Abstract
Hepatitis C virus (HCV) nonstructural protein 5B (NS5B), the viral RNA-dependent RNA polymerase (RdRp), is a tail-anchored protein with a highly conserved C-terminal transmembrane domain (TMD) that is required for the assembly of a functional replication complex. Here, we report that the TMD of the HCV RdRp can be functionally replaced by a newly identified analogous membrane anchor of the GB virus B (GBV-B) NS5B RdRp. Replicons with a chimeric RdRp consisting of the HCV catalytic domain and the GBV-B membrane anchor replicated with reduced efficiency. Compensatory amino acid changes at defined positions within the TMD improved the replication efficiency of these chimeras. These observations highlight a conserved structural motif within the TMD of the HCV NS5B RdRp that is required for RNA replication.
Hepatitis C virus (HCV) chronically infects 120 to 180 million people worldwide and is a leading cause of liver cirrhosis and hepatocellular carcinoma (6). A hallmark of HCV and of all other positive-strand RNA viruses investigated to date is their capacity to induce distinct intracellular membrane alterations which serve as a scaffold for the assembly of a viral replication complex (reviewed in reference 13). The formation of such a complex requires the concerted interplay of viral proteins, replicating RNA, intracellular membranes, and additional host factors.
HCV nonstructural protein 5B (NS5B), the viral RNA-dependent RNA polymerase (RdRp), belongs to a class of membrane proteins termed “tail-anchored proteins” (7, 16). Characteristic features of these proteins include (i) posttranslational membrane targeting via a C-terminal transmembrane domain (TMD), (ii) integral membrane association, and (iii) cytosolic orientation of the functional protein domain (reviewed in references 9 and 18). The TMD of the HCV RdRp was mapped to the C-terminal 21 amino acids and is required for viral RNA replication, similarly to the membrane segments of the other HCV nonstructural proteins (2, 14, 15). Sequence analyses and structure predictions coupled with systematic mutational analyses suggested that the TMD of the HCV RdRp may be involved in intramembrane protein-protein interactions required for the assembly of a functional replication complex (14).
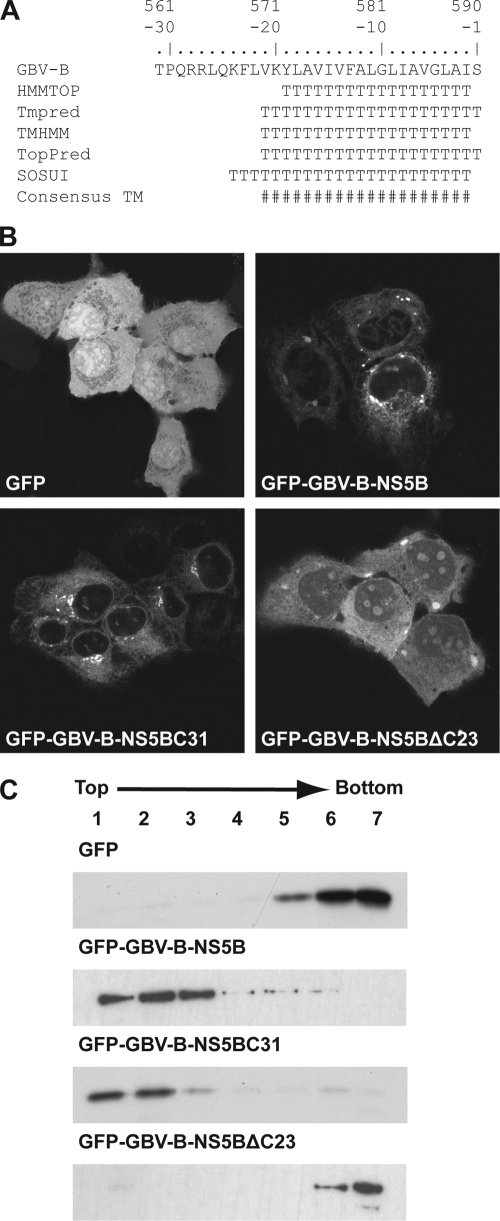
In order to gain further insight into the function and conserved features of the HCV RdRp TMD, we investigated whether GB virus B (GBV-B), the second species in the Hepacivirus genus (11), is membrane associated by a similar membrane anchor. GBV-B causes acute and chronic hepatitis in New World primates, is the closest relative of HCV, and has been used as a surrogate in vivo model virus for HCV infection (3). As shown in Fig. 1 A, the algorithms HMMTOP, Tmpred, TMHMM, TopPred, and SOSUI strongly predicted the presence of a TMD within the C-terminal 19 to 24 amino acids of GBV-B NS5B.
