant at nsP1 position 538 (isoleucine to threonine), the 18-amino-acid deletion in nsP3, or the opal-to-cysteine change near the nsP3 C terminus. Strikingly, introduction of threonine at nsP1 position 538 into the G100 background (G119) was sufficient to enhance STAT1 inhibition by G100 to the level seen during S300 infection (Fig. 6), while introduction of either nsP3 virulence determinant had no effect. G119- and S300-mediated inhibition was also comparable when we measured STAT1 activation after IFN-γ treatment (data not shown). Therefore, the virulence determinant at nsP1 position 538 is necessary and sufficient for enhanced STAT1 inhibition by the adult mouse neurovirulent S300 virus.
FIG. 6.
The presence of threonine at nsP1 position 538 is sufficient for inhibition of STAT1 activation during S300 infection. To assay for gain of function, Vero cells were infected for 6, 8, or 10 h with the indicated viruses (MOI = 15 PFU/cell) and then stimulated for 20 min with 1,000 U IFN-β/ml. Whole-cell extracts were prepared and analyzed by Western blotting as described in the legend to Fig. 1. The diagram indicates the locations of four virulence determinants of S300 origin (black) which were individually introduced into the avirulent G100 background (white). Threonine (T) replaced the G100-encoded isoleucine at nsP1 position 538, within the nsP1/2 cleavage domain; nsP3 positions 386 to 403 were deleted from wild-type G100 (Δ); and the wild-type opal termination codon at G100 nsP3 position 555 was replaced with cysteine (C). Three independent experiments yielded results consistent with the data shown.
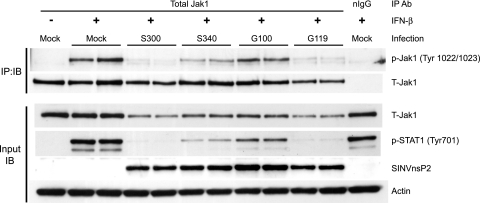
Modulation of STAT1 activation by threonine at nsP1 position 538 correlates with upstream inhibition of Jak1 activation.
The data in Fig. 5 and 6 clearly implicate threonine at nsP1 position 538 as a critical determinant of the S300-mediated STAT1 inhibition. To evaluate whether this determinant is also involved in the inhibition of Jak1 activation previously described for Fig. 3, we infected Vero cells with S300 and S340, which encodes isoleucine at nsP1 position 538, as well as with G100 and G119, which encodes threonine at nsP1 position 538. After IFN-β stimulation at 8 hpi, we found that the degree of inhibition of STAT1 activation by each virus (Fig. 7, input IB panel) correlated tightly with the relative inhibition of Jak1 tyrosine phosphorylation (Fig. 7, IP:IB panel). As discussed in relation to Fig. 3, levels of total Jak1 within S300- and G119-infected cells were lower than those measured in S340- and G100-infected cells, which may play a role in the mechanism by which viruses encoding threonine at nsP1 position 538 effectively inhibit downstream STAT1 activation, but this possibility requires further study. These data clearly indicate that a single determinant previously demonstrated to be essential for adult mouse neurovirulence is involved in the potent inhibition of tyrosine phosphorylation of both STAT1 and its upstream activating kinase, Jak1.
FIG. 7.
Modulation of STAT1 activation by threonine at nsP1 position 538 correlates with upstream inhibition of Jak1 activation. Vero cells were infected for 8 h with S300, S340 (isoleucine at nsP1 position 538), G100, or G119 (threonine at nsP1 position 538) at an MOI of 15 PFU/cell and then stimulated for 20 min with 1,000 U IFN-β/ml. Separate immunoblots of immunoprecipitated Jak1 and input lysates were then performed as described in the legend to Fig. 3. The data are representative of three separate experiments totaling 5 independent samples per infection group.
