Abstract
Two effective (vac+) and two ineffective (vac−) candidate live-attenuated influenza vaccines (LAIVs) derived from naturally selected genetically stable variants of A/TK/OR/71-delNS1[1-124] (H7N3) that differed only in the length and kind of amino acid residues at the C terminus of the nonstructural NS1 protein were analyzed for their content of particle subpopulations. These subpopulations included total physical particles (measured as hemagglutinating particles [HAPs]) with their subsumed biologically active particles of infectious virus (plaque-forming particles [PFPs]) and different classes of noninfectious virus, namely, interferon-inducing particles (IFPs), noninfectious cell-killing particles (niCKPs), and defective interfering particles (DIPs). The vac+ variants were distinguished from the vac− variants on the basis of their content of viral subpopulations by (i) the capacity to induce higher quantum yields of interferon (IFN), (ii) the generation of an unusual type of IFN-induction dose-response curve, (iii) the presence of IFPs that induce IFN more efficiently, (iv) reduced sensitivity to IFN action, and (v) elevated rates of PFP replication that resulted in larger plaques and higher PFP and HAP titers. These in vitro analyses provide a benchmark for the screening of candidate LAIVs and their potential as effective vaccines. Vaccine design may be improved by enhancement of attributes that are dominant in the effective (vac+) vaccines.
Live-attenuated vaccines are considered more effective than their inactive or single-component counterparts because they activate both the innate and adaptive immune systems and elicit responses to a broader range of antigens for longer periods of time (2, 10, 25, 28). Influenza virus variants with alterations in the reading frame of the nonstructural NS1 protein gene (delNS1), which express truncated NS1 proteins, characteristically induce enhanced yields of type I interferon (IFN) relative to the yields of their isogenic parental virus encoding full-length NS1 proteins (11, 13, 21, 33, 39). Many of these delNS1 variants have proved to be effective as live-attenuated influenza vaccines (LAIVs), providing protection against challenge virus in a broad range of species (33, 46), including chickens (39, 44). The IFN-inducing capacity of the virus is considered an important element in the effectiveness of LAIVs (33). In that context, influenza viruses are intrinsically sensitive to the antiviral action of IFN (31, 32, 36), although they may display a nongenetic-based transient resistance (36). In addition, IFN sensitizes cells to the initiation of apoptosis by viruses (42) and by double-stranded RNA (40), which may be spontaneously released in the course of influenza virus replication (14). Furthermore, IFN functions as an adjuvant to boost the adaptive immune response in mammals (3, 4, 11, 26, 41, 43, 46) and in chickens when administered perorally in the drinking water of influenza virus-infected birds (19). This raises the question: does the enhanced induction of IFN by delNS1 variants suffice to render an infectious influenza virus preparation sufficiently attenuated to function as an effective live vaccine? To address that question, we turned to a recent report that described the selection of several variants of influenza virus with a common backbone of A/TK/OR/71-SEPRL (Southeast Poultry Research Laboratory) that contained NS1 protein genes which were unusual in the length and nature of the amino acid residues at the C termini of the truncated NS1 proteins that they expressed because of the natural introduction of a frameshift and stop codon by the deletion in the NS1 protein gene (44). delNS1 variants were isolated from serial low-inoculum passages of TK/OR/71-delNS1[1-124] (H7N3) in eggs (44). Four of these genetically stable plaque-purified variants, each encoding a truncated NS1 protein of a particular length, were tested as a candidate LAIV in 2-week-old chickens. Two of the delNS1 variants were effective as live vaccines (double deletions [D-del] pc3 and pc4) (phenotypically vac+), and two were not (D-del pc1 and pc2) (phenotypically vac−) (44), despite only subtle differences in their encoded delNS1 proteins. Why were they phenotypically different?
The present study addresses this question by analyzing and comparing the different virus particles that constitute the subpopulations of these two effective (vac+) and two ineffective (vac−) live vaccine candidates. These analyses are based on recent reports in which noninfectious but biologically active particles (niBAPs) in subpopulations of influenza virus particles were defined and quantified (20, 21, 29). The study described in this report reveals several quantitative and qualitative differences between the particle subpopulations of the four candidate LAIVs, including the different types of IFN-induction dose-response curves, the quantum (maximum) yields (QY) of IFN induced, the efficacy of the interferon-inducing particles (IFPs), the replication efficiency of the virus, and the size of the plaques that they produced. Evidence is presented that the in vitro analysis of virus particle subpopulations may be useful to distinguish vac+ from vac− LAIV candidates and provide a basis for identifying and enhancing the performance of particles with desirable phenotypes.
MATERIALS AND METHODS
Cell culture and media.
Monolayers of primary chicken embryo cells (CECs) and chicken embryo kidney cells (CEKs) were prepared from cell suspensions of 9-day-old and 18-day-old embryos, respectively, obtained from Charles River SPAFAS, Inc., Storrs, CT. The cells were grown in attachment solution (AS; NCI medium plus 6% calf serum) (38) and incubated at 38.5°C. GMK-Vero and MDCK cells were grown in AS and Eagle's minimal essential medium (plus 5% calf serum), respectively, and incubated at 37.5°C.
LAIV viruses.
In a previous study, four LAIV virus variants (D-del pc1 to pc4) were generated naturally during passage of a stock of A/TK/OR/71-delNS1[1-124] (H7N3) virus in embryonated eggs (44). These viruses were confirmed to be genetically stable through plaque purification, followed by at least 5 passages in 10-day-old embryonated eggs. For the present study, these four variants and an isogenic virus encoding a full-size NS1[1-230] protein, used here and termed wild type, were reconstructed as already described using a reverse genetics system (44). The only differences among these five reconstituted viruses are the length and amino acid composition at the C terminus of the NS1 protein, as described previously (44) and shown in Table 1.
TABLE 1.
