Abstract
The biological, serological, and genomic characterization of a paramyxovirus recently isolated from rockhopper penguins (Eudyptes chrysocome) suggested that this virus represented a new avian paramyxovirus (APMV) group, APMV10. This penguin virus resembled other APMVs by electron microscopy; however, its viral hemagglutination (HA) activity was not inhibited by antisera against any of the nine defined APMV serotypes. In addition, antiserum generated against this penguin virus did not inhibit the HA of representative viruses of the other APMV serotypes. Sequence data produced using random priming methods revealed a genomic structure typical of APMV. Phylogenetic evaluation of coding regions revealed that amino acid sequences of all six proteins were most closely related to APMV2 and APMV8. The calculation of evolutionary distances among proteins and distances at the nucleotide level confirmed that APMV2, APMV8, and the penguin virus all were sufficiently divergent from each other to be considered different serotypes. We propose that this isolate, named APMV10/penguin/Falkland Islands/324/2007, be the prototype virus for APMV10. Because of the known problems associated with serology, such as antiserum cross-reactivity and one-way immunogenicity, in addition to the reliance on the immune response to a single protein, the hemagglutinin-neuraminidase, as the sole base for viral classification, we suggest the need for new classification guidelines that incorporate genome sequence comparisons.
Viruses from the Paramyxoviridae family have caused disease in humans and animals for centuries. Over the last 40 years, many paramyxoviruses isolated from animals and people have been newly described (16, 17, 22, 29, 31, 32, 36, 42, 44, 46, 49, 58, 59, 62-64). Viruses from this family are pleomorphic, enveloped, single-stranded, nonsegmented, negative-sense RNA viruses that demonstrate serological cross-reactivity with other paramyxoviruses related to them (30, 46). The subfamily Paramyxovirinae is divided into five genera: Respirovirus, Morbillivirus, Rubulavirus, Henipavirus, and Avulavirus (30). The Avulavirus genus contains nine distinct avian paramyxovirus (APMV) serotypes (Table 1), and information on the discovery of each has been reported elsewhere (4, 6, 7, 9, 12, 34, 41, 50, 51, 60, 68).
TABLE 1.
Characteristics of prototype viruses APMV1 to APMV9 and the penguin virus
| Strain | Host | Disease | Distribution | Fusion cleavagec | GI accession no. |
|---|---|---|---|---|---|
| APMV1/Newcastle disease virus | >250 species | High mortality | Worldwide | GRRQKRF | 45511218 |
| Inapparent | Worldwide | GGRQGRLa | 11545722 | ||
| APMV2/Chicken/CA/Yucaipa/1956 | Turkey, chickens, psittacines, rails, passerines | Decrease in egg production and respiratory disease | Worldwide | DKPASRF | 169144527 |
| APMV3/Turkey/WI/1968 | Turkey | Mild respiratory disease and moderate egg decrease | Worldwide | PRPSGRLa | 209484147 |
| APMV3/Parakeet/Netherlands/449/1975 | Psittacines, passerines, flamingos | Neurological, enteric, and respiratory disease | Worldwide | ARPRGRLa | 171472314 |
| APMV4/Duck/Hong Kong/D3/1975 | Duck, geese, chickens | None known | Worldwide | VDIQPRF | 210076708 |
| APMV5/Budgerigar/Japan/Kunitachi/1974 | Budgerigars, lorikeets | High mortality, enteric disease | Japan, United Kingdom, Australia | GKRKKRFa | 290563909 |
| APMV6/Duck/Hong Kong/199/1977 | Ducks, geese, turkeys | Mild respiratory disease and increased mortality in turkeys | Worldwide | PAPEPRLb | 15081567 |
| APMV7/Dove/TN/4/1975 | Pigeons, doves, turkeys | Mild respiratory disease in turkeys | United States, England, Japan | TLPSSRF | 224979458 |
| APMV8/Goose/DE/1053/1976 | Ducks, geese | None known | United States, Japan | TYPQTRLa | 226343050 |
| APMV9/Duck/NY/22/1978 | Ducks | None known | Worldwide | RIREGRIa | 217068693 |
| APMV10/Penguin/Falkland Islands/324/2007 | Rockhopper penguins | None Known | Falkland Islands | DKPSQRIa | 300432141 |
Requires the addition of an exogenous protease.
Protease requirement depends on the isolate examined.
Putative.