DISCUSSION
The importance of the type I IFN system in the immediate control of virus replication is illustrated by the quantity and diversity of mechanisms carried by most (if not all) virus families to antagonize the production of and/or cellular response to this family of cytokines. In the case of alphaviruses, this is further illustrated by the finding that avirulent SINV strains become fully virulent in mice deficient in the type I IFN response (IFNAR−/− mice) (50, 51), suggesting that SINV must downregulate IFN production and/or signaling to some extent in order to cause disease in immunocompetent mice. Previous work has suggested that alphaviruses, including SINV, antagonize type I IFNs through global shutoff of host macromolecular synthesis (3, 17, 19, 20). Recent work from our group and others demonstrated that VEEV and SINV infections result in a failure of cultured cells to respond to type I and II IFNs, as indicated by reduced STAT1 tyrosine phosphorylation (57, 63); however, in one study, inhibition of host macromolecular synthesis was proposed to play the dominant role in IFN antagonism, since STAT1 inhibition did not result in reduced ISG transcription in cultured mouse neurons infected with VRP (63). In light of these previous results, we set out to test whether there was any correlation between the in vivo virulence profiles of different SINVs and their ability to antagonize STAT activation. Importantly, we found that SINV strain AR86 (S300), which causes lethal disease in adult mice, demonstrated more complete and rapid inhibition of STAT1 activation than two avirulent SINV strains, Girdwood (G100) and TR339, which only partially inhibited STAT1 activation (as previously reported for TR339 [63]). AR86 and Girdwood each downregulated host protein synthesis, with similar kinetics, and enhanced STAT1 inhibition mapped to a single determinant, nsP1 position 538, known to modulate AR86 neurovirulence (27, 60) but not shutoff of host transcription/translation (12). These results suggest that Jak/STAT signaling inhibition, not host cell shutoff, contributes to the enhanced virulence profile of AR86.
Though we do not yet understand the mechanism by which AR86 inhibits IFN-α/β and IFN-γ signaling, analysis of the type I and type II IFN receptor complexes clearly demonstrated that defective STAT1 activation was associated with defects at the level of Jak protein activation at both receptor complexes. Decreased Jak activation could result from an inhibition of Jak kinase activity, from protein tyrosine phosphatase activity, or from a loss of receptor-ligand interaction (e.g., via decreased IFN receptor surface expression). Additional studies are required to determine whether the virus interferes with the ability of receptor complexes to associate with and/or maintain expression at the cell surface. It is interesting that total Jak1 protein levels, but not levels of Jak2 and Tyk2, were reduced in S300-infected cells relative to those measured in G100-infected cells (as much as 50%) (Fig. 3). If the moderate reduction of total Jak1 protein during S300 infection (and G119 infection) (Fig. 7) specifically represents the fraction of Jak1 that participates in signal transduction at the IFN receptor complexes, this could explain the much larger reductions in levels of tyrosine-phosphorylated Jak1, Jak2, Tyk2, STAT1, and STAT2, but further studies are required to evaluate this possibility. Previous studies with VRP infection indicated that Tyk2/Jak1 activation by IFN-β was normal at times of maximal STAT1 inhibition, but IFN-γ-mediated Jak1/Jak2 phosphorylation was reduced without a major decrease in IFNGR2 surface expression (57). These results suggest that while both VEEV and AR86 inhibit STAT1 activation by IFN-β and IFN-γ, these viruses may act through distinct mechanisms.
Mapping studies (Fig. 5 and 6) revealed that a single S300 determinant, nsP1 position 538, is necessary and sufficient for enhanced Jak/STAT signaling inhibition. This determinant has previously been shown to play an essential role in adult mouse neurovirulence by the AR86 virus (27, 60). Recently, we demonstrated that SINV nsP1 position 538 modulates type I IFN induction independent of its effects on Jak/STAT signaling (12), which suggests that this determinant regulates both the induction and signaling arms of the type I IFN response, each of which could be important for virulence. A possible link to type I IFN disruption is the ability of this determinant to modulate cleavage of the nonstructural polyprotein precursor. The nsP1 position 538 determinant lies within the nsP1/2 cleavage domain, and relative to the SINV consensus isoleucine at this position, the virulence-associated threonine residue delays the kinetics by which polyprotein intermediates are processed into mature nsPs (28). Interestingly, when we compared our various panels of mutant viruses, potent STAT1 inhibition correlated tightly with viruses that maintained expression of proteins corresponding to polyprotein precursors (data not shown). Therefore, we are currently evaluating whether enhanced STAT1 inhibition by viruses carrying threonine at nsP1 position 538 is a direct result of delayed polyprotein processing and, if so, whether inhibition might be mediated by a P123 precursor or some other cleavage intermediate.