FIG. 1.
The C-terminal 23 amino acids of GBV-B NS5B mediate membrane association. (A) Sequence analyses of the GBV-B NS5B C terminus. Amino acids are numbered with respect to the NS5B protein and negatively from its C terminus. The following methods to predict TMDs were combined: HMMTOP (http://www.enzim.hu/hmmtop/), Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html), TMHMM (http://www.cbs.dtu.dk/services/TMHMM), TopPred (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form_toppred), and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/). T, predicted transmembrane amino acid; #, consensus minimum transmembrane segment, considering all prediction methods. (B) Subcellular localization of GFP fusion constructs. Plasmids pCMVGFP, pCMVGFP-GBV-B-NS5B, pCMVGFP-GBV-B-NS5BC31, and pCMVGFP-GBV-B-NS5BΔC23 were transfected into U2-OS cells, followed by fixation and confocal laser scanning microscopy. (C) Membrane flotation analyses. Hypotonic lysates of U-2 OS cells transfected with the constructs above were analyzed by equilibrium centrifugation through 5 to 37.5% (wt/vol) Nycodenz gradients. Fractions were collected from the top and analyzed by immunoblotting using monoclonal antibody JL-8 against GFP (Clontech, Palo Alto, CA). Under these conditions, membranes and associated proteins float to the upper, low-density fractions while soluble and aggregated material remains in the lower, high-density fractions.
To experimentally validate these predictions, we fused either full-length NS5B or the C-terminal 31 amino acids of NS5B derived from an infectious GBV-B cDNA clone (4) (kindly provided by Jens Bukh, University of Copenhagen, Denmark) via a serine-glycine linker sequence to the C terminus of green fluorescent protein (GFP), yielding construct pCMVGFP-GBV-B-NS5B or pCMVGFP-GBV-B-NS5BC31, respectively. In addition, a construct was prepared in which GBV-B NS5B with a deletion of its C-terminal 23 amino acids was fused to GFP (pCMVGFP-GBV-B-NS5BΔC23). The subcellular localization of these constructs was analyzed by confocal laser scanning microscopy. As shown in Fig. 1B, GFP alone is distributed diffusely within the cell. In contrast, the fine reticular cytoplasmic fluorescence pattern with perinuclear enhancement and staining of the nuclear membrane clearly indicates that full-length GBV-B NS5B is targeted to intracellular membranes. The same pattern was observed when only the C-terminal 31 amino acids of GBV-B NS5B were fused to GFP. Importantly, deletion of the C-terminal 23 amino acids of GBV-B NS5B resulted in diffuse cytoplasmic and nuclear staining, with some accumulation in nucleoli, as described previously for an HCV NS5B construct with a deletion of its C-terminal 21 amino acids (16).
Membrane association of GBV-B NS5B through a C-terminal membrane anchor was confirmed by membrane flotation analyses. In brief, hypotonic lysates of transfected cells were analyzed by equilibrium centrifugation in Nycodenz gradients, as described previously (2, 12). Under these conditions, membranes and associated proteins float to the top of the gradient while soluble and aggregated material remains at the bottom. As shown in Fig. 1C, the fusion proteins comprising either full-length GBV-B NS5B or only its C-terminal 31 amino acids were found in the membrane fractions, while GFP alone or the GBV-B NS5BΔC23 construct remained in the bottom fractions. In conclusion, these results demonstrate that the C-terminal segment of GBV-B NS5B mediates membrane association. Therefore, the GBV-B RdRp, as is its HCV counterpart, is a tail-anchored protein.
