Abstract
Oseltamivir is routinely used worldwide for the treatment of severe influenza A virus infection, and should drug-resistant pandemic 2009 H1N1 viruses become widespread, this potent defense strategy might fail. Oseltamivir-resistant variants of the pandemic 2009 H1N1 influenza A virus have been detected in a substantial number of patients, but to date, the mutant viruses have not moved into circulation in the general population. It is not known whether the resistance mutations in viral neuraminidase (NA) reduce viral fitness. We addressed this question by studying transmission of oseltamivir-resistant mutants derived from two different isolates of the pandemic H1N1 virus in both the guinea pig and ferret transmission models. In vitro, the virus readily acquired a single histidine-to-tyrosine mutation at position 275 (H275Y) in viral neuraminidase when serially passaged in cell culture with increasing concentrations of oseltamivir. This mutation conferred a high degree of resistance to oseltamivir but not zanamivir. Unexpectedly, in guinea pigs and ferrets, the fitness of viruses with the H275Y point mutation was not detectably impaired, and both wild-type and mutant viruses were transmitted equally well from animals that were initially inoculated with 1:1 virus mixtures to naïve contacts. In contrast, a reassortant virus containing an oseltamivir-resistant seasonal NA in the pandemic H1N1 background showed decreased transmission efficiency and fitness in the guinea pig model. Our data suggest that the currently circulating pandemic 2009 H1N1 virus has a high potential to acquire drug resistance without losing fitness.
Oseltamivir resistance was rare until 2008, when resistant seasonal H1N1 viruses were found circulating in the general Scandinavian population (15). Soon after, studies from other countries in Europe also reported the isolation of oseltamivir-resistant viruses, and eventually, oseltamivir resistance was recognized as a global phenomenon (9, 27). Prior to 2008, resistant viruses were primarily isolated from patients with nonresponsive influenza virus infections or from infected patients who received a low-dose prophylaxis regiment prior to symptom onset. At the time, these resistant isolates accounted for 1% of the circulating H1N1 virus. Drug resistance mutations were identified during oseltamivir development, including a histidine-to-tyrosine mutation at position 275 (H275Y) in N1 neuraminidase (NA). This mutation in particular was shown to attenuate virus growth and pathology in ferrets (17). Additionally, oseltamivir-resistant viruses with a nearby mutation in N2 neuraminidase transmitted less efficiently than oseltamivir-sensitive viruses in the guinea pig transmission model (4). Surprisingly, the seasonal 2008 H1N1 viral isolates that spread around the world had the same tyrosine mutation, which was previously associated with iatrogenic infections and attenuation. Furthermore, epidemiological studies concluded that this resistant virus developed independently of drug selection, suggesting that compensatory adaptations allowed an attenuating mutation to become permissible (3, 18). The ability of resistant 2008 isolates to perform on par with nonresistant 2008 isolates in growth curves, in mean plaque size, and in a transmission model was recently confirmed (2). Currently, 99% of seasonal H1N1 viruses are oseltamivir resistant; however, the prevalence of these viruses is very low due to replacement by a novel reassortant H1N1 virus (6, 8). This novel reassortant was originally identified in Mexico by doctors concerned about a jump in the number of influenza cases during the month of March in 2009 (7). Later referred to as swine-origin influenza virus, novel H1N1 virus, or 2009 pandemic H1N1 virus, this virus would continue to efficiently transmit around the world, even during the summer months of the northern hemisphere. Its robust transmission was later confirmed in aerosol transmission models, in which 86% of ferrets and 100% of guinea pigs exposed to infected animals contracted pandemic influenza (22, 28, 31). Oseltamivir was used broadly during the outbreak, treating those with complications and prophylactically treating close contacts of confirmed cases. The use of oseltamivir in this manner provided ample opportunity for oseltamivir-resistant viruses to develop. More than 225 cases of oseltamivir-resistant infections have been confirmed from the beginning of the pandemic, including four incidents of suspected aerosol transmission (21, 32, 33). Fortunately, these clinical isolates never progressed into stable transmission in the general public. This study seeks to evaluate if introducing the H275Y mutation into the pandemic 2009 H1N1 virus attenuates virus replication in vitro or in vivo using the guinea pig model and the ferret model to test aerosol transmission efficiency. Furthermore, this study evaluates if a reassortant between the circulating novel H1N1 virus and seasonal neuraminidase (NA) forms a well-adapted, resistant virus capable of efficient transmission.
Currently, oseltamivir is the drug of choice for treating novel H1N1 complications and outpatient prophylaxis. Therefore, it is of great importance to study the in vitro replication and transmission phenotypes of oseltamivir-resistant novel H1N1 viruses to understand why broad oseltamivir resistance has not occurred or whether we should expect it to occur in the future.
MATERIALS AND METHODS
Viruses and cells.
MDCK cells and A549 cells were grown in Dulbecco's minimal essential medium (DMEM) or Eagle's minimum essential media (MEM), supplemented with penicillin-streptomycin and 10% fetal bovine serum. Influenza viruses were propagated in MDCK cells over 3 days at 35°C in the presence of 1 μg/ml tosylsulfonyl-phenylalanyl-chloromethyl ketone (TPCK)-treated trypsin (Sigma-Aldrich, St. Louis, MI). All in vitro or in vivo experiments involving primary isolates, recombinant A/California/04/2009 (A/Cal/04/2009) (H1N1) influenza viruses, or A/Hansa Hamburg/01/2009 (A/HH/01/2009) (H1N1) influenza viruses were conducted under enhanced biosafety level 2 (BSL2) conditions, as per institutional policy.