We cannot rule out the possibility that the threonine codon modulates Jak/STAT inhibition independent of its effects on polyprotein processing. Furthermore, although the threonine codon is essential for efficient inhibition of STAT activation, the fact that viruses lacking threonine at this position retain partial inhibitory activity suggests that other determinants contribute to STAT inhibition. Interestingly, mutating TR339 nsP1 position 538 from an isoleucine to a threonine (39ns1), which also enhances neurovirulence (27), resulted in enhanced inhibition of STAT1 phosphorylation (data not shown); however, this inhibition did not reach the magnitude seen when threonine at nsP1 position 538 was introduced into G100 (G119), suggesting that other differences between TR339 and Girdwood (G100), which is more closely related to AR86, also contribute to the inhibitory mechanisms. When combined with previous results with VEEV (57, 63), these data implicate the viral nonstructural proteins as mediators of Jak/STAT signaling inhibition. However, while we observed no role for the structural proteins in Jak/STAT inhibition, it is clear that determinants within alphavirus structural proteins (1, 59) and noncoding regions (62) also contribute to the resistance of alphaviruses to type I IFN.
Antagonism of Jak/STAT signaling is a feature of other encephalitic viruses, including the flaviviruses Japanese encephalitis virus, tick-borne encephalitis virus, and West Nile virus (WNV), all of which have been shown to inhibit responses to IFN-α/β and/or IFN-γ (4, 25, 39). Recently, expression of the NS5 protein of the virulent WNV strain NY99, but not of NS5 from the attenuated WNV strain Kunjin, was shown to effectively disrupt STAT1 activation by IFN-α/β and IFN-γ. Effective STAT1 inhibition by the Kunjin strain could be rescued by the introduction of a single amino acid encoded by the virulent NY99 strain (NS5 S653F) (35), in a manner analogous to our findings with SINV. At least in the case of SINV AR86 infection, the importance of the determinant at nsP1 position 538 for both adult mouse neurovirulence and Jak/STAT inhibition suggests that this inhibition contributes to pathogenesis. We previously reported that S300 (wild type nsP1 [Thr 538]) and S340 (mutant nsP1 [Ile 538]) establish infection and replicate within the CNS to similar levels at early times. However, by day 6 postinfection, when S300-infected mice begin to succumb to infection, S300 continues to replicate within the CNS, while the S340 virus is largely cleared (27), suggesting that the differences in virulence between these strains result in part from an ability of S300 to avoid viral clearance (27, 60). Antiviral antibody and IFN-γ mediate noncytolytic clearance of nonpathogenic SINV from the CNS (5, 8, 36, 37), and IFN-γ treatment of SINV-infected rat neuronal cultures results in the clearance of infectious virus, a reduction in viral protein synthesis, and restored cellular protein synthesis within 24 h (7), effects that were later determined to require Jak1-dependent signaling (6). Since AR86 is a potent inhibitor of IFN-γ-mediated STAT1 activation, it is possible that this virus is more resistant to mechanisms of IFN-γ-dependent viral clearance from the CNS, which ultimately leads to the induction of lethal neurologic disease. Studies are under way to directly assess this possibility.
In summary, we have demonstrated that an adult mouse neurovirulent strain of Sindbis virus exhibits an enhanced ability to interfere with Jak/STAT activation by either type I or type II IFNs and that this effect is mediated by a virulence determinant at nsP1 position 538. These studies strongly suggest that the ability to block Jak/STAT signaling contributes to alphavirus virulence. Therefore, additional studies are needed to both define the molecular mechanisms underlying Jak/STAT antagonism by alphaviruses and investigate the relative role of inhibition of either type I or type II IFN receptor signaling in the pathogenesis of alphavirus-induced disease.