Comparison of biologically active and hemagglutinating particles in allantoic fluid stocks of wild-type TK/OR/71(H7N3) and candidate delNS1 LAIV variants
| rgTK/OR/71 designationa | No. of aa residues of NS1 protein (1-X. . .Y)b | Particle titer per ml (% of total BAPse) |
|||||
|---|---|---|---|---|---|---|---|
| PFPsc | IFPsc | DIPsc | niCKPsc | BAPsc,d | HAPsc,e | ||
| Wild type | 1-230 | 0.83 × 108 (4.1) | 4.2 × 108f (20.9) | 12 × 108 (59.7) | 3.07 × 108 (15.3) | 20.1 × 108 | 1,024 × 108 |
| D-del pc2 (vac−)g | 1-115. . .125 | 0.022 × 108 (0.02) | 7.2 × 108f (7.4) | 89 × 108 (91.7) | 0.88 × 108 (0.9) | 97.1 × 108 | 70 × 108 |
| D-del pc1 (vac−)g | 1-80. . .90 | 0.0059 × 108 (0.006) | 14 × 108f (13.4) | 89 × 108 (85.5) | 1.14 × 108 (1.1) | 104.1 × 108 | 384 × 108 |
| D-del pc4 (vac+)g | 1-91. . .93 | 0.90 × 108 (0.4) | 3.3 × 108h (1.5) | 220 × 108 (97.1) | 2.30 × 108 (1) | 226.1 × 108 | 1,024 × 108 |
| D-del pc3 (vac+)g | 1-69. . .86 | 0.36 × 108 (0.2) | 1.3 × 108h (0.74) | 170 × 108 (97.9) | 1.91 × 108 (1.1) | 173.6 × 108 | 448 × 108 |
All viruses were generated using a plasmid-based reverse genetics system (44).
For example, with D-del pc2, (1-X) represents the N-terminal amino acid (aa) residues 1 to 115 found in the wild-type A/TK/OR/71-SEPRL, and (…Y) represents the C-terminal amino acid residues 116 to 125 which differ from the wild-type sequences due to the introduction of a frameshift and new stop codon in the deleted NS1 protein gene. For details, see Fig. 2 of reference 44.
Titers expressed as number of particles per milliliter of allantoic fluid. This includes PFPs (iCKPs) (20), IFPs (21), DIPs (20), niCKPs (20, 29), and HAPs (8, 20).
BAP titers ≈ the sum of the titers of PFPs, IFPs, DIPs, and niCKPs.
Estimate on the basis of the average number of physical particles in 1 hemagglutinating unit, i.e., 5 × 106 (8, 20).
IFPs calculated from type r ≥ 1 IFN induction dose-response curves (17, 18; see Materials and Methods and Fig. 1).
Arranged in the order of increasing effectiveness as LAIV on the basis of the titer of antibodies against heterologous virus that they induced (44).
For simplicity, these virus variants are referred to as phenotypically vac− (ineffective) or vac+ (effective) vaccines, on the basis of their capacity to induce an antibody response, prevent virus shedding, and elicit protection against the heterologous virus, A/CK/NJ/150383-7/02 (H7N2) (44). By these criteria, D-del pc1 and pc2 viruses were vac− and D-del pc3 and pc4 were vac+. All stocks were stored at −80°C, thawed rapidly, and held on ice for use.
Interferon induction.
Primary CECs were seeded at a high density to produce confluent monolayers after 24 h and aged for 10 days in vitro to developmentally mature the IFN system (5, 35, 38). These developmentally aged cells were used to test the IFN-inducing capacities of the viruses. The IFN induction protocol was described recently (20). Briefly, replicate monolayers of developmentally aged CECs (107 cells/50 mm dish) were infected with increasing amounts (multiplicities) of the test virus and incubated at 40.5°C (optimal for IFN induction/production [38]) in medium without serum. The medium was harvested after 20 to 22 h, and the supernatant was assayed for acid-stable type I IFN, as described earlier (38). A standard preparation of UV-irradiated avian reovirus (45) was used to assess the IFN-inducing capacity of each batch of CECs, and a standard of recombinant chicken alpha IFN (rChIFN-α) (37) was used to calibrate the sensitivity of each CEC preparation to the action of IFN (36, 38).
IFN detection and quantification.
The titers of IFN were based on a 50% cytopathic effect (CPE50) assay in 96-well trays of CECs following treatment with IFN for 24 h at 37.5°C, infection with vesicular stomatitis virus (VSV) strain HR, and incubation for 48 h, before determination of the 50% end point photometrically (34, 36, 38).
IFN-induction curves, QYs of IFN, titers of IFPs, and IFP efficiency.
Full dose (amount of virus)-response (IFN yield) curves for IFN induction were generated from developmentally aged CECs, as reported previously (18). The type of IFN-induction curves observed and calculation of IFP titers were described in part previously (17, 18, 23). They were based on a Poisson distribution of IFP among the CEC population, P(r) = (e−m mr)/r! [where P(r) is the fraction of cells that get r IFPs when exposed to a multiplicity of m, r is the actual number of IFPs that enter the cell, r! is the factorial of r, m is the average number of IFPs in the virus suspension per cell in the monolayer, and e is the base of the natural logarithm (e = 2.71828…)], and the QY of IFN produced by cells in response to infection by IFP. Type r ≥ 1 curves best fit a class of cells in which a full yield of IFN is induced in any cell infected with 1 or more IFPs, i.e., P(r ≥ 1) = (1 − e−m)QY, where e−m is the fraction of cells that get zero IFPs. Type r = 2 curves best fit a model in which cells that receive 2 and only 2 IFPs respond by producing a QY of IFN, whereas cells infected with 1 or >2 IFPs induce little or no IFN, i.e., P(r = 2) = (e2m2/4em)QY.
In a type r ≥ 1 curve, the dilution of virus that induces 0.63 of the QY of IFN contains, on average, a multiplicity of plaque-forming particles (PFPs) (mIFP) of 1, from which the IFP titer can be calculated as described previously (18). In a type r = 2 curve, the dilution of virus that induces a QY of IFN occurs when 0.27 of the cells are infected with an average mIFP of 2. Since virtually all of the virus binds to the cell monolayer during the attachment period and the number of IFN-inducing cells and the dilution of virus required to induce a QY of IFN are known, the IFP titer can be calculated (18).
Assays for PFPs, niCKPs, DIPs, and HAPs.
