Abstract
We analyzed the biochemical and ultrastructural properties of hepatitis C virus (HCV) particles produced in cell culture. Negative-stain electron microscopy revealed that the particles were spherical (∼40- to 75-nm diameter) and pleomorphic and that some of them contain HCV E2 protein and apolipoprotein E on their surfaces. Electron cryomicroscopy revealed two major particle populations of ∼60 and ∼45 nm in diameter. The ∼60-nm particles were characterized by a membrane bilayer (presumably an envelope) that is spatially separated from an internal structure (presumably a capsid), and they were enriched in fractions that displayed a high infectivity-to-HCV RNA ratio. The ∼45-nm particles lacked a membrane bilayer and displayed a higher buoyant density and a lower infectivity-to-HCV RNA ratio. We also observed a minor population of very-low-density, >100-nm-diameter vesicular particles that resemble exosomes. This study provides low-resolution ultrastructural information of particle populations displaying differential biophysical properties and specific infectivity. Correlative analysis of the abundance of the different particle populations with infectivity, HCV RNA, and viral antigens suggests that infectious particles are likely to be present in the large ∼60-nm HCV particle populations displaying a visible bilayer. Our study constitutes an initial approach toward understanding the structural characteristics of infectious HCV particles.
Hepatitis C virus (HCV) is a major cause of chronic hepatitis worldwide, with approximately 170 million humans chronically infected. Persistent HCV infection often leads to fibrosis, cirrhosis, and hepatocellular carcinoma (27). There is no vaccine against HCV, and the most widely used therapy involves the administration of type I interferon (IFN-α2Α) combined with ribavirin. However, this treatment is often associated with severe adverse effects and is often ineffective (53).
HCV is a member of the Flaviviridae family and is the sole member of the genus Hepacivirus (43). HCV is an enveloped virus with a single-strand positive RNA genome that encodes a unique polyprotein of ∼3,000 amino acids (14, 15). A single open reading frame is flanked by untranslated regions (UTRs), the 5′ UTR and 3′ UTR, that contain RNA sequences essential for RNA translation and replication, respectively (17, 18, 26). Translation of the single open reading frame is driven by an internal ribosomal entry site (IRES) sequence residing within the 5′ UTR (26). The resulting polyprotein is processed by cellular and viral proteases into its individual components (reviewed in reference 55). The E1, E2, and core structural proteins are required for particle formation (5, 6) but not for viral RNA replication or translation (7, 40). These processes are mediated by the nonstructural (NS) proteins NS3, NS4A, NS4B, NS5A, and NS5B, which constitute the minimal viral components necessary for efficient viral RNA replication (7, 40).
Expression of the viral polyprotein leads to the formation of virus-like particles (VLPs) in HeLa (48) and Huh-7 cells (23). Furthermore, overexpression of core, E1, and E2 is sufficient for the formation of VLPs in insect cells (3, 4). In the context of a viral infection, the viral structural proteins (65), p7 (31, 49, 61), and all of the nonstructural proteins (2, 29, 32, 41, 44, 63, 67) are required for the production of infectious particles, independent of their role in HCV RNA replication. It is not known whether the nonstructural proteins are incorporated into infectious virions.
The current model for HCV morphogenesis proposes that the core protein encapsidates the viral genome in areas where endoplasmic reticulum (ER) cisternae are in contact with lipid droplets (47), forming HCV RNA-containing particles that acquire the viral envelope by budding through the ER membrane (59). We along with others showed recently that infectious particle assembly requires microsomal transfer protein (MTP) activity and apolipoprotein B (apoB) (19, 28, 50), suggesting that these two components of the very-low-density lipoprotein (VLDL) biosynthetic machinery are essential for the formation of infectious HCV particles. This idea is supported by the reduced production of infectious HCV particles in cells that express short hairpin RNAs (shRNAs) targeting apolipoprotein E (apoE) (12, 30).
HCV RNA displays various density profiles, depending on the stage of the infection at which the sample is obtained (11, 58). The differences in densities and infectivities have been attributed to the presence of host lipoproteins and antibodies bound to the circulating viral particles (24, 58). In patients, HCV immune complexes that have been purified by protein A affinity chromatography contain HCV RNA, core protein, triglycerides, apoB (1), and apoE (51), suggesting that these host factors are components of circulating HCV particles in vivo.
Recent studies using infectious molecular clones showed that both host and viral factors can influence the density profile of infectious HCV particles. For example, the mean particle density is reduced by passage of cell culture-grown virus through chimpanzees and chimeric mice whose livers contain human hepatocytes (39). It has also been shown that a point mutation in the viral envelope protein E2 (G451R) increases the mean density and specific infectivity of JFH-1 mutants (70).
HCV particles exist as a mixture of infectious and noninfectious particles in ratios ranging from 1:100 to 1:1,000, both in vivo (10) and in cell culture (38, 69). Extracellular infectious HCV particles have a lower average density than their noninfectious counterparts (20, 24, 38). Equilibrium sedimentation analysis indicates that particles with a buoyant density of ∼1.10 to 1.14 g/ml display the highest ratio of infectivity per genome equivalent (GE) both in cell culture (20, 21, 38) and in vivo (8). These results indicate that these samples contain relatively more infectious particles than any other particle population. Interestingly, mutant viruses bearing the G451R E2 mutation display an increased infectivity-HCV RNA ratio only in fractions with a density of ∼1.1 g/ml (21), reinforcing the notion that this population is selectively enriched in infectious particles.
The size of infectious HCV particles has been estimated in vivo by filtration (50 to 80 nm) (9, 22) and by rate-zonal centrifugation (54 nm) (51) and in cell culture by calculation of the Stokes radius inferred from the sedimentation velocity of infectious JFH-1 particles (65 to 70 nm) (20). Previous ultrastructural studies using patient-derived material report particles with heterogeneous diameters ranging from 35 to 100 nm (33, 37, 42, 57, 64). Cell culture-derived particles appear to display a diameter within that range (∼55 nm) (65, 68).
In this study we exploited the increased growth capacity of a cell culture-adapted virus bearing the G451R mutation in E2 (70) and the enhanced particle production of the hyperpermissive Huh-7 cell subclone Huh-7.5.1 clone 2 (Huh-7.5.1c2) (54) to produce quantities of infectious HCV particles that were sufficient for electron cryomicroscopy (cryoEM) analyses. These studies revealed two major particle populations with diameters of ∼60 and ∼45 nm. The larger-diameter particles were distinguished by the presence of a membrane bilayer, characterized by electron density attributed to the lipid headgroups in its leaflets. Isopycnic ultracentrifugation showed that the ∼60-nm particles are enriched in fractions with a density of ∼1.1 g/ml, where optimal infectivity-HCV RNA ratios are observed. These results indicate that the predominant morphology of the infectious HCV particle is spherical and pleomorphic and surrounded by a membrane envelope.