Six of these serotypes were classified in the latter half of the 1970s, when the most reliable assay available to classify paramyxoviruses was the hemagglutination inhibition (HI) assay (61). However, there are multiple problems associated with the use of serology, including the inability to classify some APMVs by comparing them to the sera of the nine defined APMVs alone (2, 8). In addition, one-way antigenicity and cross-reactivity between different serotypes have been documented for many years (4, 5, 14, 25, 29, 33, 34, 41, 51, 52, 60). The ability of APMVs, like other viruses, to show antigenic drift as it evolves over time (37, 43, 54) and the wide use and availability of precise molecular methods, such as PCR and genome sequencing, demonstrate the need for a more practical classification system.
The genetic diversity of APMVs is still largely unexplored, as hundreds of avian species have never been surveyed for the presence of viruses that do not cause significant signs of disease or are not economically important. The emergence of H5N1 highly pathogenic avian influenza (HPAI) virus as the cause of the largest outbreak of a virulent virus in poultry in the past 100 years has spurred the development of surveillance programs to better understand the ecology of avian influenza (AI) viruses in aquatic birds around the globe, and in some instances it has provided opportunities for observing other viruses in wild bird populations (15, 53). In 2007, as part of a seabird health surveillance program in the Falkland Islands (Islas Malvinas), oral and cloacal swabs and serum were collected from rockhopper penguins (Eudyptes chrysocome) and environmental/fecal swab pools were collected from other seabirds.
While AI virus has not yet been isolated from penguins in the sub-Antarctic and Antarctic areas, there have been two reports of serum antibodies positive to H7 and H10 from the Adélie species (11, 40). Rare isolations of APMV1, both virulent (45) and of low virulence (8), have been reported from Antarctic penguins. Sera positive for APMV1 and AMPV2 have also been reported (21, 24, 38, 40, 53). Since 1981, paramyxoviruses have been isolated from king penguins (Aptenodytes patagonicus), royal penguins (Eudyptes schlegeli), and Adélie penguins (Pygoscelis adeliae) from Antarctica and little blue penguins (Eudyptula minor) from Australia that cannot be identified as belonging to APMV1 to -9 and have not yet been classified (8, 11, 38-40). The morphology, biological and genomic characteristics, and antigenic relatedness of an APMV recently isolated from multiple penguin colonies on the Falkland Islands are reported here. Evidence that the virus belongs to a new serotype (APMV10) and a demonstration of the advantages of a whole genome system of analysis based on random sequencing followed by comparison of genetic distances are presented. Only after all APMVs are reported and classified will epidemiological information be known as to how the viruses are moving and spreading as the birds travel and interact with other avian species.
MATERIALS AND METHODS
Sample collection.
Oropharyngeal swabs (193), cloacal swabs (193), and serum samples (99) were collected from 31 adult and 162 juvenile rockhopper penguins. In addition, fresh environmental fecal samples (150) were collected from multiple areas east of Berkeley Sound from other species: upland geese (Chloephaga picta), imperial shags (Phalacrocorax atriceps), speckled teal (Anas flavirostris), and crested ducks (Anas specularioides). In total, samples from 75 geese and 25 of each of the other species were pooled into groups of five. All swabs were placed into 1.5 ml of brain heart infusion (BHI) broth with antibiotics (10,000 U penicillin G/ml, 1,000 μg/ml gentamicin sulfate, and 5 μg amphotericin B/ml; Sigma Chemical Co., St. Louis, MO). The longitude and latitude of each location were noted with a global positioning system.
Virus detection, isolation, and characterization.
RNA was extracted from the 386 swab fluids from penguins and the 150 pooled fecal samples from the other species using the MagMAX-96 viral isolation kit (Ambion, Austin, TX) with the KingFisher processor (Thermo Scientific, Waltham, MA) and initially screened with real-time reverse transcriptase PCR (RT-PCR) for the AI virus matrix gene (M gene) (55). The BHI broth (100 μl) from each M-gene-positive sample was then inoculated into each of three 9-day-old specific-pathogen-free (SPF) embryonating chicken eggs (ECE). Fluid was harvested at 4 days postinoculation, and hemagglutination (HA) assays were performed following each of the three passages with fluid from individual eggs (10). For the second and third passages, 200 μl was inoculated into each of three eggs for each sample, which was tested as described above. The BinaxNOW influenza A&B test (Inverness Medical, Princeton, NJ) was run on the HA-positive samples. Real-time RT-PCR for APMV1 was performed on RNA from HA-positive egg fluids that were negative for AI virus to identify the NDV matrix or polymerase genes (27, 65). This material was also used as the antigen source for hemagglutination inhibition (HI) assays against each of the APMV sera and is described in detail below.