Oseltamivir-resistant virus.
In order to isolate oseltamivir-resistant mutants, the pandemic H1N1 virus A/HH/01/2009 was serially passaged on MDCK cells in the presence of increasing concentrations of oseltamivir. Briefly, confluent MDCK cell monolayers were infected with A/HH/01/2009 at a multiplicity of infection (MOI) of 0.01 and subsequently incubated for 48 h at 33°C in postinfection medium (DMEM with 100 IU/ml penicillin, 100 μg/ml streptomycin, 0.3% bovine serum albumin [BSA], 20 mM HEPES buffer, and 0.5 μg/ml TPCK-treated trypsin) supplemented with oseltamivir carboxylic acid (TRC Inc., North York, Canada). Oseltamivir concentrations were increased 5-fold with every passage, ranging from 1 nM (passage 1) to 1.95 mM (passage 10). Virus titers of harvested supernatants were determined in MDCK cells by plaque assay. Aliquots of passaged viruses were cultured once in the absence of oseltamivir before oseltamivir resistance was determined. The NA and hemagglutinin (HA) gene segments of four oseltamivir-resistant virus clones were sequenced.
Reverse-genetics plasmids.
The rescue plasmids carrying the eight genomic segments of A/Cal/04/2009 virus were described previously (12, 13). Plasmids used to rescue A/HH/01/2009 will be described elsewhere. The rescue plasmid encoding the NA segment of an oseltamivir-resistant seasonal H1N1 influenza virus (A/New York/1326/2008) was generated by cloning reverse transcription-PCR (RT-PCR)-amplified cDNA into the pPol1 expression vector (30). Mutations were introduced into the NA genomic sequences using the QuikChange XL site-directed mutagenesis kit (Stratagene, Santa Clara, CA). A H275Y mutation (C843T/C845T) and a silent PstI restriction endonuclease consensus sequence (G893C) were introduced into the NA segment of A/Cal/04/2009 virus. A H275Y mutation (C842T) was introduced into the NA segment of A/HH/01/2009 virus.
Virus rescue.
Recombinant viruses were generated by reverse genetics as described elsewhere (12, 13, 29). Recombinants of A/Cal/04/2009 virus were rescued carrying wild-type NA (rCal09-wt), NA with the H275Y mutation (rCal09-H275Y), or the NA segment of A/New York/1326/2008 virus [rCal09(7:1)NY1326]. Recombinants of A/HH/01/2009 virus carried a wild-type NA (rHH-wt) or an NA with the H275Y mutation (rHH-H275Y). Supernatants were plaque purified on MDCK cells in the presence of 1 μg/ml of TPCK-treated trypsin, and single plaques were passaged on MDCK cells to generate virus stocks (13). The HA genes from the Cal09-wt and Cal09-H275Y viral stocks were sequenced and did not contain any mutations.
Virus titrations.
Virus titers were determined by plaque assay using 10-fold serial dilutions on MDCK cells and quantified after incubation at 35°C for 2 to 3 days. Samples were diluted in postinfection medium or phosphate-buffered saline (PBS) with 0.3% BSA prior to plaque assay.
Virus growth curves.
MDCK cells were seeded at 106 cells/well in 6-well plates. After allowing the cells to attach for 4 h, cells were washed in Opti-MEM (Invitrogen, San Diego, CA) and incubated with virus at an MOI of 0.004 or 0.001 diluted in postinfection medium or Opti-MEM. Diluted virus was permitted to adsorb onto cells for 1 h before addition of 2 ml postinfection medium supplemented with 1 μg/ml of TPCK-treated trypsin. The titers of supernatants sampled at select time points were determined by plaque assay.
Plaque reduction assays.
Confluent MDCK cells seeded in 6-well plates were incubated with approximately 50 to 100 PFU of virus. At 1 hour postinoculation, the inoculum was removed and replaced by postinfection medium overlay containing 1 μg/ml of TPCK-treated trypsin and either 0.64% agar (Thermo Fisher Scientific, Waltham, MA) or 3% Avicel (Thermo Fisher Scientific, Waltham, MA). The overlay medium was supplemented with oseltamivir carboxylate (TRC Inc. or Hoffmann-La Roche, Basel, Switzerland) or zanamivir (kindly provided by Glaxo Smith Kline, Brentford, United Kingdom), as required. At 2 to 3 days postinfection, cell cultures were fixed for 1 h using 4% formaldehyde diluted in PBS. For A/HH/01/2009-derived viruses, plaques were stained with crystal violet and counted manually, and plaque size was estimated by visual observation. The lowest concentration of inhibitor that resulted in at least 10-fold reduction of plaque size was referred to as the 90% inhibitory concentration (IC90). Plaques produced by A/Cal/04/2009-derived viruses were immunostained (25) using an anti-pandemic H1N1 HA monoclonal antibody (24, 26). Plaque size was quantified by scanning immunostained plaques and measuring individual plaque areas using the “Measure and Label” plug-in automatically installed on ImageJ version 1.43 (1). Images were acquired at 1,200 dots per inch (DPI), and measurements were taken manually on images at 400% of the original size to ensure accurate measurement of each plaque.
Neuraminidase activity inhibition assay.
NA activity of viruses inactivated with Triton X-100 was determined in the absence or presence of neuraminidase inhibitors using the NA-Star influenza NA inhibitor resistance detection kit (Applied Biosystems, Foster City, CA), according to the instructions of the manufacturer. Fifty percent inhibitory concentration (IC50) values were calculated using nonlinear curve fitting with GraphPad Prism software (GraphPad Software, La Jolla, CA).
Contact transmission experiments in guinea pigs.