The methods used to quantify subpopulations of PFPs, noninfectious cell-killing particles (niCKPs), defective interfering particles (DIPs), and hemagglutinating particles (HAPs) (in which these particles are subsumed) have been described previously (20, 21, 29). Briefly, PFPs were detected by plaque formation in CEKs (trypsin was not required) or Vero cells (trypsin was required). The number of niCKPs represents the difference between the total number of CKPs, determined by the clonogenic assay (29), and the number of infectious CKPs, where PFPs are infectious CKPs (iCKPs) (thus, total CKPs − iCKPs = niCKPs). DIPs were measured by the reduction in the PFP yield of a DIP-free helper virus (20, 27). The amount of HAPs was estimated on the basis of their capacity to agglutinate chicken red blood cells using a conversion factor (5 × 106) averaged from data from several reports (8, 20) to provide a first approximation of the number of physical particles in 1 hemagglutinating unit. The HAP titers obtained in this way should accurately reflect the relative titers of the different subpopulations, although they do not necessarily represent their sums.
Virus replication.
Virus replication was determined over a 48-h period at 40.5°C, which is normothermic for chickens. Confluent monolayers containing 3.0 × 106 CEKs in 50-mm dishes were infected at an mPFP of 0.03. The virus was suspended in 300 μl of NCI medium and attached to cells for 1 h. Unattached virus was aspirated, and the cells were overlaid with fresh serum-free medium. The medium was harvested at appropriate times, cleared of cell debris by centrifugation, and assayed for PFP and HAP activities.
RESULTS
IFN-induction dose-response curves, IFN QY, IFP titers, and IFP efficiency.
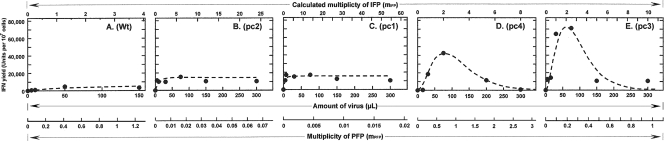
IFN-induction dose-response curves (17, 18) were generated by exposing developmentally aged primary CECs (35, 36) to increasing amounts of the four candidate LAIVs derived from TK/OR/71-delNS1 variants that expressed truncated NS1 proteins of different lengths or an isogenic virus that expressed the full-size NS1[1-230] protein (wild type) (Table 1) (44). Figure 1 shows representative responses of the wild type and each of these variants arranged by increasing QY. Two qualitatively different types of IFN-induction dose-response curves were observed. Figure 1A to C shows that wild-type and delNS1 vac− variants pc1 and pc2 generated IFN-induction dose-response curves that best fit a model based on a Poisson distribution of IFPs in which the fraction of cells infected with ≥1 IFP induces a QY of IFN, i.e., the maximum amount of IFN that a cell is capable of producing when exposed to that particular IFP. The type r ≥ 1 dose-response curve is the most commonly observed with influenza virus and many types of viruses tested (17, 18, 21). In contrast, Fig. 1D and E shows that delNS1 vac+ variants pc3 and pc4 generated dose-response curves that best fit a model in which cells infected with 2 and only 2 IFPs induce a QY of IFN and cells infected with 1 or >2 IFPs produce little or no IFN. This type r = 2 IFN-induction dose-response curve was described in theoretical terms years ago (16) but was observed experimentally for the first time in the present study. Importantly, the type r = 2 curves were observed in the same batches of aged CECs in which wild-type, pc1, and pc2 viruses induced the more common type r ≥ 1 curves. The QY of IFN observed at the plateau of the type r ≥ 1 curve for wild-type virus averaged 4,000 U/107 CECs, whereas the r ≥ 1 curves for vac− pc1 and pc2 were similar, averaging 15,500 U/107 CECs. The QY of IFN for vac+ variants pc3 and pc4 was observed at the peak of a type r = 2 curve (see Materials and Methods) and averaged 57,250 U/107 CECs (Fig. 1; Table 2).
FIG. 1.
IFN-induction dose (amount of virus)-response (IFN yield) curves generated by candidate LAIV delNS1 and wild-type (Wt) viruses in developmentally aged CECs at 40.5°C. These have been arranged in the order of increasing quantum (maximal) yields of IFN. Actual data points are represented by closed circles. The dashed lines represent the expected curves following the Poisson distribution of IFPs among cells in the monolayer, i.e., P(r) = (e−mmr)/r!. (A to C) Type r ≥ 1 curves, where P(r ≥ 1) = (1 − e−m)QY; (D and E) type r = 2 curves, where P(r = 2) = (e2m2/4em)QY (the various terms are defined in the text). Type r ≥ 1 curves (A to C) show that a cell infected with 1 or more IFPs produces a QY of IFN. Type r = 2 curves (D and E) represent the class of cells that, when infected with 2 and only 2 IFPs, produces a QY of IFN; cells infected with 1 or more than 2 IFPs produce little or no IFN.
TABLE 2.
Comparison of quantum (maximal) yields of IFN and relative efficiency of IFN induction
| rgTK/OR/71 designationa | No. of aa residues of NS1 protein (1-X. . .Y)b | QY of IFN (U/1.0 × 107 cells) | No. of IFPs inducing the QY of IFN | IFN yield (U/106 PFP) | Relative IFN induction efficiencyc |
|---|---|---|---|---|---|
| Wild type | 1-230 | 4,000 | 1.0 × 107d | 400 | 1.0 |
| D-del pc2 (vac−)e | 1-115. . .125 | 14,000 | 1.0 × 107d | 1,400 | 3.5 |
| D-del pc1 (vac−)e | 1-80. . .90 | 17,000 | 1.0 × 107d | 1,700 | 4.3 |
| D-del pc4 (vac+)e | 1-91. . .93 | 42,500 | 5.4 × 106f | 8,000 | 20.0 |
| D-del pc3 (vac+)e | 1-69. . .86 | 72,000 | 5.4 × 106f | 13,300 | 33.3 |
See footnote a in Table 1.
See footnote b in Table 1.
Calculated relative to the IFN U/106 IFP value for the wild-type virus.
The lowest number of IFPs required to infect all (i.e., 1.0 × 107) cells in order to induce a QY of IFN for type r ≥ 1 dose-response curves.
Arranged in the order of increasing effectiveness as LAIV on the basis of the titer of antibodies against heterologous virus that they induced (44).
The number of IFPs infecting 27% (i.e., 2.7 × 106) of the cells in order to induce a QY of IFN for type r = 2 dose-response curves. Each of these cells is infected with 2 IFPs. The total number of IFPs contributing to the QY equals 5.4 × 106. See Materials and Methods for calculations.