MATERIALS AND METHODS
Cells and viruses.
Hyperpermissive Huh-7.5.1 cell subclone Huh-7.5.1 clone 2 cells (54, 69) were cultured in Dulbecco's modified Eagle's medium (DMEM)-10% fetal calf serum (FCS) at 37°C and 5% CO2. Cell culture-adapted JFH-1 variant D183 virus was previously described (70).
Determination of viral infectivity titers and HCV RNA copy number.
Infectivity titers were determined in Huh-7 cells by endpoint dilution and immunofluorescence, as previously described (20, 69). Infectivity titers are expressed as the number of infection focus forming units per ml (FFU/ml).
HCV RNA levels were determined in total cellular RNA extracted by the guanidinium thiocyanate-phenol-chloroform (GTC) method (13) followed by quantitative reverse transcription-PCR (RT-qPCR), as previously described (69). To determine HCV copy numbers, standard curves were prepared by serial 10-fold dilution of a known amount of a plasmid bearing the amplified HCV sequence (69).
Virus production, concentration, and purification.
Huh-7.5.1. cells (3.5 × 106 cells/flask) were plated in T162 vented cap cell culture flasks (Corning, Lowell, MA). The next day, cells were inoculated at a multiplicity of 0.01 with D183 virus stocks and incubated at 37°C. Supernatants were collected after 3 days and replaced with 15 ml/flask of serum-free OptiMem medium (Invitrogen, Carlsbad, CA). After an additional 20 h of incubation, the supernatants were collected, the cells were replenished with 15 ml of warm OptiMem and incubated for an additional 20 h, and then the final supernatants were collected. The supernatants from days 4 and 5 were filtered through a 0.45-μm-pore-size filter and stored at −80°C.
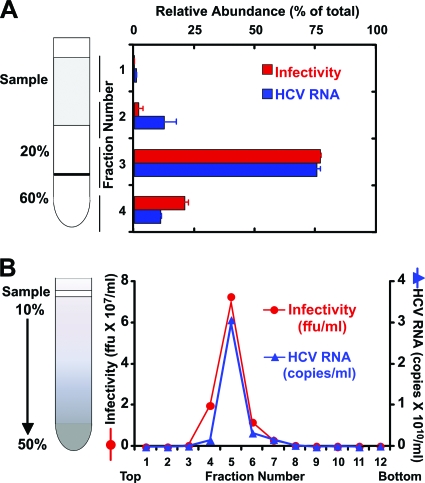
Crude virus supernatants, typically ∼300 ml (from 20 T162 flasks), were concentrated in Centricon centrifugal devices with a 100-kDa cutoff (Millipore, Temecula, CA) by centrifugation at 3,500 rpm until they reached the desired final volume (∼1 ml). Concentrated samples were overlaid onto a 1-ml 20% sucrose-TNE (10 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM EDTA) layer floating over a cushion of 1 ml of 60% sucrose-TNE (Fig. 1A). Samples were subjected to ultracentrifugation in an SW60 rotor for 2 h at 150,000 × g. Four fractions were collected by aspiration from the top, and most of the infectivity and HCV RNA were concentrated in fraction 3, which corresponded to the 20 to 60% sucrose interphase (Fig. 1A). Sucrose was removed from fraction 3 by exchange into TNE buffer by centrifugation in Microcon-100 centrifugal devices with a 100-kDa cutoff (Millipore, Temecula, CA).
FIG. 1.
Infectious HCV particles purified by sucrose gradient centrifugation. (A) Concentrated supernatants isolated from infected cells were subjected to ultracentrifugation on a sucrose step gradient. (B) The particles collected from the 20-60% interface (fraction 3) were further purified in a continuous 10 to 50% sucrose gradient. Infectivity and HCV RNA levels were determined by titration and quantitative PCR, and the results were expressed as the numbers of FFU per ml and HCV RNA genome equivalents (GE) per ml, respectively.
Purification of total virus populations by sedimentation velocity.
Cushion-purified virus samples were diluted in 200 μl of TNE buffer and overlaid onto a continuous (10 to 50%) sucrose gradient (3.6 ml) (Fig. 1B). Rate-zonal ultracentrifugation at 200,000 × g for 1 h separated particles in the supernatant by virtue of their sedimentation velocity (20). Twelve fractions (∼300 μl) were collected from the top and analyzed for viral infectivity and HCV RNA. Most of the infectivity and HCV RNA were typically located in fractions 5 and 6 (Fig. 1B), which were pooled, buffer exchanged into phosphate-buffered saline (PBS), and concentrated into small volumes (∼25 μl) by ultrafiltration through Microcon-100 filters (Millipore, Temecula, CA).
Purification of viral particles by isopycnic ultracentrifugation density.
Cushion-purified, concentrated virus samples (∼200 μl) were overlaid onto a discontinuous gradient with 20, 30, 40, 50, and 60% sucrose steps (∼700 μl) and subjected to overnight ultracentrifugation at 120,000 × g, as previously described (20). Twelve fractions were collected from the top and analyzed for buoyant density, viral infectivity, and HCV RNA content. The highest infectivity-HCV RNA ratio was detected in fractions whose density ranged from 1.10 to 1.14 g/ml. These fractions were pooled, buffer exchanged into PBS using Microcon filters as described above, and compared with pooled fractions of lower (<1.1g/ml) and higher (>1.4 g/ml) densities, which displayed substantially lower specific infectivity than the intermediate-density fractions.
Virus immunoprecipitation experiments.
Agarose beads (∼50 μl) containing the monoclonal neutralizing recombinant human IgG against HCV E2 (AR3A) (36) or the anti-HIV recombinant human IgG (B6) (52) were kindly provided by Mansun Law (The Scripps Research Institute, La Jolla, CA). The beads were equilibrated in PBS and incubated overnight at 4°C with ∼250 μl of crude virus preparations in OptiMem (diluted 1:5 in PBS). As a control, diluted virus was incubated in the absence of beads to determine the baseline infectivity and HCV RNA values. After overnight incubation, the supernatant was collected and tested for infectivity. RNA was extracted from washed (three times with PBS) beads using GTC (4.2 M guanidine thiocyanate, 0.75 M sodium citrate, pH 7.3, 0.5% sarcosyl) (13), and the HCV RNA copy number was determined as described above.