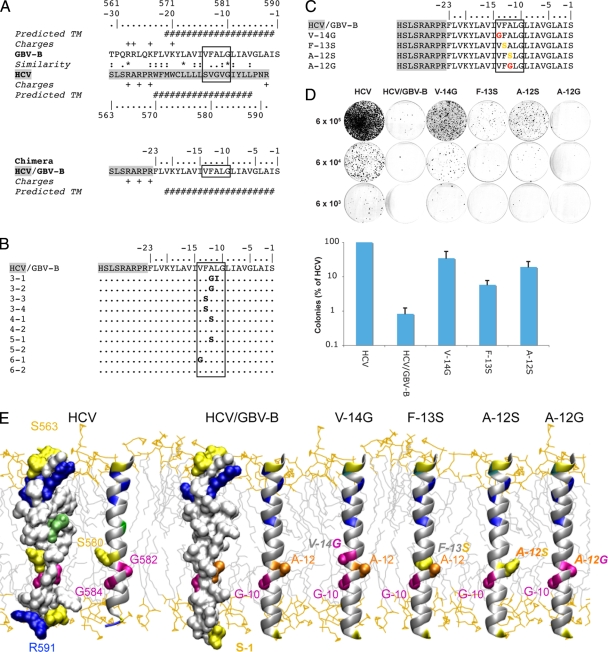
To explore whether the membrane segments of these RdRps can functionally replace each other, we generated a subgenomic genotype 1b replicon in which the HCV NS5B TMD was replaced by the corresponding newly identified segment from GBV-B NS5B. As shown in Fig. 2A, the design of the chimeric replicon was based on (i) the similarity of amino acid sequences, (ii) the predicted transmembrane segments, and (iii) the distribution of charged residues. Positively charged amino acid residues flanking the HCV NS5B TMD at the N-terminal end were preserved, as these may be involved in proper positioning of the membrane anchor. In the GBV-B sequence, an additional positively charged lysine residue can be found at position −20. We assumed that amino acids phenylalanine, leucine, and valine at positions −21 to −23 are part of the membrane anchor due to their hydrophobic character. In this setting, lysine −20 would be located inside the membrane, which is conceivable given the length of the lysine side chain that allows the charge to reach the membrane surface (17).
FIG. 2.
Chimeric HCV/GBV-B replicon. (A) Design of a chimeric replicon with the catalytic domain of HCV NS5B and the membrane anchor segment of GBV-B NS5B. This design is based on (i) the similarity of amino acid sequences (ClustalW alignment) (asterisk, invariant; colon, highly similar; dot, similar), (ii) the predicted transmembrane segments (see Fig. 1A and reference 7 for GBV-B and HCV, respectively), and (iii) the distribution of charged residues. Amino acids are numbered with respect to HCV and GBV-B NS5B proteins and are numbered negatively from the C terminus of GBV-B NS5B. The box indicates the conserved structural motif, as described in the text. (B) Sequencing of the chimeric HCV/GBV-B replicon. The C-terminal chimeric region is depicted. The first number of the clone designation represents the cell colony and the second the bacterial clone. (C and D) Identification of compensatory amino acid changes in the GBV-B NS5B TMD. (C) C-terminal amino acid sequences of the reengineered replicon constructs. (D) RNA replication of the chimeric constructs. In vitro-transcribed replicon RNA was electroporated into Huh-7.5 cells, followed by G418 selection and colony staining 3 weeks later. Cells (6 × 105, 6 × 104, and 6 × 103 per 100-mm tissue culture dish) were plated from each electroporation. Results of a representative experiment are shown in the upper panel. The lower panel illustrates the mean colony numbers ± standard deviations (SD) from four independent experiments. (E) Three-dimensional structure models of the TMDs of HCV NS5B, the HCV/GBV-B NS5B chimera, and reengineered mutants. Models were tentatively positioned within a phospholipid bilayer. For HCV NS5B and the HCV/GBV-B chimera, both amino acid surface and ribbon representations are shown to highlight the holes at the TMD surface due to the short side chain residues in the conserved structural motif (boxed in panels A, B and C). For the reengineered mutants, the side chain surfaces of key residues are represented together with the overall ribbon representations. Amino acid numbering of the HCV/GBV-B chimera from the C terminus is according to panels A, B, and C. Residues are colored according to side chain chemical properties: hydrophobic, gray; polar (Ser, Thr, Asn, Gln), yellow; basic (Arg and Lys), blue; acidic (Asp and Glu), red; Cys, green. Glycine and alanine residues within the conserved structural motif are magenta and orange, respectively. Mutated residues at positions −12, −13, and −14 in the HCV/GBV-B chimera are colored accordingly. The structure