Five- to six-week-old female Hartley strain guinea pigs (300 to 400 g) were obtained from Charles River Laboratory. For each experiment, four guinea pigs were inoculated intranasally with 104 PFU of A/HH/01/2009 virus, rHH-wt virus, or rHH-H275Y virus in 300 μl of PBS. At 24 hours postinfection, four naïve guinea pigs were placed into the same isolator, sharing bedding, water bottles, and food dishes. Nasal wash samples were collected from directly inoculated and contact-exposed animals at days 2, 4, 6, 8, and 11 postinfection. All animal procedures were conducted under BSL3 biocontainment conditions in accordance with local guidelines. Animals were anesthetized through intramuscular injection of a ketamine-xylazine mixture prior to inoculation and nasal wash treatment.
Aerosol transmission experiments in guinea pigs.
Aerosol experiments were conducted as described elsewhere (4, 19, 20, 31). In brief, four guinea pigs were inoculated intranasally on day 0 with 103 PFU of rCal09-wt virus, rCal09-H275Y virus, or rCal09(7:1)NY1326 virus in 300 μl of PBS and sequestered from four naïve guinea pigs. At 24 hours postinoculation, all 8 guinea pigs were transferred into a Caron environmental test chamber (model 6030) maintained at 20°C and 20% humidity. Animals were placed in wire-sided transmission cages with open tops to allow air exchange among all eight cages. Nasal wash samples were collected from animals on days 2, 4, 6, and 8 postinfection for virus titration on MDCK cells.
Transmission experiments in ferrets.
Six- to nine-month-old male Fitch ferrets (1,000 to 1,500 g) were obtained from Triple F Farms, Sayre, PA. Ferrets were confirmed to be seronegative to seasonal (A/Brisbane/59/2007 and A/Brisbane/10/2007) and pandemic (A/California/04/2009) influenza viruses by hemagglutination inhibition assay. Ferrets were housed in 36-in.-wide by 36-in.-deep by 36-in.-high plastic poultry isolation units (Plas Labs, Lansing, MI) with HEPA-filtered air intake and exhaust. Baseline serum, body weight, and temperature were obtained prior to the transmission experiments. Temperatures were obtained by a subcutaneous implantable temperature transponder (IPTT 300; Bio Medic Data Systems, Seaford, DE). Animals were kept on a 12-h light/12-h dark cycle and had free access to food and water. All animal procedures were conducted under enhanced BSL2 biocontainment conditions in accordance with the MSSM Institutional Biosafety Program and Institutional Animal Use and Care Committee guidelines. Animals were anesthetized through intramuscular injection of a ketamine-xylazine mixture prior to inoculation and nasal wash treatment.
Experimental infections of ferrets were conducted with the rCal09-wt, rCal09-H275Y, rHH-wt, and rHH-H275Y viruses. Each experimental setting involved one directly inoculated ferret, one physical contact ferret, and one aerosol contact ferret. Directly inoculated ferrets received 104 PFU of the indicated viruses by intranasal inoculation. At 1 day postinfection, physical and aerosol contact ferrets were introduced into the unit to initiate transmission. Aerosol contact ferrets were separated from the inoculated and physical contact ferrets by a 3/8-in.-thick fiberglass sheet with 3/8-in. holes spaced 1.5 in. apart. Ferrets were arranged such that the directly inoculated and contacted ferrets were housed on the side with air intake, while the aerosol contact ferrets were housed on the side with air exhaust. This arrangement facilitated the directional flow of aerosolized virus from the directly inoculated ferret to the aerosol contact ferret. Ferrets were monitored daily for body weight, temperature, and signs of morbidity. On days 2, 4, 6, 8, and 10 postinfection, nasal wash samples were collected from all animals for virus titration on MDCK cells.
Viral competition experiments.
Oseltamivir-resistant recombinant or reassortant viruses containing either the H275Y mutation (rCal09-H275Y) or the NA segment of strain A/New York/1326/2008 [rCal09(7:1)NY1326] were mixed at a 1:1 ratio with wild-type virus rCal09-wt. Mixtures of rCal09 viruses were tested in guinea pig aerosol transmission experiments as previously described, using a total viral inoculum of 103 PFU. In addition, rCal09-H275Y in a 1:1 mixture with rCal09-wt was tested in a ferret transmission experiment as described above. Nasal wash samples were collected and titers were determined by plaque assay. Virus-positive samples were selected for quantification of wild-type and mutant NA segment sequences by either pyrosequencing-based single nucleotide polymorphism (SNP) analysis or restriction enzyme analysis.
Pyrosequencing-based SNP analysis.
SNP analysis was applied to quantify the proportions of wild-type and NA-mutant sequences in the samples collected from viral competition experiments with H275Y mutants and wild-type viruses. RNA was isolated and reverse transcribed using random hexamer primers. Further processing was performed by varionostic GmbH (Ulm, Germany). The PCR product (∼40 μl) was immobilized on 3 μl streptavidin-Sepharose HP beads (GE Healthcare, Waukesha, WI), followed by annealing of the sequencing primer (5 μM) for 2 min at 80°C. Primer sequences are available upon request. Pyrosequencing was performed using the PSQ ID system (Qiagen, Hilden, Germany) with the nucleotide dispensation orders assigned for the assay. SNP quantification was done with PyroMark ID software (Biotage AB, Uppsala, Sweden).
NA mutant analysis using a restriction enzyme.