Negative-stain electron microscopy.
Samples (25 μl) were fixed by addition of 1 volume of 8% paraformaldehyde (PFA) in 0.1 M cacodylate buffer (pH 7.4). After incubation for 20 min, aliquots (4 to 5 μl) were placed on Parafilm (Alcan Packaging-Neenah, WI). Parlodion-coated nickel grids were subjected to glow discharge and then immediately inverted onto the droplets. After incubation for 3 min at room temperature (RT), the grids were blotted, and negative staining was performed by application of 3% uranyl acetate (pH 4) for 2 min at RT. The stain was removed by blotting, and the grids were air dried. Grids were examined using a transmission electron microscope (Philips FEI CM100; Eindhoven, Netherlands) operating at 100 kV. Digital images were generated by direct recording using a charge-coupled-device (CCD) camera (SIS Megaview III camera; Olympus, Munster, Germany) or by scanning micrographs (Finescan 2750 high resolution scanner; Fujifilm, Edison, NJ).
Immunolabeling.
Sample grids were inverted onto a PBS-5% bovine serum albumin (BSA) droplet for 5 min at RT and then incubated for 1 h at RT with human monoclonal anti-E2 AR3A antibody (36) and/or a mouse monoclonal anti-apoE antibody (clone 3D12; Meridian Life Science, Inc., Memphis, TN) diluted 1:20 and 1:10 in PBS-1% BSA, respectively. The grids were then washed three times (2 min each time) with PBS-1% BSA, followed by incubation for 1 h at RT with the corresponding secondary antibodies conjugated to 6- or 12-nm gold particles (Jackson Labs, Watson Grove, PA) at various dilutions in PBS-1% BSA. The grids were then washed with PBS, fixed for 5 min in PBS-1% glutaraldehyde, washed with H2O for 5 min, and stained and imaged as described above.
Detergent treatment.
Purified particles were fixed with 1% glutaraldehyde for 20 min at RT. One aliquot was treated with one volume of 0.4% NP-40 in PBS for 5 min, and a second aliquot was treated with detergent-free PBS. Both samples were then purified through a 20% sucrose cushion, buffer exchanged into PBS, and then negatively stained as described above.
Viral antigen quantification.
Relative core protein levels were determined by enzyme-linked immunosorbent assay (ELISA) as previously described (20). Relative E2 levels were quantified by ELISA using a recombinant anti-E2 monoclonal IgG (AR3A) (36). Briefly, gradient fractions were adsorbed overnight at 4°C onto a Nunc Maxisorp ELISA plate (Thermo Fisher Scientific, Rochester, NY). Wells were washed twice with 200 μl of PBS and fixed for 20 min at RT with 4% PFA in PBS (pH 7). Wells were washed twice and blocked with 5% nonfat milk in PBS for 1 h at RT. Wells were then washed twice with 200 μl of PBS and replenished with 50 μl of a 2 μg/ml dilution of the recombinant anti-E2 IgG (AR3A) (36) in binding buffer (3% BSA-0.3% Triton X-100 in PBS). Wells were washed four times with 200 μl of PBS and replenished with 50 μl of an 80 ng/ml dilution of horseradish peroxidase (HRP)-conjugated goat anti-human antibodies (Pierce, Rockford, IL). After incubation for 1 h at RT, wells were washed four times with PBS before the developing reagent was added (Pierce, Rockford, IL). The colorimetric reaction was stopped by adding 1 volume of 1 M H2SO4. Serial 2-fold dilutions of the peak fraction were used to generate a standard curve that was used for conversion of the values of the optical density 450 nm (OD450).
Electron cryomicroscopy.
For biosafety, highly concentrated virus preparations were fixed in PFA-cacodylate buffer as described above to inactivate viral infectivity. Purified HCV samples were vitrified by standard methods for cryoEM (66). In brief, an aliquot (∼3 μl) was applied to a glow-discharged, perforated carbon-coated grid (either 2/4 Cu-Rh Quantifoil or 2/2-4C C-flat), blotted with filter paper, and rapidly plunged into liquid ethane. Low-dose images were recorded at a magnification of ×50,000 on an FEI Tecnai F20 Twin transmission electron microscope operating at 120 kV, with a nominal underfocus ranging from 1.5 to 3.5 μm and a pixel size of 0.271 nm at the specimen level. All images were recorded with a Gatan 4,000- by 4,000-pixel CCD camera utilizing the manual mode of Leginon data collection software (62). The grids were maintained at −180°C using a Gatan 626 cryo-stage.
Image analysis.
Image analysis was performed using the NIH open source software Image J (available at: http://rsbweb.nih.gov/ij/). Particle diameters were measured on digitized images, both point to point by ruler and by comparison with circles of defined radii. Analysis was performed only on intact particles, with fewer than 1% of the particles appearing to be broken.
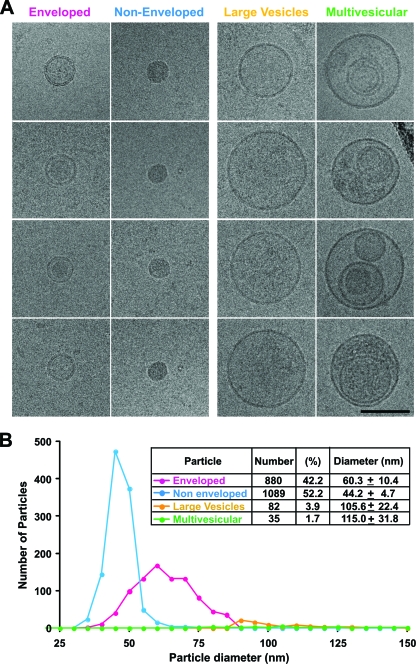
Classification of particle types into zero, one, or multiple membrane bilayers was based on morphological analysis of the cryoEM images by two or more independent observers. Only particles displaying a distinct electron-dense bilayer were considered enveloped (E). Nonenveloped (NE) particles did not display a visible bilayer. A minor population of large vesicular (LV) particles was observed that consisted of enveloped particles whose diameters were larger than 85 nm. Finally, a minor multivesicular (MV) particle population was observed that displayed two or more clearly distinguishable bilayers.