In addition, APMV1 (Chicken/NJ/LaSota/1946), APMV2 (Chicken/CA/Yucaipa/1956 and Finch/North Ireland-Bangor/1975), APMV3 (Turkey/WI/1968 and Parakeet/Netherlands/449/1975), APMV4 (Duck/Hong Kong/D3/1975), APMV6 (Duck/Hong Kong/199/1977), APMV7 (Dove/TN/4/1975), APMV8 (Goose/DE/1053/1976), and APMV9 (Duck/NY/22/1978) were obtained from the Southeast Poultry Research Laboratory (SEPRL) repository and used as antigens for HI assays against serum from chickens infected with the unidentified penguin virus. APMV5 (Budgerigar/Japan/Kunitachi/1975) antigen was obtained from the Veterinary Laboratories Agency (VLA), Weybridge, United Kingdom. African green monkey cells (VERO) and transformed chicken embryo fibroblast (DF1) cells were obtained from the American Type Culture Collection (Manassas, VA). An intracerebral pathogenicity index (ICPI) test was performed in day-old chicks using standard methods (10).
Serology.
To detect influenza A virus antibodies, the agar gel immunodiffusion (AGID) assay was performed as described previously (13) and a commercial blocking enzyme-linked immunosorbent assay (ELISA) for multiple species (IDEXX, Westbrook, ME) was conducted by following the manufacturer's recommendations with the 99 serum samples harvested during the surveillance. Reference antisera for APMV1 to -4 and APMV6 to -9 for the HI assays against the penguin virus antigen were produced at the National Veterinary Services Laboratory (NVSL) (Ames, IA) (10). APMV5 antiserum was produced and obtained from VLA.
Serum directed to the novel penguin virus was produced using the NVSL standard protocol. Briefly, 4 ml of clarified infectious allantoic fluid was inoculated intravenously into adult SPF leghorn chickens (Gallus gallus). At 18 days postinoculation, birds were sedated and serum was harvested. Serum from individual birds was not pooled. The Institutional Animal Care and Use Committee at SEPRL approved all animal experiments, which were performed in biosafety level 3 (BSL-3) enhanced animal rooms due to the exotic origin of the penguin paramyxovirus.
Hemagglutination inhibition tests using reference APMV sera against unknown antigens and vice versa were performed as described previously (10) except with modifications to follow the NVSL standard testing protocol (AVPRO0807.04). Briefly, in addition to the antigen being diluted to a HA titer of 8 HA units per 25 μl, the reference serum is diluted to reach an endpoint titer between 1:16 and 1:64 determined with 8 HA units of the homologous antigen (positive control) per 25 μl.
Electron microscopy.
Penguin virus and APMV8 were prepared as follows: 3 ml of infectious allantoic fluid was concentrated through a 22% sucrose cushion, and the pellet was resuspended in 0.75 ml of SPF allantoic fluid overnight at 4 C. This virus was diluted 1:2 with freshly made, buffered 5% (para)formaldehyde and incubated for 40 min. Uninfected allantoic fluid was used as a negative control. Allantoic fluid containing virus was safety tested to ensure complete inactivation using a procedure approved by the SEPRL Institutional Biosafety Committee prior to removal from BSL-3 enhanced containment. Briefly, 5% of the fluid was diluted 1:8 with brain heart infusion broth containing antibiotics and inoculated via the chorioallantoic sac into ECE and monitored daily for embryonic deaths. After 4 days, fluid was harvested, tested for HA activity, and passed into ECE for a second passage. Appropriate negative and positive controls were included. Electron microscopy was performed using a negative stain, differential centrifugation, and conventional biological processing with chemical fixations containing (para)formaldehyde and glutaraldehyde in the primary fixative and osmium tetroxide in the secondary fixative. All samples were viewed with a JEOL JEM-1210 transmission electron microscope at an accelerating voltage of 120 kV.
RNA isolation and cDNA amplification.