Acknowledgments
We thank Thomas Morrison for continued interest, scientific discussions, and editorial commentary on the manuscript. Martha Collier, Bianca Trollinger, Lance Blevins, and Andrew Morgan provided expert technical support with VRP production, cell culture, and reagent preparation. Mehul Suthar was involved with the generation of the AR86/Girdwood chimeric virus panel.
This study was supported by NIH grant R01 AI067641 to M.T.H. J.D.S. was supported by NIH grant T32 GM008719.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Aguilar, P. V., A. P. Adams, E. Wang, W. Kang, A. S. Carrara, M. Anishchenko, I. Frolov, and S. C. Weaver. 2008. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 82:4920-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. V., L. W. Leung, E. Wang, S. C. Weaver, and C. F. Basler. 2008. A five-amino-acid deletion of the eastern equine encephalitis virus capsid protein attenuates replication in mammalian systems but not in mosquito cells. J. Virol. 82:6972-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, P. V., S. C. Weaver, and C. F. Basler. 2007. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 81:3866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binder, G. K., and D. E. Griffin. 2001. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science 293:303-306. [DOI] [PubMed] [Google Scholar]
- 6.Burdeinick-Kerr, R., D. Govindarajan, and D. E. Griffin. 2009. Noncytolytic clearance of Sindbis virus infection from neurons by gamma interferon is dependent on Jak/STAT signaling. J. Virol. 83:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdeinick-Kerr, R., and D. E. Griffin. 2005. Gamma interferon-dependent, noncytolytic clearance of Sindbis virus infection from neurons in vitro. J. Virol. 79:5374-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdeinick-Kerr, R., J. Wind, and D. E. Griffin. 2007. Synergistic roles of antibody and interferon in noncytolytic clearance of Sindbis virus from different regions of the central nervous system. J. Virol. 81:5628-5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrnes, A. P., J. E. Durbin, and D. E. Griffin. 2000. Control of Sindbis virus infection by antibody in interferon-deficient mice. J. Virol. 74:3905-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles, P. C., J. Trgovcich, N. L. Davis, and R. E. Johnston. 2001. Immunopathogenesis and immune modulation of Venezuelan equine encephalitis virus-induced disease in the mouse. Virology 284:190-202. [DOI] [PubMed] [Google Scholar]
- 11.Colamonici, O. R., H. Uyttendaele, P. Domanski, H. Yan, and J. J. Krolewski. 1994. p135tyk2, an interferon-alpha-activated tyrosine kinase, is physically associated with an interferon-alpha receptor. J. Biol. Chem. 269:3518-3522. [PubMed] [Google Scholar]
- 12.Cruz, C. C., M. S. Suthar, S. A. Montgomery, R. Shabman, J. Simmons, R. E. Johnston, T. E. Morrison, and M. T. Heise. 2010. Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein. Virology 399:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J., S. Lustig, N. Ruggli, Y. Akov, and C. M. Rice. 1997. Genetic determinants of Sindbis virus neuroinvasiveness. J. Virol. 71:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman, R. L., S. P. Manly, M. McMahon, I. M. Kerr, and G. R. Stark. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38:745-755. [DOI] [PubMed] [Google Scholar]
- 16.Friedman, R. L., and G. R. Stark. 1985. Alpha-interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature 314:637-639. [DOI] [PubMed] [Google Scholar]