For illustration, raw images were resized and processed using the Gaussian blur algorithm (radius, 2 to 3 pixels) in Adobe Photoshop CS2 (version 9.0.2) for Mac.
Statistical analysis.
Statistical significance of the difference between the mean diameters of particles with different buoyant densities was calculated using a one tailed, two-sample t test assuming unequal variance (heteroscedastic).
RESULTS
Preparation of highly purified HCV particles.
The low yield of infectious HCV in cell culture has impeded ultrastructural and biochemical analysis. This barrier has been partially overcome by the development of a robust cell culture system. By the use of a cell culture-adapted (D183) virus with vigorous growth characteristics (70) and a hyperpermissive cell clone derived from Huh-7.5.1 cells (54), the yield of infectious virus was increased by two logs from a titer of ∼104 to ∼106 FFU/ml. Infection of Huh-7.5.1 clone 2 (Huh-7.5.1 c2) cells at a low multiplicity of infection ([MOI] 0.01) produced stocks that routinely contained 2 × 106 to 5 × 106 FFU/ml 3 days after infection. Substitution of complete medium with serum-free OptiMem (Invitrogen, Carlsbad, CA) 20 h before collection of the supernatants did not reduce the yield of infectious virus (data not shown), and high-titer (>106 FFU/ml) virus stocks with reduced serum content could be generated. This enabled efficient concentration of infectious supernatants by ultrafiltration through anisotropic membranes (Centricon filters with a 100-kDa cutoff; Millipore, Temecula, CA), resulting in virus preparations containing ∼108 FFU/ml and nearly 1011 HCV RNA GEs/ml.
We purified the concentrated viral particles by ultracentrifugation as described in Materials and Methods. Briefly, concentrated supernatants were pelleted through a 20% sucrose cushion onto a 60% cushion (Fig. 1A). Partially purified particles were collected from the 20-60% interface, where most (>75%) of the infectivity and HCV RNA were recovered (Fig. 1A, fraction 3). This material was subjected to ultracentrifugation in a continuous 10 to 50% sucrose gradient to purify viral particles based on their sedimentation velocities (Fig. 1B). This procedure permitted recovery of virtually all the infectivity (>85%) and HCV RNA in a single peak (Fig. 1B, fraction 5), which migrated with the expected sedimentation velocity (20). In contrast to what we observed for the parental JFH-1 virus (20), the majority of both the infectivity and the HCV RNA of the D183 virus comigrated in the same fraction (Fig. 1B, fraction 5), probably due to the higher average density of the infectious D183 virus particles reflecting a point mutation (G451R) in the viral E2 protein (70).
By negative-stain EM, virus-like particles (VLPs) were observed only in the samples from infected cells (Fig. 2) and not in parallel, control samples from noninfected cells (data not shown). These preparations revealed a pleomorphic particle population with various diameters (58.7 ± 19.3 nm; n = 450) (Fig. 2), similar to those previously described for JFH-1 virus produced in cell culture (23, 65) and from infected patients (42, 57, 64). The particles displayed a smooth surface with no visible surface projections (Fig. 2).
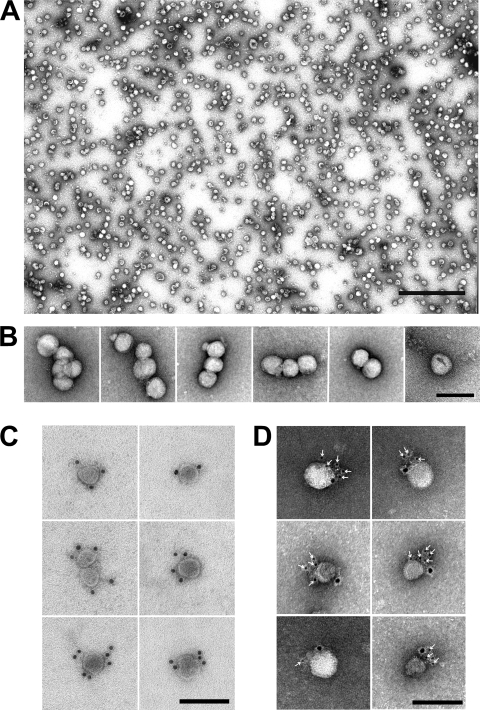
FIG. 2.
HCV particles are pleomorphic and display E2 and apolipoprotein E on their surface. (A) Electron micrograph of highly purified, negatively stained HCV particles demonstrates their fairly uniform size but pleomorphic shape. Particles were purified as described in the legend of Fig. 1. Scale bar, 500 nm. (B) Close-up views show the heterogeneous sizes and shapes. (C) Representative images show specific decoration of a subset (5 to 20%) of particles with an antibody directed against the viral envelope protein E2. (D) Representative images show specific decoration of E2-positive particles (6-nm gold) with anti-apoE antibodies (12-nm gold). Scale bar, 100 nm (B, C, and D).
In order to confirm that these preparations contained HCV particles, we incubated them with a well-characterized E2-specific neutralizing antibody (AR3A) (36) known to specifically immuno-deplete viral infectivity and immunoprecipitate HCV RNA from infectious supernatants (see Fig. S1 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS1.tif). A small but significant fraction of the particles (5 to 20%) displayed specific decoration with 12-nm gold-conjugated secondary antibody only after incubation with anti-E2 antibodies (Fig. 2C). It is noteworthy that the antibody-labeled particles were morphologically indistinguishable from those not decorated (see Fig. S2 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS2.tif). No decoration was observed with isotype control antibody (data not shown) directed against the HIV envelope (52), confirming the specificity of the interaction and the viral nature of the purified particles. Since we failed to specifically decorate the particles with different antibodies against the core protein, the major structural component of HCV particles, we set out to determine if the purified particles could be specifically decorated with antibodies against apolipoprotein E, a cellular factor shown to be associated with HCV particles in vivo (51) and in vitro (12). Virus-like particles could be specifically decorated with anti-apoE (12-nm gold) (Fig. 2D) and anti-E2 (6 nm-gold) (Fig. 2D, white arrows) and not with an isotype control antibody, reinforcing the notion that the purified particles display antigens previously found in HCV virions.