Selected nasal wash samples were used for viral RNA extraction and RT-PCR. The primers used targeted consensus sequences in neuraminidases of A/New York/1326/2008 and A/California/04/2009 (5′ primer starting at the 33rd nucleotide, CAAAAGATAATAACCATTGG; 3′ primer starting at the 1,403rd nucleotide, AACTCAGCACCGTCTGGCCAAGACC). The amplified NA segment was gel purified and digested with PstI endonuclease. The NA sequence of A/New York/1326/2008 contained a unique PstI restriction site, allowing for resistant virus detection in a heterogeneous virus population.
Statistics.
Statistical analysis was performed using GraphPad Prism 4 and GraphPad's online QuickCalcs statistical calculators. Student's t test was used to evaluate plaque area statistical significance, and Fisher's exact test was used to evaluate transmission rate statistical significance.
RESULTS
Neuraminidase inhibitor resistance in 2009 pandemic H1N1 influenza virus.
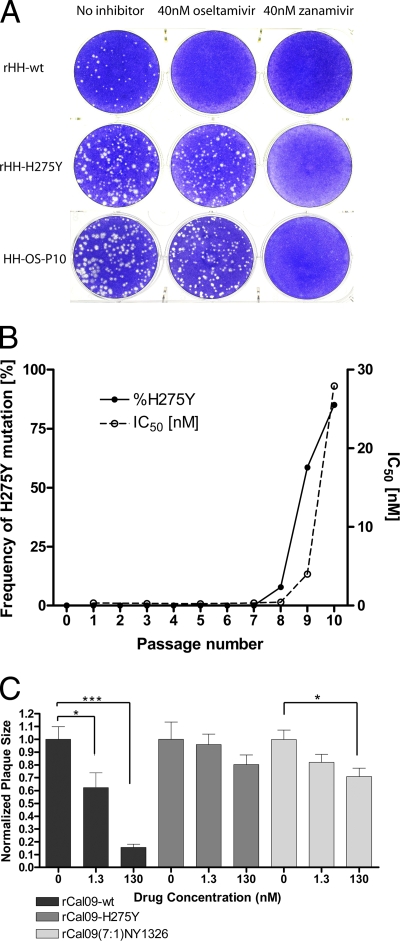
Widespread oseltamivir resistance could arise in 2009 pandemic H1N1 influenza viruses by point mutations in the NA segment or by reassortment with seasonal H1N1 viruses. We experimentally assessed the first possibility by subjecting the A/HH/01/2009 isolate of the pandemic H1N1 virus to 10 serial passages on MDCK cells in the presence of escalating doses of oseltamivir carboxylate. Unlike wild-type virus, passage 10 virus (HH-Os-P10) grew equally well in the presence or absence of oseltamivir but remained susceptible to zanamivir (Fig. 1A). Virus from each passage was analyzed for enhanced oseltamivir resistance of the virus-bound NA enzyme. The 50% inhibitory concentration (IC50) remained below 1 nM until passage 8, before this value started to rise and eventually reached 28 nM at passage 10 (Fig. 1B).
FIG. 1.
Oseltamivir-resistant pandemic H1N1 mutants are selected in cell culture and carry the substitution H275Y. (A) Oseltamivir inhibited plaque formation of wild-type pandemic H1N1 virus (rHH-wt). Mutants selected by 10 serial passages on MDCK cells in the presence of increasing oseltamivir concentrations (HH-Os-P10) or generated by introduction of the H275Y substitution into NA (rHH-H275Y) were not affected by oseltamivir. Zanamivir inhibited plaque formation of all three viruses. (B) Passaging of pandemic A/HH/01/2009 on MDCK cells in the presence of escalating oseltamivir concentrations resulted in the occurrence of a H275Y mutation in the NA gene, starting at passage 8. This was paralleled by increased resistance to oseltamivir-mediated inhibition of NA activity. (C) Pandemic H1N1 with wild-type NA (rCal09-wt) showed significant reductions in plaque size, with increasing drug concentrations supplemented to the agar overlay. No significant differences in plaque size were observed for the rCal09-H275Y mutant and a rCal09(7:1)NY1326 virus at an oseltamivir concentration of 1.3 nM. No statistical difference in plaque sizes was observed between the rCal09-H275Y and the rCal09(7:1)NY1326 viruses at both the 1.3 nM (P value = 0.17) and 130 nM (P value = 0.34) drug concentrations. The mean plaque area is normalized to the untreated plaque area, and error bars denote the standard errors of the means (SEM). Statistical significance was determined using Student's t test. *, P < 0.05; ***, P < 0.001.
Oseltamivir-resistant clinical isolates of the pandemic H1N1 virus usually carry a mutation in NA that converts histidine 275 to tyrosine (H275Y) (33). Sequencing of the passage 10 virus showed that the H275Y mutation was the only genetic alteration in the NA segment. The passaged virus further carried an N142D substitution in HA. Virus from each passage was subjected to a pyrosequencing-based single nucleotide polymorphism (SNP) analysis in order to determine at which stage the H275Y mutation had become dominant. A substantial fraction (7.8%) of the virus population carried this mutation at passage 8, and in passage 10, the proportion of mutant virus was 85% (Fig. 1B). To confirm that the resistance phenotype was associated with the H275Y mutation, we generated recombinant viruses using plasmids derived from the A/HH/01/2009 and the A/California/04/2009 virus isolates. As predicted, the H275Y recombinant viruses formed plaques in the presence of oseltamivir but not zanamivir (Fig. 1A and C). A more detailed analysis revealed that the IC50s of pandemic H1N1 viruses carrying the H275Y mutation were in the 100 nM range, whereas the IC50 of the wild-type virus was well below 1 nM (Table 1). This degree of resistance corresponds to that of a seasonal H1N1 strain, A/Berlin/59/08 (BLN59/08), that served as a control. Of note, the H275Y mutants remained fully susceptible to zanamivir (Table 1). The IC50s for the inhibition of different neuraminidases, determined using a biochemical assay, were consistent with the IC90 values of viral inhibition, determined by plaque reduction assays (Table 1).