- 17.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner, C. L., C. W. Burke, M. Z. Tesfay, P. J. Glass, W. B. Klimstra, and K. D. Ryman. 2008. Eastern and Venezuelan equine encephalitis viruses differ in their ability to infect dendritic cells and macrophages: impact of altered cell tropism on pathogenesis. J. Virol. 82:10634-10646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garmashova, N., S. Atasheva, W. Kang, S. C. Weaver, E. Frolova, and I. Frolov. 2007. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J. Virol. 81:13552-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garmashova, N., R. Gorchakov, E. Volkova, S. Paessler, E. Frolova, and I. Frolov. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil, M. P., E. Bohn, A. K. O'Guin, C. V. Ramana, B. Levine, G. R. Stark, H. W. Virgin, and R. D. Schreiber. 2001. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. U. S. A. 98:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, D. E. 2010. Recovery from viral encephalomyelitis: immune-mediated noncytolytic virus clearance from neurons. Immunol. Res. 47:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. Sindbis-group alphavirus replication in periosteum and endosteum of long bones in adult mice. J. Virol. 74:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise, M. T., D. A. Simpson, and R. E. Johnston. 2000. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J. Virol. 74:4207-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heise, M. T., L. J. White, D. A. Simpson, C. Leonard, K. A. Bernard, R. B. Meeker, and R. E. Johnston. 2003. An attenuating mutation in nsP1 of the Sindbis-group virus S.A.AR86 accelerates nonstructural protein processing and up-regulates viral 26S RNA synthesis. J. Virol. 77:1149-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heise, M. T., A. Whitmore, J. Thompson, M. Parsons, A. A. Grobbelaar, A. Kemp, J. T. Paweska, K. Madric, L. J. White, R. Swanepoel, and F. J. Burt. 2009. An alphavirus replicon-derived candidate vaccine against Rift Valley fever virus. Epidemiol. Infect. 137:1309-1318. [DOI] [PubMed] [Google Scholar]
- 30.Hidmark, A. S., G. M. McInerney, E. K. Nordstrom, I. Douagi, K. M. Werner, P. Liljestrom, and G. B. Karlsson Hedestam. 2005. Early alpha/beta interferon production by myeloid dendritic cells in response to UV-inactivated virus requires viral entry and interferon regulatory factor 3 but not MyD88. J. Virol. 79:10376-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson, A. C., T. R. Moench, B. D. Trapp, and D. E. Griffin. 1988. Basis of neurovirulence in Sindbis virus encephalomyelitis of mice. Lab. Invest. 58:503-509. [PubMed] [Google Scholar]
- 32.Johnson, K. E., and D. M. Knipe. 2010. Herpes simplex virus-1 infection causes the secretion of a type I interferon-antagonizing protein and inhibits signaling at or before Jak-1 activation. Virology 396:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimstra, W. B., K. D. Ryman, K. A. Bernard, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 1999. Infection of neonatal mice with Sindbis virus results in a systemic inflammatory response syndrome. J. Virol. 73:10387-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurent-Rolle, M., E. F. Boer, K. J. Lubick, J. B. Wolfinbarger, A. B. Carmody, B. Rockx, W. Liu, J. Ashour, W. L. Shupert, M. R. Holbrook, A. D. Barrett, P. W. Mason, M. E. Bloom, A. Garcia-Sastre, A. A. Khromykh, and S. M. Best. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, B., and D. E. Griffin. 1992. Persistence of viral RNA in mouse brains after recovery from acute alphavirus encephalitis. J. Virol. 66:6429-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine, B., J. M. Hardwick, B. D. Trapp, T. O. Crawford, R. C. Bollinger, and D. E. Griffin. 1991. Antibody-mediated clearance of alphavirus infection from neurons. Science 254:856-860. [DOI] [PubMed] [Google Scholar]
- 38.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 39.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lustig, S., M. Halevy, D. Ben-Nathan, and Y. Akov. 1992. A novel variant of Sindbis virus is both neurovirulent and neuroinvasive in adult mice. Arch. Virol. 122:237-248. [DOI] [PubMed] [Google Scholar]