model of the HCV RdRp TMD was deduced from nuclear magnetic resonance analyses of a synthetic peptide comprising NS5B amino acids 563 to 591 (F. Penin, unpublished data). To construct the model structure of GBV-B NS5B TMD, similar α-helical transmembrane segments of known three-dimensional structure were used as templates. The chimera model was assembled by combining the structures of segment 563 to 569 of HCV NS5B (F. Penin, unpublished data), segment 46 to 72 of F1F0 ATP synthase subunit c (Protein Data Bank [PDB] entry 1C0V), and segment 26 to 48 of phospholamban (PDB entry 2KB7) using the Swiss-PdbViewer program (http://spdbv.vital-it.ch/). The figure was generated from three-dimensional atom coordinates using Visual Molecular Dynamics (http://www.ks.uiuc.edu/Research/vmd) and rendered with POV-Ray (http://www.povray.org).
Since the C-terminal HCV NS5B coding region overlaps with an essential cis-acting replication element (CRE) (5, 19), this NS5B CRE was duplicated after the stop codon in order to avoid effects on RNA replication resulting from an alteration of RNA secondary structures (5, 14). Plasmid pBSK8499-9605/dv1-3, harboring an NS5B CRE duplication (kindly provided by Ralf Bartenschlager, University of Heidelberg, Heidelberg, Germany), was used to construct wild-type and chimeric replicons designated pHCVrep/neo-S2201I-dsl3 and pHCVrep/neo-S2201I-GBV-B-NS5BC23-dsl3, respectively. In vitro-transcribed replicon RNA was electroporated into Huh-7.5 cells (1) (kindly provided by Charles M. Rice, Rockefeller University, New York, NY), followed by G418 selection and colony staining 3 weeks later. Interestingly, the HCV/GBV-B chimeric construct was viable, albeit with an approximately 120-fold-reduced efficiency compared to that of the wild-type HCV replicon harboring the NS5B CRE duplication (Fig. 2D).
The significantly reduced colony formation efficiency suggested that viable chimeric clones had acquired compensatory changes. In order to identify such changes, selected clones were expanded, followed by RNA extraction, reverse transcription, PCR amplification, and sequencing of the HCV/GBV-B chimeric reading frame. Strikingly, amino acid changes were found to cluster in a narrow region in the center of the GBV-B NS5B TMD and represented primarily changes to amino acids with small and polar side chains, such as glycine and serine (Fig. 2B). Additional changes, but without any consistent pattern or clustering, were found in the HCV polyprotein (data not shown).
To test whether the amino acid changes identified within the GBV-B NS5B TMD were indeed compensatory changes, we reengineered them into the parental pHCVrep/neo-S2201I-GBV-B-NS5BC23-dsl3 replicon construct (Fig. 2C). The leucine-to-isoleucine change observed in clone 3-1 was not analyzed further, because this substitution preserves the amino acid physicochemical properties. As shown in Fig. 2D, the change from valine at position −14 (V−14G) to glycine and from alanine at position −12 to serine (A−12S) resulted in markedly enhanced replication efficiency (approximately 42- and 23-fold, respectively). Furthermore, the change from phenylalanine at position −13 to serine (F−13S) enhanced the replication efficiency approximately 7-fold. In contrast, the change from alanine at position −12 to glycine (A−12G) (clones 3-1 and 3-2 in Fig. 2B) did not increase the number of G418-resistant colonies, suggesting that changes elsewhere in the polyprotein compensated for the replication efficiency of this clone. Taken together, compensatory amino acid changes in the center of the GBV-B NS5B TMD partially restored replication of the HCV/GBV-B chimeric replicon.