TABLE 1.
NA IC50 and viral IC90 values for H1N1 viruses
| Virus | Inhibitory concn (nM) |
|||
|---|---|---|---|---|
| 50%a |
90%b |
|||
| Oseltamivir | Zanamivir | Oseltamivir | Zanamivir | |
| rHH-wt | 0.33 | 0.37 | 40 | 40 |
| HH-Os-P10 | 98.30 | 0.49 | 4,000 | 40 |
| rHH-H275Y | 164.30 | 0.45 | 4,000 | 40 |
| BLN59/08 (H1N1) | 111.60 | 0.31 | 4,000 | 40 |
Determined by NA-Star assay.
Determined by plaque reduction assay.
To experimentally mimic the possibility that the pandemic H1N1 virus might recombine with a seasonal H1N1 virus, we generated a 7+1 reassortant virus in which the NA-encoding segment of the pandemic virus was replaced by the corresponding segment of the seasonal virus A/NewYork/1326/2008 (H1N1). The resulting virus, rCal09(7:1)NY1326, was viable and, as expected, exhibited a high degree of oseltamivir resistance (Fig. 1C).
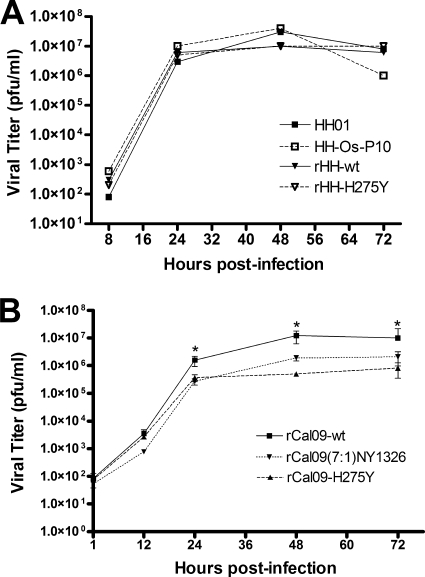
Viruses carrying the H275Y mutation (HH-Os-P10, rHH-H275Y, and rCal09-H275Y) as well as the reassortant virus rCal09(7:1)NY1326 replicated on MDCK cells, with kinetics comparable to those of the respective parental strains (A/HH/01/2009, rHH-wt, and rCal09-wt), and reached similar peak titers (Fig. 2).
FIG. 2.
Oseltamivir-resistant pandemic H275Y mutants and a reassortant with a seasonal H1N1 virus are not attenuated in the MDCK cell culture. H275Y mutants of pandemic A/HH/01/2009 (HH-Os-P10 and rHH-H275Y) and A/California/04/2009 (rCal09-H275Y) showed multicycle growth kinetics comparable to those of the respective wild-type viruses (A/HH/01/2009, rHH-wt, and rCal09-wt). (A) Similar growth kinetics of the A/HH/01/2009 virus and H275Y mutant derivatives in MDCK cells infected at an MOI of 0.001. (B) Similar growth kinetics of the A/California/04/2009 virus and H275Y mutant derivatives in MDCK cells infected at an MOI of 0.004. Additionally, similar kinetics were also observed for a reassortant virus carrying NA derived from an oseltamivir-resistant seasonal H1N1 virus [rCal09(7:1)NY1326] in MDCK cells infected at an MOI of 0.004. Statistical significance was determined by using Student's t test. *, P < 0.05.
Oseltamivir-resistant viruses are readily transmitted in the guinea pig model system.
To determine if oseltamivir-resistant viruses can spread among guinea pigs, nasal wash samples were collected from inoculated and exposed animals at various times postinfection in order to monitor viral transmission. In the first series of experiments, guinea pigs were infected with 10,000 PFU of the natural isolate A/HH/01/2009 (Fig. 3A), its recombinant equivalent (rHH-wt [Fig. 3B]), or the mutant rHH-H275Y (Fig. 3C). These viruses all grew well in inoculated animals and were efficiently transmitted by direct contact to exposed animals, although the H275Y mutant virus appeared delayed in nasal wash samples obtained from two of the four exposed animals (Fig. 3C).
FIG. 3.
Wild-type and oseltamivir-resistant pandemic H1N1 viruses are transmitted efficiently by physical and aerosol contact in the guinea pig model. The pandemic H1N1 isolate (A/HH/01/2009) (A), its recombinant wild-type equivalent (rHH-wt) (B), and a H275Y NA-mutant (rHH-H275Y) (C) were transmitted from guinea pigs intranasally (i.n.) inoculated with 104 PFU to naïve guinea pigs exposed by physical contact, with transmission rates of 100%. Wild-type (rCal09-wt) (D) and H275Y mutant (rCal09-H275Y) (E) recombinants of a second pandemic H1N1 virus were transmitted with comparable efficacies from inoculated guinea pigs (103 PFU, i.n.) to aerosol-exposed animals. A reassortant virus carrying NA of an oseltamivir-resistant seasonal H1N1 strain [rCal09(7:1)NY1326] (F) was transmitted to only 50% of aerosol-exposed guinea pigs. Virus transmission was assessed by determining viral titers in nasal wash samples collected at the indicated times. Transmission rates for aerosol experiments were determined by two independent experiments performed at 20°C and 20% humidity in environmental chambers.