The IFPs averaged for the two vac− variants, though present in 4.6-fold excess compared to the amount of IFPs in the vac+ variants (Table 1), were less efficient in inducing IFN on an IFP-per-cell basis. The IFPs in the wild-type subpopulation were even less efficient (Table 2).
Sensitivity of LAIV delNS1 variants to action of interferon.
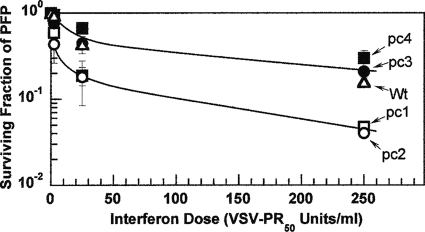
The sensitivities of the four candidate LAIV delNS1 variants and the wild type to the action of IFN were determined by exposing monolayers of primary CEKs to increasing doses of rChIFN-α (37) and measuring PFP survival at each dose, as was described earlier for other isolates of influenza virus (6, 36). Figure 2 shows that a single curve accommodates the data points for the two vac− variants, pc1 and pc2 (lower curve), and that the data points for the wild-type virus and the two vac+ variants, pc3 and pc4, can also be accommodated by a single curve (upper curve). From the initial slopes of these curves at low doses of IFN, variants pc1 and pc2 appear to be about 4 times more sensitive to the action of IFN than pc3, pc4, and wild-type virus. At higher doses of IFN, a comparison of the slopes of the PFP survival curves and extrapolation to the ordinate revealed that both sets of viruses contained a large fraction of PFPs that appear to be more refractory to the action of IFN. These biphasic survival curves resemble the triphasic curves observed in two earlier studies in which the resistance to high doses of IFN of a high-pathogenicity virus proved to be transient and was lost upon passage of the virus (36) and in the two original nonisogenic low-pathogenicity viruses, TK/OR/71-NS1[1-230] (H7N3) and the related virus delNS1[1-124] (6).
FIG. 2.
Sensitivity of candidate LAIV delNS1 and wild-type (Wt) viruses to the action of IFN. Following the exposure of 1-day-old monolayers of primary CEKs to rChIFN-α for 24 h at 38.5°C, the cell monolayers were challenged with PFPs of the respective viruses. An overlay of agarose in medium lacking serum or trypsin was incubated for 3 days before the surviving fraction of PFPs was determined. All viruses revealed biphasic survival curves, with the highest sensitivity occurring at low doses of IFN and a more resistant fraction being found at higher doses. The vac+ pc3 and pc4 variants, along with the wild-type virus, were experimentally indistinguishable (upper curve) and were less sensitive than the vac− pc1 and pc2 variants (lower curve). Error bars represent the standard deviations of two determinations. PR50, the amount of IFN that reduces the PFP titer by 50%.
DIP titers.
Standard PFP yield reduction assays (27), as reported earlier (20), were used to assess the DIP content of the four candidate LAIV variants and the wild type. The initial exponential rate of loss of PFP yield at low multiplicities of DIPs was used to calculate the titers of DIPs, as described previously (20). The four candidate LAIVs contained unusually high titers of DIPs, with the average for vac+ variants pc3 and pc4 being marginally higher (2.2-fold) than that for the vac− variants pc1 and pc2 (Table 1). In contrast, the DIP titer of the wild-type variant was about 12-fold less, similar to the titers observed for several low-pathogenicity type A influenza viruses (LPAIs), including the original TK/OR/71-SEPRL virus that had been subjected to three serial high-multiplicity passages (20).
Titers of infectious and niCKPs.
The clonogenic assay was used to determine the total CKP titer, as described previously (29). The titer of niCKPs represents the difference between the total CKP population and the infectious CKP population (iCKPs = PFPs); i.e., the number of niCKPs is equal to the number of CKPs minus the number of PFPs (29). Table 1 shows that the niCKP titers of all four delNS1 variants and the wild type were relatively high, with the average of the titers of vac− variants pc1 and pc2 being only 2-fold less than that of vac+ variants pc3 and pc4 and the wild-type virus. Further analysis revealed that the niCKP/PFP ratios of vac− pc1 and pc2 were at least 1 order of magnitude higher than those of vac+ pc3 and pc4 and the wild-type virus (Table 3). The high niCKP/PFP ratios indicate that virtually all cell killing by the vac− variants was due to niCKPs, as was the majority for the vac+ variants and wild-type virus. Thus, the extensive cytopathic effects noted early upon infection of cells with the delNS1 variants were due almost exclusively to the onset of apoptosis initiated by niCKPs and not PFPs, as was observed earlier with the original TK/OR/71-delNS1[1-124] virus (29).
TABLE 3.
Comparison of niBAP subpopulations with infectious particles (PFPs) and total physical particles (HAPs)
| rgTK/OR/71 designationa | No. of aa residues of NS1 protein (1-X. . .Y)b | Ratio of niBAPs/PFPs |
Estimated ratiod |
||||||
|---|---|---|---|---|---|---|---|---|---|
| IFPs/PFPsc | DIPs/PFPsc | niCKPs/PFPsc | HAPs/PFPs | HAPs/IFPs | HAPs/DIPs | HAPs/niCKPs | HAPs/BAPs | ||
| Wild type | 1-230 | 5 | 14 | 4 | 1,230 | 240 | 85 | 330 | 51 |
| D-del pc2 (vac−)e | 1-115. . .125 | 330 | 4,040 | 40 | 3,200 | 10 | 0.8 | 80 | 0.7 |
| D-del pc1 (vac−)e | 1-80. . .90 | 2,370 | 15,080 | 193 | 65,100 | 27 | 4.3 | 340 | 3.7 |
| D-del pc4 (vac+)e | 1-91. . .93 | 4 | 240 | 3 | 1,140 | 310 | 4.6 | 440 | 4.5 |
| D-del pc3 (vac+)e | 1-69. . .86 | 4 | 470 | 5 | 1,240 | 340 | 2.6 | 230 | 2.6 |
See footnote a in Table 1.
See footnote b in Table 1.
Based on the titers in Table 1. The ratios are rounded to the nearest whole number.