RNA was extracted from the oropharyngeal and cloacal swabs using Trizol LS (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After extraction, RNA was eluted in 100 μl of RNase-free water and stored at −80°C. PCR amplification of the penguin virus RNA from sample 324 was performed using the Qiagen one-step RT-PCR kit (Qiagen, Valencia, CA) according to the manufacturer's instructions with the following additions to the master mix: 1.5 μl of 25 mM MgCl2, 0.25 μl Hi-Fi enzyme (ABgene, Epsom, United Kingdom), and random priming techniques (1). The primers used in this shotgun approach were FR26RV-N/FR40RV (19). The following cycling conditions were used: two cycles of 10°C for 10 min, adding 0.5°C/s up to 40°C, and 40°C for 30 min; one cycle of 95°C for 15 min, followed by 39 cycles of 94°C for 45 s, 58°C for 30 s, and 72°C for 1 min 45 s, and one cycle of 72°C for 10 min; and holding at 4°C. Amplified products were separated on a 1% agarose gel. Bands above 500 bp were extracted and purified from the gel as follows: one extraction of bands from 500 bp to 1,000 bp and another extraction for bands above 1,001 bp, using the QuickClean DNA gel extraction kit (GenScript, Piscataway, NJ). The products were eluted into 30 μl elution buffer, and the samples were quantified using a standard spectrophotometer. These products were cloned with a TOPO TA cloning kit (Invitrogen) using the standard protocol with the following exceptions: 3 μl of cloning reaction mixture was added to the competent cells, and the room temperature incubation period was extended by 10 min. Penguin virus clones were sequenced using primers provided with the TOPO TA kit: M13F (GTA-AAA-CGA-CGG-CCAG) and M13R (CAG-GAA-ACA-GCT-ATG-AC).
Primers (Peng-4327F [CTC-TAT-CTC-TTA-ATG-TTA-TAC-TAT-ACC-C] and Peng 4979R [CCG-AGT-TGA-AGG-ACT-GGA-CC]) made from the penguin virus sequence obtained were used in a conventional RT-PCR to amplify a short region of the fusion protein, identifying additional swab fluids containing penguin virus. Briefly, RNA was extracted from oropharyngeal and cloacal swabs using the Qiagen RNeasy kit standard protocol, and RT-PCR was performed with the Superscript III one step-PCR kit using 40 cycles at 56°C for annealing (30 s) and 68°C for extension (1 min) and a final extension at 68°C for 5 min. Amplified cDNA products were purified by gel electrophoresis and sequenced directly using the same primers.
DNA sequencing.
All double-stranded nucleotide-sequencing reactions were performed with fluorescent dideoxynucleotide terminators in an automated Applied Biosystems International (ABI) sequencer as previously reported (26). Random genome nucleotide sequencing was done as reported previously for AI virus (1). Nucleotide sequence assembly and editing were conducted with the LaserGene sequence analysis software package (DNASTAR, Madison, WI) or with Codon Aligner using Phred and Phrap for complete genomes (CodonCode Corporation, Dedham, MA). The novel APMV10 sequence for the complete coding and intergenic regions of virus isolated from sample 324 and partial sequences of the fusion genes from samples 323, 437, and 539 were submitted to GenBank (see below).
Alignment analysis of nucleotide and deduced amino acid sequences and comparison at the amino acid level.
Alignments were performed with the ClustalW (57) program, followed by manual editing using Molecular Evolutionary Genetics Analysis (MEGA) (56). Phylogenetic trees were compiled based on the derived amino acid sequences of each gene using the maximum likelihood (as implemented in PHYML and in ProteinML) or the neighbor-joining method, as implemented in MEGA. For the HN gene the Jones, Taylor, and Thornton (JTT) substitution model for amino acids (23) and the discrete gamma distribution model (66), with estimates of shape parameter and proportions of invariable sites, were utilized.
Estimation of evolutionary distances at the nucleotide level.
The evolutionary distance between sequences was measured by the number of nucleotide substitutions occurring between the sequences after using the indicated parameters. Table 2 illustrates the number of base substitutions per site from the analysis. Estimates of evolutionary divergence between the nucleotide sequences of different polymerase genes were estimated for individual viruses as follows. The number of base substitutions per site was calculated based on the pairwise analysis of nine sequences. Standard error estimates were obtained by a bootstrap procedure (500 replicates) using the Kimura 2-parameter method in MEGA4 (28, 56). The rate of variation among sites was modeled with a gamma distribution (shape parameter = 1). All coding and noncoding positions were analyzed. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 6,581 positions in the final data set. GI numbers and serotypes are indicated.