Aerosol transmission experiments were performed with the A/California/04/2009-derived viruses. The parental virus rCal09-wt transmitted to all eight exposed guinea pigs in two different experiments (Fig. 3D), confirming our earlier result (31). The oseltamivir-resistant virus rCal09-H275Y also transmitted efficiently among aerosol-exposed guinea pigs, with seven of eight guinea pigs becoming virus positive in their nasal wash specimens by day 8 postinfection (Fig. 3E). In contrast, the reassortant virus rCal09(7:1)NY1326 was transmitted to only four out of eight aerosol-exposed guinea pigs over the course of two experiments (Fig. 3F). This rate of transmission is significantly reduced compared to that of the rCal09-wt virus (P < 0.05).
High fitness of the H275Y mutant virus in the guinea pig transmission model.
The relative fitness of oseltamivir-resistant viruses was assessed under conditions in which rCal09-wt virus and rCal09-H275Y virus were mixed at a 1:1 ratio before inoculation of guinea pigs. To monitor the fate of the two viruses, RNA was extracted from virus-positive nasal wash samples before cDNA was prepared and analyzed using a diagnostic PstI restriction site which was engineered into the NA segment of the resistant virus. These analyses indicated that the rCal04-H275Y mutant competed well with the parental virus, with wild-type and mutant virus detected in 3 of the 4 aerosol-exposed, naïve guinea pigs (Fig. 4A and C). These results were confirmed by a different detection assay in which the frequency of the NA mutation in the cDNA mixtures was assessed by pyrosequencing-based SNP analysis (data not shown). In contrast, the reassortant rCal09(7:1)NY1326 virus showed reduced fitness in the competition experiment, as only wild-type virus was detected in substantial amounts in nasal wash samples obtained from the exposed animals (Fig. 4B and D).
FIG. 4.
rCal09-H275Y virus is able to transmit as efficiently as rCal09-wt virus, whereas rCal09(7:1)NY1326 virus cannot transmit in competition experiments. Guinea pigs were intranasally inoculated with the indicated viruses mixed in a 1:1 ratio (103 PFU). (A, B) Nasal wash specimens collected from inoculated and aerosol-exposed animals were investigated for viral titers. (C, D) The presence or absence of each virus contained in the original inoculum was determined by restriction enzyme analysis. Digest assay-based banding patterns from the rCal09-wt versus rCal09-H275Y experiments illustrate that all of the inoculated guinea pigs and 3 of the 4 exposed guinea pigs were shedding a heterogeneous mixture of viruses. Restriction enzyme analysis of nasal wash samples obtained from exposed guinea pigs in the rCal09(7:1)NY1326 experiment revealed banding patterns corresponding to predominant transmission of the wild-type virus. GPs, guinea pigs.
These results suggest that the 2009 pandemic H1N1 virus with the H275Y mutation can effectively compete with wild-type virus in aerosol transmission. In contrast, the 7+1 reassortant virus was unable to compete with wild-type virus, presumably due to suboptimal compatibility of the NA gene derived from the seasonal strain, with the other viral genes derived from the pandemic H1N1 virus.
Transmission experiments with ferrets support the high degree of fitness observed with the H275Y mutant virus.
Ferrets are an accepted experimental model for human influenza viruses, as various aspects of the human disease are mimicked in these animals, including transmission and general malaise. We thus set out to test the ability of the H275Y virus mutants to transmit in this model system. In the first experiment, we compared the contact and aerosol transmissibility of the A/California/04/2009 and A/HH/01/2009 wild-type viruses to the H275Y mutant viruses by inoculating 10,000 PFU into the upper respiratory tract of one ferret per virus strain. These inoculated ferrets, which were segregated for 24 h in individual isolators, were then exposed to two naïve ferrets, of which one had physical contact and the other had aerosol contact with the inoculated animal. Under these conditions the wild-type and H275Y mutant viruses transmitted equally well by either contact or aerosol transmission (Fig. 5A). All of the inoculated and exposed ferrets showed signs of disease, and no gross differences in pathology between the two pandemic strains were observed (data not shown). In a second experiment, we inoculated 1:1 mixtures of mutant and wild-type virus and determined the virus composition in the nasal wash samples by using the pyrosequencing-based SNP assay described above. This analysis showed that the composition of the viral mixtures did not change substantially during contact and aerosol transmission (Fig. 5B), supporting the conclusion from the guinea pig experiments that the wild-type and H275Y mutant 2009 pandemic H1N1 viruses possess similar fitness.
FIG. 5.
Wild-type and oseltamivir-resistant pandemic H1N1 viruses are transmitted efficiently by physical and aerosol contact in the ferret transmission model. (A) Wild-type viruses (rHH01-wt and rCal09-wt) and H275Y mutants (rHH01-H275Y and rCal09-H275Y) reached comparable titers in nasal wash samples following intranasal inoculation with 104 PFU. They were readily transmitted to ferrets exposed by physical or aerosol contact. (B) Ferrets were intranasally inoculated with wild-type and H275Y mutant viruses mixed in a 1:1 ratio (104 PFU). (Top) Nasal wash samples collected from directly inoculated, contact-exposed, and aerosol-exposed animals were investigated for viral titers. (Bottom) Pyrosequencing-based single nucleotide polymorphism (SNP) analysis shows that wild-type and mutant viruses were transmitted by both routes.