Estimated on the basis of the average number of physical particles in 1 hemagglutinating unit, i.e., 5 × 106 (8, 20): niBAPs ≈ sum of IFPs, DIPs, and niCKPs. Total BAPs ≈ niBAPs plus PFPs.
Arranged in the order of effectiveness as LAIV on the basis of the titer of antibodies they induced against a heterologous virus (44).
Comparison of PFP and HAP titers, growth rates, and plaque morphology of vac− and vac+ delNS1 variants.
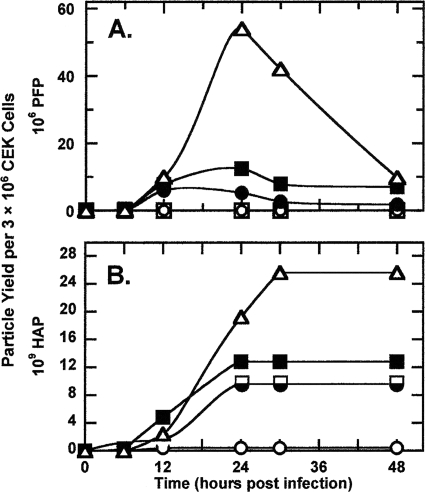
Table 1 shows the PFP titers of the four candidate LAIVs used in the vaccine study (44), along with the titer of the isogenic wild type. The average PFP titer in the allantoic fluid of the two vac− variants, pc1 and pc2, was low (1.4 × 106/ml) compared to that of the two vac+ variants, pc3 and pc4 (63 × 106/ml) (Table 1). The ratio of the PFP titer for vac+/PFP titer for vac− was 45. The PFP titer of the wild type was 83 × 106/ml. The low PFP titers of the vac− variants could not be attributed to the accompanying high DIP content because the vac+ variants, with their much higher PFP titers, had even more DIPs in the allantoic fluid (Table 1). Consequently, we compared the growth rates of the different delNS1 variants and the wild type (Fig. 3). Confluent monolayers of primary CEKs were infected at 40.5°C, which is normothermic for chickens and the optimal temperature for IFN production in chicken cells (38). CEKs do not require trypsin in the overlay to maintain plaque formation, and as observed from the abundant growth of wild-type virus in the kidneys of 1-day-old chicks (1.33 × 109 50% embryo lethal doses/g of tissue) (6), CEKs are excellent host cells. The highest efficiency of plaque formation was obtained when the cells were not allowed to age in vitro (35, 38). In Fig. 3A, the yield of PFPs per 3 × 106 cells is reported as a function of time. The rapid decline in PFP titer once peak yields are reached is attributed to the cessation of new infectious particle formation and the relatively short half-life (≈7 h) of PFPs at 40.5°C (data not shown). Both vac− variants produced enough infectious particles in CEKs to sustain the formation of small plaques (Fig. 4). In contrast, the vac+ variants consistently formed larger plaques, though not as large as the plaques formed by the wild type. Figure 4 shows the plaque size and morphology characteristic of each of the variants as they appeared after incubation at 37.5°C for 3 days in CEKs. Although the appearance of plaques means that the yield of PFP per CEK is high enough to sustain plaque formation, the low average yield of PFPs per cell suggests that not all cells in a primary CEK monolayer may serve as hosts. This view is borne out by the intact fibroblast-like cells observed within a plaque and the increase in their number with the age of the monolayer. The small plaques observed for pc2 and pc1 viruses and at a low frequency for pc3 viruses do not appear to reflect the action of IFN, since the number and size of the plaques did not change when 2-aminopurine (30 mM), which is known to block the induction of IFN in CECs (24), was present in the agarose overlay.
FIG. 3.
Growth curves of candidate LAIV delNS1 and wild-type viruses in CEK cells at 40.5°C. Confluent monolayers of primary CEKs were infected with each virus at an mPFP of 0.03, and serum-free NCI medium was added. Replicate dishes were harvested following incubation for the times indicated, and samples were clarified of cell debris and then assayed for PFP (A) and HAP (B) activity as described in Materials and Methods. Symbols: ▵, wild type; ○, pc2; □, pc1; ▪, pc4; •, pc3.
FIG. 4.
Characterization of plaque phenotypes of candidate LAIV-delNS1 and wild-type (Wt) viruses. Monolayers of primary CEK cells were infected with each virus and incubated at 37.5°C for 3 days, fixed with formalin, and stained with Giemsa. Plaque sizes or numbers did not change when 2-aminopurine, an inhibitor of IFN induction in chicken embryonic cells (24), was included in the overlay, indicating that any differences in plaque size were not attributable to the IFN system (24). The plaque plates have been arranged in the order of increasing diameter (mean ± standard deviation mm; n = 55).
Figure 3B shows that the average peak number of HAPs produced per CEK as a function of time at 40.5°C was in the order wild type > delNS1 pc4 > pc3 = pc1 ≫ pc2. This order is essentially the same as that observed for the egg-derived stocks of these candidate LAIVs shown in Table 1. Once they were produced, the HAPs were stable (Fig. 3B). D-del pc2 was the least efficient in producing HAP. Table 3 shows that relative to wild-type virus, a higher fraction of the HAPs in populations of D-del variants is assembled into BAPs.
DISCUSSION
Four candidate LAIVs with different deletions in the NS1 protein gene were naturally selected from serial passages of A/TK/OR/71-delNS1[1-124] in embryonated eggs (44). These deletions in the NS1 protein gene were stable when they were rescued in reassortant viruses (44). Two of the delNS1 variants functioned as effective (vac+) LAIVs, as reflected by the high titer of antibodies induced against challenge with a heterologous virus and a significant reduction in virus shedding. The other two candidate LAIVs were ineffective as vaccines (vac−) (44). The in vitro analyses of these four delNS1 variants for their content and the activities of their BAPs, namely, PFPs, IFPs, DIPs, and niCKPs, along with the activity of the HAPs (20, 21, 22, 29), show that they can be used to distinguish vac+ from vac− candidate LAIVs. Thus, the appropriate in vitro tests can be used to screen large numbers of potential LAIVs and provide insight into what viral phenotypes should be enhanced in a search for a more effective vaccine.