- 41.Lustig, S., A. C. Jackson, C. S. Hahn, D. E. Griffin, E. G. Strauss, and J. H. Strauss. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKnight, K. L., D. A. Simpson, S. C. Lin, T. A. Knott, J. M. Polo, D. F. Pence, D. B. Johannsen, H. W. Heidner, N. L. Davis, and R. E. Johnston. 1996. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strains which affect cell culture and in vivo phenotypes. J. Virol. 70:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearse, R. N., R. Feinman, K. Shuai, J. E. Darnell, Jr., and J. V. Ravetch. 1993. Interferon gamma-induced transcription of the high-affinity Fc receptor for IgG requires assembly of a complex that includes the 91-kDa subunit of transcription factor ISGF3. Proc. Natl. Acad. Sci. U. S. A. 90:4314-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pichlmair, A., O. Schulz, C. P. Tan, J. Rehwinkel, H. Kato, O. Takeuchi, S. Akira, M. Way, G. Schiavo, and C. Reis e Sousa. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 47.Platanias, L. C., S. Uddin, P. Domanski, and O. R. Colamonici. 1996. Differences in interferon alpha and beta signaling. Interferon beta selectively induces the interaction of the alpha and beta L subunits of the type I interferon receptor. J. Biol. Chem. 271:23630-23633. [DOI] [PubMed] [Google Scholar]
- 48.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryman, K. D., and W. B. Klimstra. 2008. Host responses to alphavirus infection. Immunol. Rev. 225:27-45. [DOI] [PubMed] [Google Scholar]
- 50.Ryman, K. D., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryman, K. D., K. C. Meier, C. L. Gardner, P. A. Adegboyega, and W. B. Klimstra. 2007. Non-pathogenic Sindbis virus causes hemorrhagic fever in the absence of alpha/beta and gamma interferons. Virology 368:273-285. [DOI] [PubMed] [Google Scholar]
- 52.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53-76. [DOI] [PubMed] [Google Scholar]
- 53.Schindler, C., D. E. Levy, and T. Decker. 2007. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 282:20059-20063. [DOI] [PubMed] [Google Scholar]
- 54.Shabman, R. S., T. E. Morrison, C. Moore, L. White, M. S. Suthar, L. Hueston, N. Rulli, B. Lidbury, J. P. Ting, S. Mahalingam, and M. T. Heise. 2007. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J. Virol. 81:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shuai, K., A. Ziemiecki, A. F. Wilks, A. G. Harpur, H. B. Sadowski, M. Z. Gilman, and J. E. Darnell. 1993. Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature 366:580-583. [DOI] [PubMed] [Google Scholar]
- 56.Silvennoinen, O., J. N. Ihle, J. Schlessinger, and D. E. Levy. 1993. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature 366:583-585. [DOI] [PubMed] [Google Scholar]
- 57.Simmons, J. D., L. J. White, T. E. Morrison, S. A. Montgomery, A. C. Whitmore, R. E. Johnston, and M. T. Heise. 2009. Venezuelan equine encephalitis virus disrupts STAT1 signaling by distinct mechanisms independent of host shutoff. J. Virol. 83:10571-10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson, D. A., N. L. Davis, S. C. Lin, D. Russell, and R. E. Johnston. 1996. Complete nucleotide sequence and full-length cDNA clone of S.A.AR86 a South African alphavirus related to Sindbis. Virology 222:464-469. [DOI] [PubMed] [Google Scholar]
- 59.Spotts, D. R., R. M. Reich, M. A. Kalkhan, R. M. Kinney, and J. T. Roehrig. 1998. Resistance to alpha/beta interferons correlates with the epizootic and virulence potential of Venezuelan equine encephalitis viruses and is determined by the 5′ noncoding region and glycoproteins. J. Virol. 72:10286-10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suthar, M. S., R. Shabman, K. Madric, C. Lambeth, and M. T. Heise. 2005. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J. Virol. 79:4219-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watling, D., D. Guschin, M. Muller, O. Silvennoinen, B. A. Witthuhn, F. W. Quelle, N. C. Rogers, C. Schindler, G. R. Stark, J. N. Ihle, et al. 1993. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature 366:166-170. [DOI] [PubMed] [Google Scholar]
- 62.White, L. J., J. G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 75:3706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin, J., C. L. Gardner, C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2009. Similarities and differences in antagonism of neuron alpha/beta interferon responses by Venezuelan equine encephalitis and Sindbis alphaviruses. J. Virol. 83:10036-10047. [DOI] [PMC free article] [PubMed] [Google Scholar]