To compare the positions of compensatory changes relative to the HCV TMD, structure models were generated by molecular modeling using homologous TMDs of known structure as template and the Swiss-PdbViewer program as described in the legend to Fig. 2E. As shown in Fig. 2E, amino acid changes in the GBV-B RdRp TMD clustered within a narrow region that corresponds to the position of an absolutely conserved GVG motif in the HCV RdRp TMD. We have reported previously that replacement of the glycine residues within this motif by leucine (LVL mutant) disrupts HCV RNA replication (14). These observations, together with the physicochemical characteristics of the compensatory changes, suggest that small polar and flexible amino acids are required at this position within the HCV TMD to generate a structural element involved in essential intramembrane protein-protein interactions. In this context, the compensatory changes of helix-stabilizing large residues valine and phenylalanine into helix-breaker residues serine and glycine are expected to provide more flexibility to transmembrane α-helix folding, which should favor further interactions with other TMDs. Therefore, serine and glycine residues could directly contribute to the formation of a specific interaction site or modify specific interactions indirectly. These different features may explain the distinct phenotypes observed for the A−12S and A−12G substitutions. Taken together, these data strongly suggest that a conserved structural element within the HCV and GBV-B NS5B TMDs is essential for the establishment of a functional replication complex.
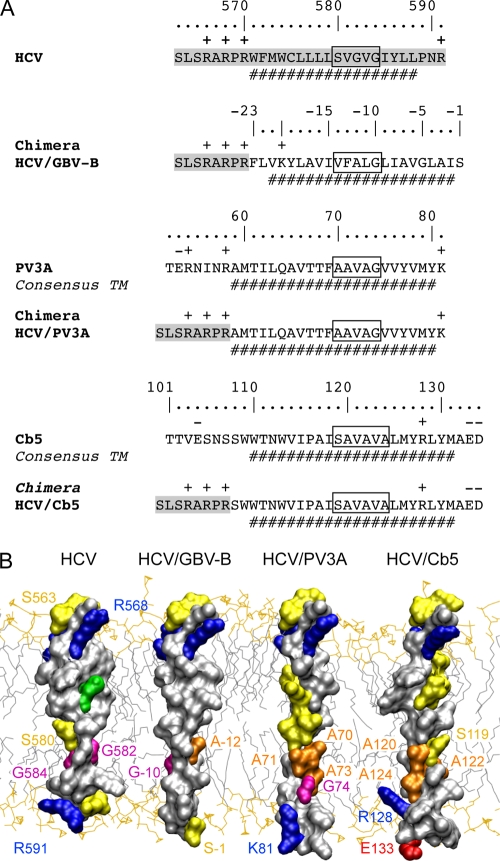
It has been reported previously that the HCV RdRp TMD can be functionally replaced by the membrane segment of poliovirus protein 3A (PV3A) (10). In accordance with this report, we found that an HCV-PV3A chimera replicated almost as efficiently as the wild type in the context of subgenomic replicons with an NS5B CRE duplication (Fig. 3A and data not shown). In contrast, a chimeric construct harboring the C-terminal membrane segment of human cytochrome b5 (Cb5), a prototype tail-anchored protein (8), did not yield any viable clones (Fig. 3A and data not shown). As shown in Fig. 3A, the same HCV sequence was present on the N-terminal side of the different TMDs in all chimeras, allowing comparable N-terminal positioning in the phospholipid bilayer of all resulting three-dimensional models. Interestingly, the putative interaction site identified above consists primarily of amino acids with small side chains in HCV and PV3A (Fig. 3A, boxes). A possible difference in the Cb5 TMD at the position of the putative interaction site could be the more rigid organization due to the presence of helix-stabilizing alanine residues and the absence of glycine helix-breaker residues, as shown in the three-dimensional model in Fig. 3B. One can speculate, therefore, that the introduction of glycine and serine residues allows the flexible adaptation of a structural interaction element in the GBV-B segment that is present in HCV and PV3A. In contrast, this element required for RNA replication is absent in Cb5 or impeded by its rigid structural organization. Even though this hypothesis of a defined interaction site composed of small amino acids remains speculative at this point, our data clearly highlight a conserved structural motif within the HCV RdRp TMD that is critical for HCV replication and is conserved to some extent in the TMDs of the GBV-B RdRp and of PV3A.