The composition of BAPs in subpopulations of influenza virus (20, 21, 29) was measured in vitro to determine whether the phenotypes that they express can be used to distinguish vac+ from vac− LAIVs. Data presented herein define several parameters measured in vitro that differentiate vac+ from vac− LAIVs. Prominent are three parameters associated with the IFN system: (i) the capacity to induce IFN, as measured by the quantum yield from full IFN-induction dose-response curves; (ii) the regulation of IFN induction, as revealed by the nature of the type of IFN-induction curve; and (iii) the efficiency of the IFPs. The order of decreasing expression of these parameters is ranked pc3 > pc4 ≫ pc1 ≈ pc2 ≫ wild type, placing the two vac+ variants first, the two vac− variants next, and the wild-type virus a distant last (Fig. 1; Table 2).
In a seemingly anomalous situation, the IFP subpopulation in the vac− variants was, on average, 4.6 times greater than that of the vac+ variants (Table 1), yet the relative efficiency of IFN induction by the IFPs of the vac+ variants was, on average, 6.8 times that of the vac− variants (Table 2). Furthermore, relative to wild-type virus, the efficiency of IFN induction by the D-del variants increases in the same order as the QY of IFN (Fig. 1, Table 2). Notably, the unusual type r = 2 nature of the IFN-induction dose-response curves of the two vac+ variants differed markedly from the usual type r ≥ 1 nature of the curves that characterized the two vac− variants and the wild type (Fig. 1), indicating that IFN induction by vac+ variants was highly downregulated at an mIFP of >2.
The data also demonstrate that the vac+ variants are less sensitive to the action of IFN than the vac− variants (Fig. 2), suggesting that the vac+ variants, with their increased capacity to induce IFN, may persist longer in the vaccinated host, thereby continuing to induce IFN and prolong the time that it can function as a natural adjuvant to enhance the production of antibody (4, 11, 19) and to help with clearance of the virus. In addition, there is a distinct replication advantage of the vac+ variants over the vac− variants (Fig. 3), with the former displaying on average a 45-fold greater PFP titer than the latter in allantoic fluid (Table 1). These differences in replication are reflected in the larger size of the plaques which develop from vac+ variants in primary CEKs (Fig. 4).
The presence and sizes of the subpopulations of DIPs and niCKPs do not of themselves help to distinguish vac+ and vac− variants. However, the contribution of these subpopulations to the vac+ phenotype has not been ruled out. For example, the unusually high titers of DIPs, which make up over 90% of the BAP population (Table 1), may exert a protective effect through the action of blocking virus replication (9, 20, 27) independent of the virus subtype (9), making them invaluable additions to the LAIVs. The demonstration that DIPs may render virulent avian influenza virus avirulent (1, 7) adds credence to a possible ameliorating role of DIPs in LAIVs. Whether the substantial numbers of niCKPs in a vac+ LAIV contribute to survival of the host because of the widespread destruction of cells through apoptosis (29) is not known. However, it is clear that DIPs do not function as niCKPs, nor do they block their action (see Fig. 5 in reference 20).
Quantitative estimates of the HAP population in which all of the above-mentioned BAPs are subsumed were based on published data which compare by various means the numbers of physical particles of influenza virus at the end point of a standardized hemagglutination (HA) test (8, 20). The conversion factor used herein may not be correct in absolute numbers because it has been applied to the native unpurified virus suspended in allantoic fluid used to vaccinate birds (44). However, at the least, the HAP titers shown here should be constant in relative terms for all of the populations of virus described. Although 96% of the HA activity of these allantoic fluid preparations is found in the pellet upon high-speed centrifugation (data not shown), indicative of the absence of soluble HA, it is possible that a small portion of the HA activity resides in the apoptosis-induced cell debris in the size range of influenza virus particles. Nonetheless, it is clear that the sum of the niBAP titers constitutes a significant fraction of the total HAPs in excess of that accounted for by PFPs. Does each BAP represent a particle of a distinct subpopulation, or do some BAP populations display overlapping phenotypes? The answer to this question requires further study. However, data reported thus far, including those obtained by UV target analysis, for the various phenotypes show that BAPs vary in the minimum gene requirements for their expression (20). For some BAPs, like DIPs, it is clear that the vast majority of the particles do not function as niCKPs, nor do they interfere with cell killing (20).
Current studies show that IFN-induction-suppressing particles (ISPs) (16, 21) have the potential to act as IFPs following truncation of the NS1 protein gene (a small gene target) or exposure to low doses of UV radiation (a large gene target) (15, 21). The titers of the DIP subpopulations in the 4 stocks of candidate LAIVs were unusually high, averaging 11.8-fold higher than the titer for the isogenic wild-type virus (Table 1) and 9.9-fold higher than the titer for the original TK/OR/71-delNS1[1-124] virus serially passed 3 times in MDCK cells to maximize the content of DIPs (20). The DIPs of the D-delNS1 variants constituted from 86 to 98% of the BAP population, making them the predominant BAPs (Table 1). On that basis, it is tempting to infer a causal relationship between the truncated NS1 proteins common to the D-del variants and the propensity to generate DIPs, perhaps akin to that reported for the NS2/NEP protein (30) and in keeping with the generally recognized role of the NS1 protein in replication.
Regardless of the absolute number of physical particles measured as HAPs or the extent to which the activities of BAPs represent discrete or overlapping functions, the biological assays used to define BAP subpopulations of LAIVs in vitro can collectively distinguish vac+ from vac− populations. On the basis of the findings of the present study, the successful vac+ LAIV might be expected to display collectively the following characteristics: (i) highly efficient IFPs, (ii) induction of high QYs of IFN, (iii) generation of IFN-induction dose-response curves that reflect tightly regulated IFN induction as a function of mIFP, (iv) enhanced resistance to IFN action, (v) robust levels of virus replication and yields of PFPs, (vi) production of relatively large plaques, and (vii) high titers of DIPs and niCKPs relative to PFP titers.
Because IFN plays a dominant role in successful LAIVs (11, 12, 13, 33), ameliorates pathogenesis (6), and functions as a natural adjuvant (3, 4, 11, 19, 41, 43), it may be possible to augment the levels of IFN induced endogenously by the vaccine through the exogenous delivery of IFN in drinking water, a procedure used in chickens that allowed the precise control of IFN doses and that is both tactically and economically feasible (19). Peroral use of IFN has also been shown to boost the adaptive immune system in other hosts (26, 41, 43, 46). Low doses of UV radiation enhance even further the IFN-inducing capacity of influenza viruses that express truncated NS1 protein (21) and hence might serve as a second means of augmenting the IFN levels in vaccinated hosts. Assays of BAPs and the attributes of the subpopulations that they generate provide a fingerprint of effective LAIVs, and more will be tested to determine the extent to which their detection and interaction with host cells in vitro is predictive of the vac+/vac− phenotypes.