TABLE 2.
Estimates of evolutionary divergence at the nucleotide level between APMV10 and other viruses
| APMV straina | Divergence or SEb |
|||||||
|---|---|---|---|---|---|---|---|---|
| APMV10 (penguin) | APMV2 (169133527) | APMV8 |
APMV7 (224979458) | APMV6 |
||||
| 226343057 | 225350515 | 147574365 | 15081567 | 193506926 | ||||
| 10 (penguin) | 0.0274 | 0.0271 | 0.0273 | 0.0492 | 0.0644 | 0.0636 | 0.0612 | |
| 2 (169133527) | 0.8433 | 0.0292 | 0.0296 | 0.0661 | 0.0649 | 0.0659 | 0.0633 | |
| 8 (226343057) | 0.8927 | 0.9407 | 0.0005 | 0.0574 | 0.0644 | 0.0662 | 0.0602 | |
| 8 (225350515) | 0.8919 | 0.9443 | 0.0018 | 0.0584 | 0.0645 | 0.0663 | 0.0612 | |
| 7 (224979458) | 1.428 | 1.5348 | 1.3841 | 1.3898 | 0.0495 | 0.049 | 0.0543 | |
| 6 (147574365) | 1.5035 | 1.5569 | 1.5066 | 1.5066 | 1.4621 | 0.0013 | 0.0039 | |
| 6 (15081567) | 1.5005 | 1.553 | 1.5268 | 1.5268 | 1.4683 | 0.013 | 0.0036 | |
| 6 (193506926) | 1.4644 | 1.546 | 1.5167 | 1.52 | 1.4702 | 0.0612 | 0.0601 | |
GI numbers of isolates are given for each APMV.
Standard errors are shown in italics. Bold values are distances of viruses of the same serotype.
Estimates of evolutionary divergence between the hemagglutinin-neuraminidase (HN) proteins.
The evolutionary distance between proteins was measured by the number of amino acid substitutions occurring between the sequences after using the indicated parameters. Table 5 illustrates the number of amino acid substitutions per site from the analysis. In total, 254 sequences of the HN protein were aligned using the ClustalW program (57) and separated into groups representing the nine defined APMV serotypes (Table 3; see also the supplemental material). The average distances between groups and standard errors were determined using MEGA4 software (over 500 bootstrap replicates). All sites (n = 499), except for the gaps, were included. A homogeneous substitution pattern among lineages with gamma-distributed rates and the Dayhoff matrix were used to compare groups (18). Estimates of distances for the HN protein between selected viral sequences from individual viruses were also compared using the above settings in MEGA. Further details of these mathematical models and general guidelines for the use of these methods are given at http://www.megasoftware.net.
TABLE 3.
Estimates of HN protein evolutionary divergence between AMPV10 and individual viruses of APMV2 and APMV8 serotypes
| APMV straina | Divergence or SEb |
||||||
|---|---|---|---|---|---|---|---|
| APMV10 (penguin) | APMV2 |
Murayamac (538243) | APMV8 |
||||
| 169144533 | 21541576 | 16326624 | 226343055 | 226343062 | |||
| 10 (penguin) | 0.0857 | 0.0856 | 0.0860 | 0.0967 | 0.0915 | 0.0916 | |
| 2 (169144533) | 1.029 | 0.0017 | 0.0023 | 0.0360 | 0.1000 | 0.0986 | |
| 2 (21541576) | 1.0268 | 0.0018 | 0.0016 | 0.0359 | 0.0996 | 0.0982 | |
| 2 (16326624) | 1.0371 | 0.0037 | 0.0018 | 0.0363 | 0.1004 | 0.0991 | |
| Murayama (538243) | 1.1624 | 0.3205 | 0.3176 | 0.3228 | 0.0969 | 0.0955 | |
| 8 (226343055) | 1.0779 | 1.1389 | 1.1338 | 1.145 | 1.1741 | 0.0048 | |
| 8 (226343062) | 1.0998 | 1.1158 | 1.1108 | 1.1218 | 1.1529 | 0.0147 | |
GI numbers of isolates are given for each APMV.
Standard errors are shown in italics. Bold values are distances of viruses of the same serotype.
Murayama virus is a monkey-adapted APMV2.
Evidence of recombination.