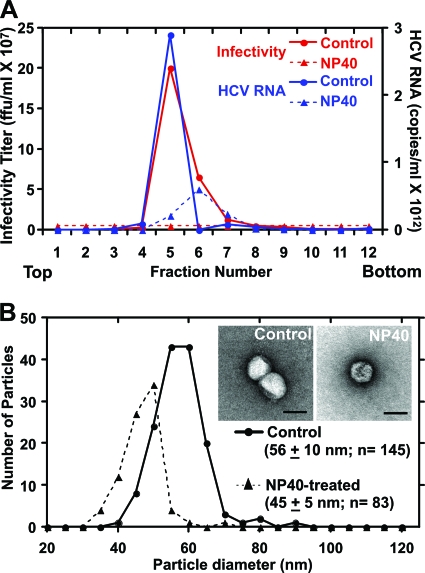
When the virus preparation was treated with detergent (0.2% NP-40), all of the infectivity and most (∼80%) of the HCV RNA content were lost (Fig. 3A), suggesting that the detergent removed the envelope from the virions and destabilized most of the capsids (46) (Fig. 3A). Negative-stain EM showed that the detergent-treated particles were smaller in diameter (45 ± 5 nm; n = 83) than untreated particles (56.1 ± 10 nm; n = 145) (Fig. 3B), which recapitulated the decrease in Stokes radii calculated by sedimentation velocity for patient-derived particles before and after detergent treatment (51). Detergent-treated particles also displayed a rough, angular surface (Fig. 3B, inset) that contrasts with the relatively smooth surface observed in untreated particles (Fig. 3B, inset).
FIG. 3.
Particles treated with 0.2% NP-40 are noninfectious and display a decrease in diameter. Detergent-treated and untreated particles were subjected to ultracentrifugation in a continuous 10 to 50% sucrose gradient. (A) Detergent treatment eliminated infectivity and resulted in a substantial loss of HCV RNA. (B) Particle diameter decreased from ∼60 to ∼45 nm with detergent treatment. Inset shows representative negatively stained particles. Scale bar, 50 nm.
Negatively stained particles were pleomorphic with an average diameter of ∼55 nm. However, the use of heavy metal stain (uranyl acetate), a solid support (carbon), and subsequent drying would likely result in deformation and loss of native structure. Thus, we examined the virus preparations by electron cryomicroscopy, where particles are vitrified in solution and observed in a close-to-native state.
cryoEM of purified particles.
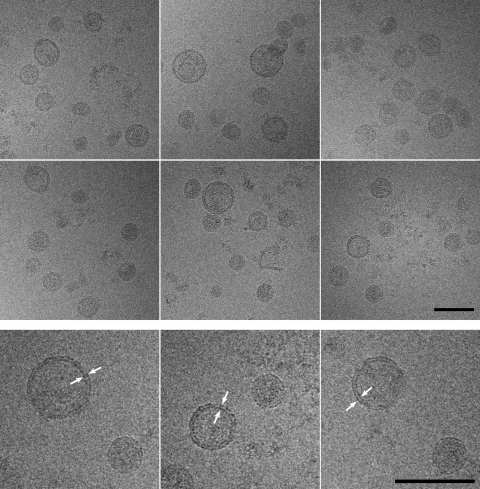
Frozen hydrated particles (Fig. 4) were pleomorphic and similar in appearance to the negatively stained particles (Fig. 2), with an average diameter of 54.6 ± 12.6 nm (range, 31 to >100 nm; n = 2,087). The predominant morphology was spherical, with a relatively smooth surface devoid of visible projections (Fig. 4). cryoEM revealed that many of the particles displayed a 5- to 6-nm-thick electron-dense bilayer (Fig. 4, arrowheads), compatible with a lipid membrane (25). This feature was used to differentiate the particle populations into four distinct classes (Fig. 5). Particles that displayed a visible bilayer were designated enveloped (E), and particles devoid of this feature were designated nonenveloped (NE) (Fig. 5A). A small number of large vesicles displayed one bilayer (LV), and multivesicular particles (MV) displayed multiple bilayers.
FIG. 4.
cryoEM showed that purified HCV particles are predominantly spherical and consist of subpopulations with different diameters. Note that the large-diameter particles are surrounded by an envelope (arrowheads), which is not detectable in the small-diameter particles. Scale bar, 100 nm.
FIG. 5.
Purified virus preparations contained two major (enveloped and nonenveloped) and two minor (large vesicle and multivesicular) subpopulations. (A) Gallery of representative electron cryomicrographs showing the four particle classes. Scale bar, 100 nm. (B) Histogram and table of 2,086 particles from four independent virus preparations revealed roughly similar numbers of enveloped and nonenveloped particles, with minor populations of large vesicles having a uniform interior density, and a few multivesicular particles that are probably nonviral and may represent exosomes.
The enveloped and nonenveloped populations were most abundant in these preparations (94%) and had similar distributions (E, 46%; NE, 53%) while the LV and MV particles represented only 4% and 1.7%, respectively. Importantly, the E particles displayed smooth surfaces, and some displayed clearly discernible internal structures that were separated from the lipid bilayers. This capsid-like object (Fig. 5) was similar in size to the NE particles, which were compact and electron dense, devoid of any internal features or surface projections (Fig. 5A). LV particles were electron lucent and occasionally displayed surface projections (Fig. 5A) while MV particles displayed variable sizes and morphologies and contained heterogeneous structures, including internal vesicles and amorphous material (Fig. 5A).
These four types of particles displayed different mean diameters (Fig. 5B). The E particles (n = 880) had a mean diameter of 60.3 ± 10.4 nm. In contrast, the NE particles had a smaller diameter with a narrower size distribution (44.24 ± 4.74 nm; n = 1,089). Finally, LV and MV particles were much larger (>85 nm), with extremely variable diameters of 105.6 ± 22.4 nm (n = 82) and 114.9 ± 31.8 nm (n = 36), respectively. Four different virus preparations displayed similar frequencies and size distributions of the four different particle populations (see Fig. S3 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS3.tif), which attests to the consistency of the purification method and analysis.
cryoEM, infectivity, and viral RNA analysis of particles of different buoyant densities.