Acknowledgments
This research was supported in part by USDA grants 58-1940-0-07 through the Center of Excellence in Vaccine Research at the University of Connecticut, a USDA SCA Award 58-6612-7-157, and a donation to the University of Connecticut's Laboratory for Virus and Interferon Research. This study benefited from the services of the Animal Cell Culture Facility of the Biotechnology-Bioservices Center of the University of Connecticut.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Bean, W. J., Y. Kawaoka, J. M. Wood, J. E. Pearson, and R. G. Webster. 1985. Characterization of virulent and avirulent A/chicken/Pennsylvanyia/83 influenza viruses: potential role of defective interfering RNAs in nature. J. Virol. 54:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belshe, R. B., K. M. Edwards, T. Vesikari, S. V. Black, R. E. Walker, M. Hultquist, G. Kemble, and E. M. Connor. 2007. Live attenuated versus inactivated influenza vaccine in infants and young children. CAIV-T Comparative Efficacy Study Group. N. Engl. J. Med. 356:685-696. [DOI] [PubMed] [Google Scholar]
- 3.Bracci, L., I. Canini, S. Puzelli, P. Sestili, M. Vanditti, M. Spada, I. Donatelli, F. Belardelli, and E. Proietti. 2005. Type I IFN is a powerful mucosal adjuvant for a selective intranasal vaccination against influenza virus in mice and affects antigen capture at mucosal level. Vaccine 23:2994-3004. [DOI] [PubMed] [Google Scholar]
- 4.Bracci, L., I. Canini, M. Vanditti, M. Spada, S. Puzelli, I. Donatelli, F. Belardelli, and E. Proietti. 2006. Type I IFN as a vaccine adjuvant for both systemic and mucosal vaccination against influenza virus. Vaccine 24(Suppl. 2):56-57. [DOI] [PubMed] [Google Scholar]
- 5.Carver, D. H., and P. I. Marcus. 1967. Enhanced interferon production from chick embryo cells aged in vitro. Virology 32:247-257. [DOI] [PubMed] [Google Scholar]
- 6.Cauthen, A. N., D. E. Swayne, M. J. Sekellick, P. I. Marcus, and D. L. Suarez. 2007. Amelioration of influenza virus pathogenesis in chickens is attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 gene. J. Virol. 81:1838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, T. M., and R. G. Webster. 1987. Defective interfering virus associated with A/chicken/Pennsylvania/83 influenza virus. J. Virol. 61:1517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desselberger, U. 1975. Relation of virus particle counts to the hemagglutinating activity of influenza virus suspensions measured by the HA pattern test and by the photometric HCU method. Arch. Virol. 49:365-372. [DOI] [PubMed] [Google Scholar]
- 9.Dimmock, N. J., E. W. Rainsford, P. E. Scott, and A. C. Marriott. 2008. Influenza virus protecting RNA: an effective prophylactic and therapeutic antiviral. J. Virol. 82:8570-8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorse, G. J., M. J. Campbell, E. E. Otto, D. C. Powers, G. W. Chambers, and F. K. Newman. 1995. Increased anti-influenza A virus cytotoxic T cell activity following vaccination of the chronically ill elderly with live attenuated or inactivated influenza virus vaccine. J. Infect. Dis. 172:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Hai, R., L. Martínez-Sobrido, K. A. Fraser, J. Ayllon, A. García-Sastre, and P. Palese. 2008. Influenza B virus NS1-truncated mutants: live-attenuated vaccine approach. J. Virol. 82:10580-10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochs, G., I. Koerner, L. Theil, S. Kothlow, B. Kaspers, N. Ruggli, A. Summerfield, J. Pavlovic, J. Stech, and P. Staeheli. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 88:1403-1409. [DOI] [PubMed] [Google Scholar]
- 13.Kochs, G., L. Martinez-Sobrido, S. Lienenklaus, S. Weiss, A. García-Sastre, and P. Staeheli. 2009. Strong interferon-inducing capacity of a highly virulent variant of influenza A virus strain PR8 with deletions in the NS1 gene. J. Gen. Virol. 90:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majde, J. A., Z. Guha-Thakurta, S. Chen, S. Bredow, and J. M. Krueger. 1998. Spontaneous release of stable viral double-stranded RNA into the extracellular medium by influenza virus-infected MDCK endothelial cells: implications for the viral acute-phase response. Arch. Virol. 143:2371-2380. [DOI] [PubMed] [Google Scholar]
- 15.Malinoski, C. P., and P. I. Marcus. 2010. Influenza virus: can a single particle express two different phenotypes—IFN induction, or suppression of IFN induction?, p. 280. Abstr. 29th Annu. Meet. Am. Soc. Virol.