FIG. 3.
Comparison of the TMDs of HCV NS5B, GBV-B NS5B, PV3A, and Cb5. (A) HCV/PV3A and HCV/Cb5 TMD chimeras were designed in analogy to the HCV/GBV-B construct, as detailed in the legend to Fig. 2A. Amino acids are numbered with respect to PV3A and Cb5 proteins. For clarity, the ClustalW sequence comparisons are not reported. (B) Amino acid surface representations. Amino acid color coding and labeling are as described in the legend to Fig. 2E. The structure models of the HCV/PV3A and HCV/Cb5 TMDs were constructed as described in the legend to Fig. 2E. In the case of the HCV/PV3A chimeric protein, segment 16 to 44 of the Pf1 major coat protein (PDB entry 2KLV) and segment 21 to 31 of photosystem I subunit PSAX (PDB entry 1JB0) served as templates. The HCV/Cb5 three-dimensional model is based on comparison with segment 198 to 228 of aquaporin (PDB entry 3LLQ). The N termini of the chimeras were positioned at the same location with respect to the membrane in order to simplify comparisons.
In conclusion, we identified the GBV-B RdRp as a new member of and the second polymerase within the tail-anchored protein family. Moreover, analyses of an HCV/GBV-B chimera allowed the identification of a conserved structural motif within the HCV RdRp TMD that is essential for the assembly of a functional replication complex. This motif appears to be shared among HCV, GBV-B, and PV3A but not Cb5, a prototype tail-anchored protein. Further exploring the function and interactions of this motif may yield new insights into the functional architecture of the HCV replication complex and may identify novel targets for therapeutic intervention.
Acknowledgments
We gratefully acknowledge Jens Bukh, Ralf Bartenschlager, and Charles M. Rice for reagents.
This work was supported by the Deutsche Forschungsgemeinschaft (Br 3440/2-1 and 3440/3-1), the Swiss National Science Foundation (3100A0-107831 and 3100A0-122447), the Novartis Foundation, the French Centre National de la Recherche Scientifique (CNRS), the Agence Nationale pour la Recherche sur le SIDA et les Hépatites Virales (ANRS), and the program Biotherapeutics of Lyon Biopole.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brass, V., J. M. Berke, R. Montserret, H. E. Blum, F. Penin, and D. Moradpour. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. U. S. A. 105:14545-14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bright, H., A. R. Carroll, P. A. Watts, and R. J. Fenton. 2004. Development of a GB virus B marmoset model and its validation with a novel series of hepatitis C virus NS3 protease inhibitors. J. Virol. 78:2062-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh, J., C. L. Apgar, and M. Yanagi. 1999. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470-478. [DOI] [PubMed] [Google Scholar]
- 5.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghany, M. G., D. B. Strader, D. L. Thomas, and L. B. Seeff. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivashkina, N., B. Wölk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, P. K., F. Janiak-Spens, W. S. Trimble, B. Leber, and D. W. Andrews. 1997. Evidence for multiple mechanisms for membrane binding and integration via carboxyl-terminal insertion sequences. Biochemistry 36:8873-8882. [DOI] [PubMed] [Google Scholar]
- 9.Kutay, U., E. Hartmann, and T. A. Rapoport. 1993. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3:72-75. [DOI] [PubMed] [Google Scholar]
- 10.Lee, H., Y. Liu, E. Mejia, A. V. Paul, and E. Wimmer. 2006. The C-terminal hydrophobic domain of hepatitis C virus RNA polymerase NS5B can be replaced with a heterologous domain of poliovirus protein 3A. J. Virol. 80:11343-11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindenbach, B. D., H.-J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott-Raven, Philadelphia, PA.
- 12.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835-40843. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 17.Strandberg, E., and J. A. Killian. 2003. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 544:69-73. [DOI] [PubMed] [Google Scholar]
- 18.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 19.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]