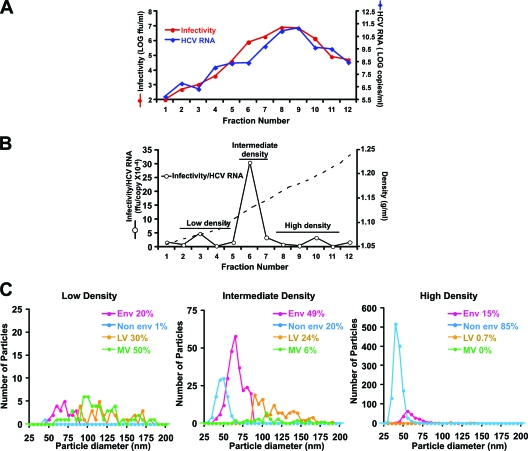
The previous results show that the predominant E and NE particle classes isolated from infection supernatants were associated with most of the HCV RNA and infectivity. To confirm the viral nature of these particles, we modified the purification scheme to separate the different particles based on their buoyant densities. Cushion-purified particles were separated by ultracentrifugation in isopycnic sucrose density gradients. Infectivity, HCV RNA, envelope (E2 protein), core protein content, and density were determined for each fraction as described in Materials and Methods and Gastaminza et al. (20). The G451R mutation is known to produce a distinct increase in the average density of infectious D183 virus particles relative to the parental JFH-1 virus (70). As expected for the D183 virus, infectivity and HCV RNA were most abundant in fractions whose density was ∼1.15 g/ml (Fig. 6 A, fractions 8 and 9). Also as expected from previous experiments (16, 20, 21), fractions around 1.1 g/ml (Fig. 6A, fractions 6 and 7) that preceded the peak of HCV RNA and infectivity consistently displayed the highest infectivity-HCV RNA ratio, suggesting that they were relatively enriched in infectious versus noninfectious particles (Fig. 6B; see also Fig. S4 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS4.tif). Therefore, the intermediate fractions preceding the peak of infectivity likely contain the largest proportion of infectious particles, albeit in lower numbers.
FIG. 6.
Enrichment of different particle populations by isopycnic sucrose density gradient ultracentrifugations. (A) Representative plot of the gradient fractions showing density, infectivity, and HCV RNA content. (B) The infectivity-HCV RNA ratio calculated from the data in panel A highlights the fractions with increased specific infectivity. (C) Plots of the size distribution and frequency of each population in pooled fractions having low, intermediate, and high densities. Population distributions were obtained from the combined analysis of two independent viral preparations.
cryoEM was then used to evaluate particle morphology in three density pools. The low-density pool (density of <1.1g/ml) (low specific infectivity) typically contained less than 2% of the infectivity and HCV RNA of the preparation. The intermediate-density pool (∼1.10 to 1.14 g/ml) displayed the highest specific infectivity and contained ∼15% of the infectivity and 5% of the total HCV RNA. Finally, the high-density pool (density of >1.14g/ml) (low specific infectivity) contained most of the infectivity (∼80%) and HCV RNA (∼94%) and most of the virus-like particles.
As shown in Fig. 6C, the low-density pool that had a low specific infectivity contained a mixture of E, LV, and MV particles, with no particular class dominating the population. In contrast, the intermediate-density pool that had a high specific infectivity was greatly enriched in E particles, which constituted 49% of the total particle population. The NE, LV, and MV particles were relatively underrepresented (20, 24, and 6%, respectively). Finally, NE particles dominated the high-density pool that had a low specific infectivity, representing 85% of the particles in that population, while E particles were much less frequent (15%). The correlation between the relative abundance of enveloped particles and the enrichment of highly infectious particles with the highest infectivity-HCV RNA ratio was verified in each of the gradients used for the analysis shown in Fig. 6C (see Fig. S5 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS5.tif), reinforcing the notion that enveloped particles are enriched in samples with higher specific infectivity. Furthermore, since the fractions enriched in enveloped particles contained high envelope (E2)/core ratios (see Fig. S6 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS6.tif), it is likely that their characteristic bilayer corresponds to the viral envelope.
Since infectious HCV particles display heterogeneous buoyant densities (Fig. 6A) and diameters (Fig. 5), we examined whether the diameters of the enveloped particles were correlated with their buoyant densities. We measured the diameters of enveloped particles with different densities and observed small, although statistically significant, differences in the diameters of high-density (57.6 ± 10.1 nm, n = 225) and intermediate-density (63.6 ± 4.5 nm, n = 260) particles (P = 2.10−13) (see Fig. S7A posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS7.tif). These differences in diameters reflected a differential size distribution in which there was an inverse relationship between particle diameter and buoyant density (see Fig. S7B posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS7.tif). Indeed, internal capsid-like structures could be visualized in enveloped particles of significantly different diameters in our preparations (see Fig. S7C posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS7.tif).
DISCUSSION
The development of a cell culture system for HCV and its optimization permitted production of high-titer virus stocks. Purification of secreted particles by sequential ultracentrifugation yielded virus preparations containing most of the infectious and noninfectious HCV RNA-containing particles.
Negative-stain EM and cryoEM analysis revealed a heterogeneous particle population with a mean diameter of ∼55 nm (Fig. 2), similar to patient-derived virus analyzed by rate-zonal ultracentrifugation (51) and negative-stain EM (33, 37, 42, 57, 64). Detergent treatment resulted in total loss of infectivity and a measurable reduction in the average virus particle diameter from ∼56 to ∼45 nm (Fig. 3), again similar to detergent-treated, patient-derived particles (51).
cryoEM analysis revealed an electron-dense bilayer around some particles (Fig. 4). This feature allowed us to distinguish two morphologically distinct particle classes (enveloped and nonenveloped) that had different diameters (∼60 versus ∼45 nm, respectively), were approximately equal in frequency, and accounted for over 90% of the total particle population (Fig. 5). Previous EM studies of patient sera described particles with similar diameters (64), indicating that the cell culture-derived virus recapitulates the morphologies of wild-type virus from infected patients.
The ∼60-nm E-type particles display a 5- to 6-nm thick surface bilayer (Fig. 4) that most likely represents a lipid membrane (25) and is consistent with removal of the envelope and reduction in particle diameter with detergent treatment (Fig. 3B). The E particles were present in all of the infectious samples we analyzed, and they were the dominant population in preparations that displayed the highest specific infectivity (i.e., infectivity-RNA ratios) and E2-core protein ratio (Fig. 6; see also Fig. S6 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS6.tif). Collectively, our results suggest that infectious HCV particles are likely present within the E particle population in these preparations. This is reinforced by the fact that the average diameter of these D183 particles is similar to the diameter of infectious JFH-1 virus particles (65 to 70 nm), which we previously calculated from their sedimentation coefficient (20). In addition, it is compatible with the diameter of infectious HCV estimated by size-based filtration and inoculation into chimpanzees (8, 22).