- 16.Marcus, P. I. 1982. Interferon induction by viruses. IX. Antagonistic activities of virus particles modulate interferon production. J. Interferon Res. 2:511-518. [DOI] [PubMed] [Google Scholar]
- 17.Marcus, P. I. 1983. Interferon induction by viruses: one molecule of dsRNA as the threshold for interferon induction, p. 115-180. In I. Gresser (ed.), Interferon 5. Academic Press, New York, NY. [PubMed]
- 18.Marcus, P. I. 1986. Interferon induction dose-response curves. Methods Enzymol. 119:106-114. [DOI] [PubMed] [Google Scholar]
- 19.Marcus, P. I., T. Girshick, L. van der Heide, and M. J. Sekellick. 2007. Super-sentinel chickens and detection of low-pathogenicity influenza virus. Emerg. Infect. Dis. 13:1608-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcus, P. I., J. M. Ngunjiri, and M. J. Sekellick. 2009. Dynamics of biologically active subpopulations in influenza virus: plaque-forming, noninfectious cell-killing, and defective-interfering particles. J. Virol. 83:8122-8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcus, P. I., J. M. Rojek, and M. J. Sekellick. 2005. Interferon induction and/or production and its suppression in influenza viruses. J. Virol. 79:2880-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcus, P. I., and M. J. Sekellick. 1976. Cell killing by viruses. III. The interferon system and the inhibition of cell killing by vesicular stomatitis virus. Virology 69:378-393. [DOI] [PubMed] [Google Scholar]
- 23.Marcus, P. I., and M. J. Sekellick. 1977. Defective-interfering particles with covalently linked [±] RNA induce interferon. Nature 266:815-819. [DOI] [PubMed] [Google Scholar]
- 24.Marcus, P. I., and M. J. Sekellick. 1988. Interferon induction by viruses. XVI. 2-Aminopurine blocks selectively and reversibly an early stage in interferon induction. J. Gen. Virol. 69:1637-1645. [DOI] [PubMed] [Google Scholar]
- 25.Mueller, S. N., W. A. Langley, E. Carnero, A. García-Sastre, and R. Ahmed. 2010. Immunization with live-attenuated influenza viruses expressing altered NS1 proteins results in potent and protective memory CD8+ T cells responses. J. Virol. 84:1847-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagao, Y., K. Yamashiro, N. Hara, Y. Horisawa, K. Kato, and A. Uemura. 1998. Oral-mucosal administration of IFN-α potentiates immune response in mice. J. Interferon Cytokine Res. 18:661-666. [DOI] [PubMed] [Google Scholar]
- 27.Nayak, D. P., T. M. Chambers, and R. K. Akkina. 1985. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr. Top. Microbiol. Immunol. 114:103-151. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, K. M., B. R. Schram, M. W. McGregor, A. S. Sheoran, C. W. Olsen, and D. P. Lunn. 1998. Local and systemic isotype-specific antibody responses to equine influenza virus infection versus conventional vaccination. Vaccine 16:1306-1313. [DOI] [PubMed] [Google Scholar]
- 29.Ngunjiri, J. M., M. J. Sekellick, and P. I. Marcus. 2008. Clonogenic assay of type A influenza viruses reveals noninfectious cell-killing (apoptosis-inducing) particles. J. Virol. 82:2673-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odagiri, T., K. Tominaga, K. Tobita, and S. Ohta. 1994. An amino acid change in the non-structural NS2 protein of an influenza A virus is responsible for the generation of defective interfering (DI) particles by amplifying DI RNAs and suppressing complementary RNA synthesis. J. Gen. Virol. 75:43-53. [DOI] [PubMed] [Google Scholar]
- 31.Portnoy, J., and T. C. Merigan. 1971. The effect of interferon and interferon inducers on avian influenza virus. J. Infect. Dis. 124:545-552. [DOI] [PubMed] [Google Scholar]
- 32.Richman, D. D., B. R. Murphy, S. Baron, and C. Uhlendorf. 1976. Three strains of influenza A virus (H3N2): interferon sensitivity in vitro and interferon production in volunteers. J. Clin. Microbiol. 3:223-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richt, J. A., and A. García-Sastre. 2009. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top. Microbiol. Immunol. 333:177-195. [DOI] [PubMed] [Google Scholar]
- 34.Rubinstein, S., P. C. Familletti, and S. Pestka. 1981. Convenient assay for interferons. J. Virol. 37:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekellick, M. J., W. J. Biggers, and P. I. Marcus. 1990. Development of the interferon system. I. In chicken cells development in ovo continues on time in vitro. In Vitro Cell. Dev. Biol. 26:997-1003. [DOI] [PubMed] [Google Scholar]
- 36.Sekellick, M. J., S. A. Carra, A. Bowman, D. A. Hopkins, and P. I. Marcus. 2000. Transient resistance of influenza virus to interferon action attributed to random multiple packaging and activity of NS genes. J. Interferon Cytokine Res. 20:963-970. [DOI] [PubMed] [Google Scholar]
- 37.Sekellick, M. J., A. F. Ferrandino, D. A. Hopkins, and P. I. Marcus. 1994. Chicken interferon gene: cloning, expression, and analysis. J. Interferon Res. 14:71-79. [DOI] [PubMed] [Google Scholar]
- 38.Sekellick, M. J., and P. I. Marcus. 1986. Induction of high titer chicken interferon. Methods Enzymol. 119:115-125. [DOI] [PubMed] [Google Scholar]
- 39.Steel, J., A. C. Lowen, L. Pena, M. Angel, A. Solórzano, R. Albrecht, D. R. Perez, A. Garcia-Sastre, and P. Palese. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart, W. E., II, E. De Clerq, and P. De Somer. 1973. Specificity of interferon-induced enhancement of toxicity for double-stranded ribonucleic acids. J. Gen. Virol. 18:237-246. [DOI] [PubMed] [Google Scholar]
- 41.Takayama, S., K. Iwaki, Y. Nishida, M. Tanaka, M. Fujii, K. Ohashi, M. Ikeda, and M. Kurimoto. 1999. Effects of oral administration of interferon-α on antibody production in mice with induced tolerance. J. Interferon Cytokine Res. 19:895-900. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, N., M. Sato, M. S. Lamphier, H. Nozawa, E. Oda, S. Nogucho, R. E. Schreiber, Y. Tsujimoto, and T. Taniguchi. 1998. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells 3:29-37. [DOI] [PubMed] [Google Scholar]
- 43.Tough, D. F., P. Borrow, and J. Sprent. 1996. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science 272:1947-1950. [DOI] [PubMed] [Google Scholar]
- 44.Wang, L., D. L. Suarez, M. Pantin-Jackwood, M. Mibayashi, A. García-Sastre, Y. M. Saif, and C.-W. Lee. 2008. Characterization of influenza virus variants with different sizes of the non-structural (NS) genes and their potential as a live influenza vaccine in poultry. Vaccine 26:3580-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winship, T. R., and P. I. Marcus. 1980. Interferon induction by viruses. VI. Reovirus: virion genome dsRNA as the interferon inducer in aged chick embryo cells. J. Interferon Res. 1:155-167. [DOI] [PubMed] [Google Scholar]
- 46.Wolff, T., and S. Ludwig. 2009. Influenza viruses control the vertebrate type I interferon system: factors, mechanisms, and consequences. J. Interferon Cytokine Res. 29:549-557. [DOI] [PubMed] [Google Scholar]