An ungapped alignment of 14,048 nucleotides (entire coding regions) from the genomes of APMV10, APMV2, and APMV8 was performed using the MUSCLE V2.3.6 program (20) along with the RDP3.24 program (35), which implements the analysis of recombination using seven different algorithms.
Nucleotide sequence accession numbers.
Sequences retrieved from GenBank public databases were used to generate alignments; accession numbers for viruses used in each table and figure can be found in the supplemental material. In general, except for the F and HN genes of APMV1, the complete sequences for the other APMV1 genes and for the other APMV serotypes available in GenBank are limited. The accession number for APMV10, isolate 324, is 300432141. The accession numbers for the smaller nucleotide sequences of isolates 323, 437, and 539 are 304435629, 304435631, and 304435633, respectively.
RESULTS
Virus isolation and identification.
As part of routine surveillance, 386 oropharyngeal (OP) and cloacal (CL) swabs from 193 rockhopper penguins and 150 environmental fecal samples from other wild bird species were tested for AI virus by real-time RT-PCR (RRT-PCR) and virus isolation (VI) in SPF ECE. Two samples from rockhopper penguins, 324 and 257, produced suspect AI virus-positive results for the matrix gene by RRT-PCR, and therefore, VI was conducted on them. Sample 324 from an OP swab collected on Eagle Hill 3 produced positive HA results after the 2nd and 3rd passages. However, the HA-positive allantoic fluid from sample 324 was negative for AI virus based on the BinaxNOW influenza A&B test, which qualitatively detects influenza A and B nucleoprotein antigens, and by RRT-PCR. RNA from this sample was analyzed with the matrix/polymerase gene multiplex RRT-PCR for APMV1 (27) and found to be negative. In addition, the 150 environmental samples and fecal swabs collected and pooled from additional species, upland geese, imperial shags, speckled teal, and crested ducks, were negative for hemagglutinating viruses after three egg passages.
The RNA from sample 324 was copied into cDNA, cloned into plasmids, and sequenced using random priming methods as indicated in Material and Methods. After an initial backbone was assembled, conventional RT-PCR was done to close gaps using clone 324-specific primers. In addition, a set of primers specifically designed to recognize a highly variable region of the fusion protein was designed based on the 324 sequence. These were used to perform additional PCR tests on all the oral and cloacal swabs from the penguins. The amplification produced 500-base-pair products from three additional cloacal swabs obtained from the rockhopper fledglings. The three new positive samples were obtained from Rugged Hill 1 (isolate 437), Rugged Hill 2 (isolate 539), and Eagle Hill 3 (isolate 323). Interestingly, there were clear nucleotide differences among these isolates, which separated them into two different phylogenetic groups that corresponded with the locations that covered two miles of coastline. Isolates 323, 437, and 539 had identities of 100%, 98.4% and 97.8%, respectively, with isolate 324. From the five locations where penguins were sampled, a total of four samples, representing three of the locations, in addition to 324OP, were positive by at least one test for the novel APMV, APMV10. Of these five, only four could be recovered through VI in SPF ECE: samples 324OP, 323CL, 437CL, and 539CL. All four samples were negative for APMV1 when tested against LaSota antiserum in an HI test. The fourth PCR-positive, but VI-negative, sample, 528CL, was also from Rugged Hill 2 (Table 4). All of the nonpenguin/environmental samples were negative for AI virus by RRT-PCR, and none of these samples produced hemagglutination after three serial passages in SPF ECE; therefore, RRT-PCR for APMV1 was not performed on these samples.
TABLE 4.
Serum sample identification and characterization
| Bird no. | Resulta of antigen experiment |
Location | Ageb | ||||
|---|---|---|---|---|---|---|---|
| ELISA AIV | HI |
||||||
| APMV10 | APMV8 | APMV1 | APMV2 | ||||
| 302 | + | 8 | − | − | − | Eagle Hill 3 | Jv |
| 400 | − | 32 | − | − | − | Rugged Hill 1 | Ad |
| 416 | − | − | 8 | − | − | Rugged Hill 1 | Jv |
| 514 | − | 128 | 64 | − | − | Rugged Hill 2 | Ad |
| 558 | + | 32 | − | − | − | Diamond Cove | Ad |
| 612 | − | 8 | − | − | − | Diamond Cove | Ad |
−, negative; +, positive. Serotypes of the antigens used for the HI assay are shown.
Jv, juvenile; Ad, adult.