The second major particle population (nonenveloped particles) is devoid of an observable bilayer (Fig. 5) and displays the average diameter of detergent-treated particles (∼45 nm), as determined by EM (Fig. 3) and rate-zonal ultracentrifugation (51). Unfortunately, detergent treatment reduced the number of particles observable by cryoEM so that it was not possible to compare their morphologies to those of untreated NE particles. In contrast to the E particles, which are relatively more abundant in the highly infectious intermediate-density fractions (Fig. 6), the NE particles are relatively more abundant in high-density fractions that contain most of the viral RNA but display relatively low infectivity (Fig. 6). Collectively, these results suggest that the NE particles contain HCV RNA but are unlikely to be infectious.
The multivesicular (MV) particles and large vesicles are infection related since they are not found in control samples, but they barely contribute to total HCV infectivity. Interestingly, they display a striking resemblance to human exosomes observed by cryoEM (56). Consistent with this hypothesis, HCV structural proteins are associated with circulating exosomes in HCV-infected patients (45). Although MV particles display very low infectivity in cell culture (Fig. 6), they could conceivably play a role in natural infections. Additional experiments aiming to determine the nature, biochemical composition, and functional properties of these exosome-like structures will be required to evaluate their functional relevance during natural HCV infection.
The results presented in this study suggest that the bilayer that differentiates E from NE particles constitutes the viral envelope. First, the E particles are found predominantly in preparations containing the highest E2-core protein ratio (see Fig. S6 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS6.tif). Second, treatment of purified HCV particles with detergent, which causes loss of viral envelope and infectivity, results in a diameter shift from 60 to 45 nm (Fig. 3), coinciding with the diameters of the E and NE particles, respectively (Fig. 5). If this is the case, infectious HCV particles appear to contain an ∼45-nm nonicosahedral capsid that is not tightly associated with the viral envelope. These features are clearly different from those of other members of the family. In other members of the Flaviviridae family, the viral envelope is usually not as distinct by cryoEM, where the lipid bilayer is tightly associated with the capsid and appears buried under a thick, smooth glycoprotein layer (34, 35). The heterogeneous enveloped population with various diameters surrounding an internal capsid resembles porcine reproductive and respiratory virus ([PRRSV] Arteriviridae) particles (60). Given the similarity in the sizes of the internal capsid and the population of nonenveloped particles, it is possible that the NE particles are capsids lacking an envelope. In fact, NE particles accumulate in high-density fractions where core antigen levels are the highest and the E2-core ratio is the lowest (see Fig. S6 posted at http://www.scripps.edu/wieland/data/Gastaminza/FigureS6.tif).
It is unlikely, but formally possible, that the NE particles display a viral envelope tightly associated with the capsid and not visible in individual images. Indeed, a recent three-dimensional reconstruction of noninfectious HCV virus-like particles produced in insect cells suggested that ∼50-nm particles have an envelope tightly associated with the capsid (68). In that study, these particles were compared with JFH-1 particles purified from isopycnic gradient fractions that contain the peak of HCV RNA, have a diameter of 50 nm, and are devoid of any evident bilayer (similar to particles in our high-density fractions) (Fig. 6). Unfortunately, the morphological heterogeneity, relatively low particle yield, and lack of measurable symmetry of both E and NE particles precluded the image analysis and reconstruction necessary to determine if the 45-nm NE particles display a lipid bilayer.
Acknowledgments
We are grateful to Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for kindly providing the infectious JFH-1 molecular clone, Dennis Burton (The Scripps Research Institute, La Jolla, CA) for providing the recombinant human anti-E2 and anti-HIV IgGs, and Michael Houghton (Chiron) for providing the MS3 antiserum against HCV core. We thank Marlene Dreux, Urtzi Garaigorta, Ken Takahashi, Stefan Wieland, and Barbie Ganser-Pornillos for their expert advice and useful discussions. We are grateful to Erick Giang, Christina Whitten, and Josan Chung for excellent technical assistance.
This work was supported by NIH grants R01-CA108304 (F.V.C.), R01-AI079043 (F.V.C.), R01-A79031 (M.L.), and R01-GM066087 (M.Y.).
This is manuscript number 20607 from the Scripps Research Institute.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Andre, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atshaves, B. P., A. L. McIntosh, H. R. Payne, A. M. Gallegos, K. Landrock, N. Maeda, A. B. Kier, and F. Schroeder. 2007. SCP-2/SCP-x gene ablation alters lipid raft domains in primary cultured mouse hepatocytes. J. Lipid Res. 48:2193-2211. [DOI] [PubMed] [Google Scholar]
- 4.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117:1397-1407. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 76:4073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 8.Bradley, D., K. McCaustland, K. Krawczynski, J. Spelbring, C. Humphrey, and E. H. Cook. 1991. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J. Med. Virol. 34:206-208. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, D. W., K. A. McCaustland, E. H. Cook, C. A. Schable, J. W. Ebert, and J. E. Maynard. 1985. Posttransfusion non-A, non-B hepatitis in chimpanzees. Physicochemical evidence that the tubule-forming agent is a small, enveloped virus. Gastroenterology 88:773-779. [PubMed] [Google Scholar]
- 10.Bukh, J. 2004. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology 39:1469-1475. [DOI] [PubMed] [Google Scholar]
- 11.Carabaich, A., M. Ruvoletto, E. Bernardinello, N. Tono, L. Cavalletto, L. Chemello, A. Gatta, and P. Pontisso. 2005. Profiles of HCV core protein and viremia in chronic hepatitis C: possible protective role of core antigen in liver damage. J. Med. Virol. 76:55-60. [DOI] [PubMed] [Google Scholar]
- 12.Chang, K. S., J. Jiang, Z. Cai, and G. Luo. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783-13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 14.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coller, K. E., K. L. Berger, N. S. Heaton, J. D. Cooper, R. Yoon, and G. Randall. 2009. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5:e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gastaminza, P., G. Cheng, S. Wieland, J. Zhong, W. Liao, and F. V. Chisari. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove, J., S. Nielsen, J. Zhong, M. F. Bassendine, H. E. Drummer, P. Balfe, and J. A. McKeating. 2008. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J. Virol. 82:12020-12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, L. F., D. Alling, T. Popkin, M. Shapiro, H. J. Alter, and R. H. Purcell. 1987. Determining the size of non-A, non-B hepatitis virus by filtration. J. Infect. Dis. 156:636-640. [DOI] [PubMed] [Google Scholar]