DISCUSSION
During the 2008-2009 influenza season, most of the seasonal H1N1 viral isolates surveyed were resistant to oseltamivir, limiting its use as a treatment option (5). This changed in April 2009, as an antigenically divergent, oseltamivir-sensitive H1N1 virus spread through the northern hemisphere during the summer months, displaying a lack of seasonality normally associated with influenza viruses. This enhanced transmission capability may have been due to virus-specific factors or the presence of a large naïve population. Heightened surveillance during the summer months revealed an increase in confirmed influenza cases, but a low percentage of these isolates were seasonal, oseltamivir-resistant H1N1 isolates (5). Even less prevalent were sensitive seasonal H1N1 infections.
Over 200 oseltamivir-resistant, 2009 pandemic H1N1 viruses have been isolated from patients, and in all of these cases, the viruses contained the H275Y mutation (33). These isolates were also likely amantadine/rimantadine resistant, making them of great public health concern (6). Where data were available, 90% of the resistant novel H1N1 viruses were isolated from patients refractory to oseltamivir treatment, and 44% of the patients in this data set were classified as immunocompromised (33). Only 4 clusters of person-to-person transmission of oseltamivir-resistant virus have been reported, and 2 of these cases occurred in hospital wards among immunocompromised individuals (11, 16, 21, 23). These data, along with the broad use of oseltamivir during the early stages of the pandemic, would suggest that novel H1N1 viruses with the H275Y mutation are attenuated or otherwise incapable of sustained human-to-human transmission. An attenuated transmission phenotype for these H1N1 viruses would be consistent with studies of drug-resistant N2 neuraminidases, in which oseltamivir-resistant H3N2 viruses transmit less efficiently than wild-type viruses without attenuated growth in vitro (4). Surprisingly, we show that oseltamivir resistance due to the H275Y mutation mildly attenuates replication without affecting transmission efficiency. Growth kinetics for the mutant 2009 pandemic H1N1 viruses were similar to those for wild-type viruses, and we observed no significant differences in the transmission rates between the sensitive and resistant recombinant viruses derived from A/HH/01/2009 and A/California/04/2009 viral isolates. Furthermore, when rCal09-wt and rCal09-H275Y viruses were put into direct competition in the upper respiratory tract of a guinea pig, both viruses were detectable in 3 out of 4 exposed guinea pigs, suggesting that the resistant virus is readily transmissible and equivocally fit. This result is in direct contrast to the rCal09(7:1)NY1326 competition experiment, in which the wild-type and reassortant viruses grew to approximately a 1:1 ratio in the inoculated guinea pigs, but only the wild-type virus was detected in the exposed animals. Incidentally, oseltamivir-resistant or -sensitive reassortant viruses produced by coinfection with 2009 pandemic H1N1 and seasonal H1N1 reassortant viruses have not been isolated from humans (33). One explanation for the lower fitness of the reassortant virus is that NA of the seasonal strain is not as effective in the genetic background of the rCal09 virus, and this results in an attenuated transmission phenotype. The transmissibility and fitness of the rHH-H275Y and rCal09-H275Y 2009 pandemic viruses were further studied in the ferret transmission model, in which ferrets inoculated with a 1:1 mixture of oseltamivir-sensitive and -resistant viruses transmitted both wild-type and mutant viruses by contact and aerosol transmission. These studies were performed using a small sample size, but the fact that the H275Y mutant viruses transmit by contact, aerosol, and in competition with the wild-type virus in ferrets supports the conclusion of the more rigorous guinea pig transmission data set. It should be noted that results similar to ours were recently reported by Hamelin et al. (14). In contrast, by studying a different pandemic H1N1 strain (A/Demark/528/09), Duan et al. found that the emergence of oseltamivir resistance was associated with a loss of viral fitness (10). This suggests that the genetic background into which oseltamivir resistance is introduced could very much influence the fitness/transmissibility of the particular virus.
Oseltamivir-resistant and -sensitive 2008 isolates were recently tested and shown to be similar in growth kinetics and transmission, suggesting that these viruses may have evolved to allow the H275Y mutation without compromising viral fitness (2, 3). Oseltamivir-resistant, 2009 pandemic H1N1 viruses are currently not circulating in the community at large, but our data and that of Hamelin et al. suggests that they certainly could (14). The H275Y mutation is clearly not as debilitating for the novel H1N1 viruses A/Cal/04/09 and A/HH/01/09, as it was for seasonal H1N1 viruses circulating prior to 2008 (17).
Acknowledgments
We thank John Steel, Anice C. Lowen, Marcus Panning, Martin Schwemmle, and Otto Haller for advice and many helpful discussions. We also acknowledge Lily Ngai and Richard Cadagan for excellent technical assistance over the course of the study. Finally, we thank GlaxoSmithKline for providing a sample of zanamivir.
This work was supported by the German Ministry of Education and Research (FluResearchNet), the W. M. Keck Foundation (grant 062009), and by CRIP, an NIH/NIAID-funded Center of Excellence on Influenza Research and Surveillance (CEIRS) (contract HHSN266200700010C).
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Baz, M., Y. Abed, P. Simon, M. E. Hamelin, and G. Boivin. 2010. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J. Infect. Dis. 201:740-745. [DOI] [PubMed] [Google Scholar]
- 3.Bloom, J. D., L. I. Gong, and D. Baltimore. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier, N. M., A. C. Lowen, and P. Palese. 2008. Oseltamivir-resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J. Virol. 82:10052-10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. 2009. FluView: 2008-2009 influenza season week 34 ending August 29, 2009. CDC, Atlanta, GA.
- 6.CDC. 2010. FluView: 2009-2010 influenza season week 16 ending April 24 2010. CDC, Atlanta, GA.
- 7.CDC. 2009. Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:467-470. [PubMed] [Google Scholar]
- 8.CDC. 2009. Update: influenza activity—United States, September 28, 2008-April 4, 2009, and composition of the 2009-10 influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 58:369-374. [PubMed] [Google Scholar]
- 9.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, A. M. Fry, and for the Oseltamivir-Resistance Working Group. 2009. Infections with oseltamivir-resistant influenza A (H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 10.Duan, S., D. A. Boltz, P. Seiler, J. Li, K. Bragstad, L. P. Nielsen, R. J. Webby, R. G. Webster, and E. A. Govorkova. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 6:e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duke Medicine News and Communications. 2009. CDC confirms four new cases of oseltamivir (Tamiflu)-resistant H1N1. Duke Medicine News and Communications, Durham, NC.
- 12.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hai, R., M. Schmolke, Z. T. Varga, B. Manicassamy, T. T. Wang, J. A. Belser, M. B. Pearce, A. Garcia-Sastre, T. M. Tumpey, and P. Palese. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamelin, M.-Ã., M. Baz, Y. Abed, C. Couture, P. Joubert, Ã. Beaulieu, N. Bellerose, M. Plante, C. Mallett, G. Schumer, G. P. Kobinger, and G. Boivin. 2010. Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoS Pathog. 6:e1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauge, S. H., S. Dudman, K. Borgen, A. Lackenby, and O. Hungnes. 2009. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 15:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.HPA. 2009. HPA statement on possible transmission. Health Protection Agency, London, England.
- 17.Ives, J. A. L., J. A. Carr, D. B. Mendel, C. Y. Tai, R. Lambkin, L. Kelly, J. S. Oxford, F. G. Hayden, and N. A. Roberts. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 55:307-317. [DOI] [PubMed] [Google Scholar]
- 18.Kramarz, P., D. Monnet, A. Nicoli, C. Yilmaz, and B. Ciancio. 2009. Use of oseltamivir in 12 European countries between 2002 and 2007—lack of association with the appearance of oseltamivir-resistant influenza A (H1N1) viruses. Euro Surveill. 14:19112. [DOI] [PubMed] [Google Scholar]
- 19.Lowen, A. C., S. Mubareka, J. Steel, and P. Palese. 2007. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 3:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowen, A. C., S. Mubareka, T. M. Tumpey, A. García-Sastre, and P. Palese. 2006. The guinea pig as a transmission model for human influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 103:9988-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mai, L. Q., H. F. L. Wertheim, T. N. Duong, H. R. van Doorn, N. Tran Hien, and P. Horby. 2010. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N. Engl. J. Med. 362:86-87. [DOI] [PubMed] [Google Scholar]
- 22.Maines, T. R., A. Jayaraman, J. A. Belser, D. A. Wadford, C. Pappas, H. Zeng, K. M. Gustin, M. B. Pearce, K. Viswanathan, Z. H. Shriver, R. Raman, N. J. Cox, R. Sasisekharan, J. M. Katz, and T. M. Tumpey. 2009. Transmission and pathogenesis of swine-origin 2009 A (H1N1) influenza viruses in ferrets and mice. Science 325:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandelboim, M., M. Hindiyeh, T. Meningher, and E. Mendelson. 2010. Possible transmission of pandemic (H1N1) 2009 virus with oseltamivir resistance. Emerg. Infect. Dis. 16:873-874. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manicassamy, B., R. A. Medina, R. Hai, T. Tsibane, S. Stertz, E. Nistal-Villáin, P. Palese, C. F. Basler, and A. Garcia-Sastre. 2010. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 6:e1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matrosovich, M., T. Matrosovich, W. Garten, and H.-D. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina, R. A., B. Manicassamy, S. Stertz, C. W. Seibert, R. Hai, R. B. Belshe, S. E. Frey, C. F. Basler, P. Palese, and A. García-Sastre. 15 June 2010, posting date. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nat. Comm. doi: 10.1038/ncomms1026. [DOI] [PMC free article] [PubMed]
- 27.Meijer, A., A. Lackenby, O. Hungnes, B. Lina, S. van der Werf, B. Schweiger, M. Opp, J. Paget, J. van de Kassteele, A. Hay, and M. Zambon. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg. Infect. Dis. 15:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munster, V. J., E. de Wit, J. M. A. van den Brand, S. Herfst, E. J. A. Schrauwen, T. M. Bestebroer, D. van de Vijver, C. A. Boucher, M. Koopmans, G. F. Rimmelzwaan, T. Kuiken, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2009. Pathogenesis and transmission of swine-origin 2009 A (H1N1) influenza virus in ferrets. Science 325:481-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleschka, S., R. Jaskunas, O. Engelhardt, T. Zurcher, P. Palese, and A. Garcia-Sastre. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 70:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steel, J., P. Staeheli, S. Mubareka, A. Garcia-Sastre, P. Palese, and A. C. Lowen. 2009. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J. Virol. 84:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tramontana, A. R., B. George, A. C. Hurt, J. S. Doyle, K. Langan, A. B. Reid, J. M. Harper, K. Thursky, L. J. Worth, D. E. Dwyer, C. Morrissey, P. D. Johnson, K. L. Buising, S. J. Harrison, J. F. Seymour, P. E. Ferguson, B. Wang, J. T. Denholm, A. C. Cheng, and M. Slavin. 2010. Oseltamivir resistance in adult oncology and hematology patients infected with pandemic (H1N1) 2009 virus, Australia. Emerg. Infect. Dis. 16:1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.WHO. 2009. Update on oseltamivir-resistant pandemic A (H1N1) 2009 influenza virus: January 2010. Wkly. Epidemiol. Rec. 85:37-48. [PubMed] [Google Scholar]