- 23.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. U. S. A. 102:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hijikata, M., Y. K. Shimizu, H. Kato, A. Iwamoto, J. W. Shih, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1993. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J. Virol. 67:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 28.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, Jr., and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes, M., S. Griffin, and M. Harris. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J. Gen. Virol. 90:1329-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, J., and G. Luo. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83:12680-12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, D. M., A. H. Patel, P. Targett-Adams, and J. McLauchlan. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaito, M., S. Watanabe, K. Tsukiyama-Kohara, K. Yamaguchi, Y. Kobayashi, M. Konishi, M. Yokoi, S. Ishida, S. Suzuki, and M. Kohara. 1994. Hepatitis C virus particle detected by immunoelectron microscopic study. J. Gen. Virol. 75:1755-1760. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann, B., G. E. Nybakken, P. R. Chipman, W. Zhang, M. S. Diamond, D. H. Fremont, R. J. Kuhn, and M. G. Rossmann. 2006. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 103:12400-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law, M., T. Maruyama, J. Lewis, E. Giang, A. W. Tarr, Z. Stamataki, P. Gastaminza, F. V. Chisari, I. M. Jones, R. I. Fox, J. K. Ball, J. A. McKeating, N. M. Kneteman, and D. R. Burton. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25-27. [DOI] [PubMed] [Google Scholar]
- 37.Li, X., L. J. Jeffers, L. Shao, K. R. Reddy, M. de Medina, J. Scheffel, B. Moore, and E. R. Schiff. 1995. Identification of hepatitis C virus by immunoelectron microscopy. J. Viral Hepat. 2:227-234. [DOI] [PubMed] [Google Scholar]
- 38.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 39.Lindenbach, B. D., P. Meuleman, A. Ploss, T. Vanwolleghem, A. J. Syder, J. A. McKeating, R. E. Lanford, S. M. Feinstone, M. E. Major, G. Leroux-Roels, and C. M. Rice. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 41.Ma, Y., J. Yates, Y. Liang, S. M. Lemon, and M. Yi. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maillard, P., K. Krawczynski, J. Nitkiewicz, C. Bronnert, M. Sidorkiewicz, P. Gounon, J. Dubuisson, G. Faure, R. Crainic, and A. Budkowska. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8240-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maniloff, J. 1995. Identification and classification of viruses that have not been propagated. Arch. Virol. 140:1515-1520. [DOI] [PubMed] [Google Scholar]
- 44.Masaki, T., R. Suzuki, K. Murakami, H. Aizaki, K. Ishii, A. Murayama, T. Date, Y. Matsuura, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masciopinto, F., C. Giovani, S. Campagnoli, L. Galli-Stampino, P. Colombatto, M. Brunetto, T. S. Yen, M. Houghton, P. Pileri, and S. Abrignani. 2004. Association of hepatitis C virus envelope proteins with exosomes. Eur. J. Immunol. 34:2834-2842. [DOI] [PubMed] [Google Scholar]
- 46.Miyamoto, H., H. Okamoto, K. Sato, T. Tanaka, and S. Mishiro. 1992. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J. Gen. Virol. 73:715-718. [DOI] [PubMed] [Google Scholar]
- 47.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 48.Mizuno, M., G. Yamada, T. Tanaka, K. Shimotohno, M. Takatani, and T. Tsuji. 1995. Virion-like structures in HeLa G cells transfected with the full-length sequence of the hepatitis C virus genome. Gastroenterology 109:1933-1940. [DOI] [PubMed] [Google Scholar]
- 49.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nahmias, Y., J. Goldwasser, M. Casali, D. van Poll, T. Wakita, R. T. Chung, and M. L. Yarmush. 2008. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47:1437-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen, S. U., M. F. Bassendine, A. D. Burt, C. Martin, W. Pumeechockchai, and G. L. Toms. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel, K., and J. G. McHutchison. 2004. Initial treatment for chronic hepatitis C: current therapies and their optimal dosing and duration. Cleve. Clin. J. Med. 71(Suppl. 3):S8-S12. [DOI] [PubMed] [Google Scholar]
- 54.Pedersen, I. M., G. Cheng, S. Wieland, S. Volinia, C. M. Croce, F. V. Chisari, and M. David. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penin, F., J. Dubuisson, F. A. Rey, D. Moradpour, and J. M. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 56.Poliakov, A., M. Spilman, T. Dokland, C. L. Amling, and J. A. Mobley. 2009. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate 69:159-167. [DOI] [PubMed] [Google Scholar]
- 57.Prince, A. M., T. Huima-Byron, T. S. Parker, and D. M. Levine. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J. Viral Hepat. 3:11-17. [DOI] [PubMed] [Google Scholar]
- 58.Pumeechockchai, W., D. Bevitt, K. Agarwal, T. Petropoulou, B. C. Langer, B. Belohradsky, M. F. Bassendine, and G. L. Toms. 2002. Hepatitis C virus particles of different density in the blood of chronically infected immunocompetent and immunodeficient patients: implications for virus clearance by antibody. J. Med. Virol. 68:335-342. [DOI] [PubMed] [Google Scholar]
- 59.Roingeard, P., C. Hourioux, E. Blanchard, D. Brand, and M. Ait-Goughoulte. 2004. Hepatitis C virus ultrastructure and morphogenesis. Biol. Cell 96:103-108. [DOI] [PubMed] [Google Scholar]
- 60.Spilman, M. S., C. Welbon, E. Nelson, and T. Dokland. 2009. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J. Gen. Virol. 90:527-535. [DOI] [PubMed] [Google Scholar]
- 61.Steinmann, E., F. Penin, S. Kallis, A. H. Patel, R. Bartenschlager, and T. Pietschmann. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suloway, C., J. Pulokas, D. Fellmann, A. Cheng, F. Guerra, J. Quispe, S. Stagg, C. S. Potter, and B. Carragher. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41-60. [DOI] [PubMed] [Google Scholar]
- 63.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trestard, A., Y. Bacq, L. Buzelay, F. Dubois, F. Barin, A. Goudeau, and P. Roingeard. 1998. Ultrastructural and physicochemical characterization of the hepatitis C virus recovered from the serum of an agammaglobulinemic patient. Arch. Virol. 143:2241-2245. [DOI] [PubMed] [Google Scholar]
- 65.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeager, M., J. A. Berriman, T. S. Baker, and A. R. Bellamy. 1994. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, X., M. Qiao, I. Atanasov, Z. Hu, T. Kato, T. J. Liang, and Z. H. Zhou. 2007. Cryo-electron microscopy and three-dimensional reconstructions of hepatitis C virus particles. Virology 367:126-134. [DOI] [PubMed] [Google Scholar]
- 69.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]