Characterization of APMV10.
Infective allantoic fluid of APMV10/penguin/Falkland Islands/324/2007 had a titer of 109.3 median embryo infectious doses (EID50) per ml and an HA titer of 2,048 when the virus was inoculated into the chorioallantoic sac of 9-day-old SPF ECE and harvested at 7 days postinoculation. This strain was able to grow in VERO and DF1 cells only with the addition of trypsin. Mean death time in ECE was greater than 90 h, with no mortality after 7 days, suggesting a virus that is of low virulence for chickens. When virus 324 was inoculated into the cerebrum of day-old SPF chickens, the ICPI value obtained was 0.00, with no clinical signs of disease. In addition, after one intravenous inoculation of live virus (324) into adult birds for the production of antisera, the HA titer of the serum after 18 days was 1,024 and the birds remained clinically normal with no signs of disease.
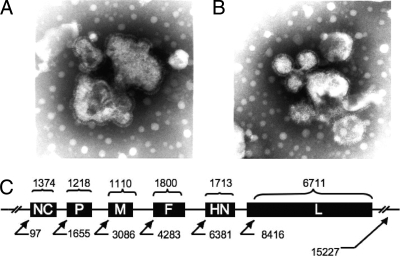
Electron microscopy of purified virus showed characteristics typical of a paramyxovirus. Pleomorphic, enveloped virions with glycoprotein projections ranged between 90 and 300 nm in diameter, some with an extruded herringbone nucleoprotein (Fig. 1B). As a comparison, we fixed purified APMV8 under identical conditions and compared the size and morphology of this virus with those of APMV10 (Fig. 1A).
FIG. 1.
Electron microscopy morphology of APMV8 (A) and APMV10 (B). (C) Schematic of APMV10 indicating gene start site, gene lengths, and length of available sequence.
Serology.
Of the 99 serum samples collected from rockhopper penguins (the same animals that provided swab samples), 5 were positive for APMV10 and all were negative for APMV1. Coincidentally, two of these birds were also weakly positive for AI antibodies using a blocking ELISA, but the subtype of AI virus was not further characterized (Table 4). Serum from an adult penguin, 514, was positive for both APMV10 (1:128) and APMV8 (1:64) (Table 4).
The penguin virus antigen from sample 324 was tested for hemagglutination inhibition against reference antisera to the prototype viruses for all of the APMVs listed in Materials and Methods using the NVSL standard testing protocol which identifies an isolate as a specific serotype if the endpoint of inhibition (HI endpoint titer) is within two well dilutions (4-fold difference) from the homologous APMV-positive control. Although sample 324 had incomplete inhibition in the first two wells in the APMV2, APMV7, and APMV8 HI tests and a single well of incomplete inhibition in the APMV3, APMV6, and APMV9 HI tests, these results are interpreted as negative, as they were not sufficiently strong to comply with international standards used for serological classification of viruses. In addition, negative results for APMV1 were obtained. Sample 324 had two wells of inhibition or a HI titer of 1:4 for APMV1, and the APMV1-positive control had 5 wells of inhibition or a HI titer of 1:32.
The reverse HI assay testing antiserum to the penguin virus, prepared following international serology guidelines from SPF chickens, against the 11 prototype APMV antigens listed in the methods showed inhibition only to the homologous penguin antigen, 324 (data not shown). Sample 324 was also tested against a panel of broadly reactive monoclonal antibodies that recognize APMV1, and no inhibition was found using these antibodies. Most significant was the lack of reaction against the APMV1 B79 monoclonal antibody that is known to recognize most APMV1 strains, including pigeon paramyxovirus serotype 1 (PPMV1), which is an APMV1 variant known to bind to different monoclonal antibodies.
Sequencing and phylogenetic analysis.
Assembly of random generated sequences produced a contig of 15,226 bp, representing an incomplete genome longer than the reported sequences of APMV2/Chicken/California/Yucaipa/1956 (14,904 bp) and shorter than those of APMV8/Goose/Delaware/1053/1976 (15,342 bp). Partially overlapping, randomly generated sequences (n = 502) were obtained, providing 11-fold coverage of the genome. The sequence contained all expected coding sequences for an APMV, including the coding regions and the noncoding terminal sequences (95 noncoding nucleotides at the 3′ end and 101 noncoding nucleotides after the polymerase gene stop signal) (Fig. 1C).
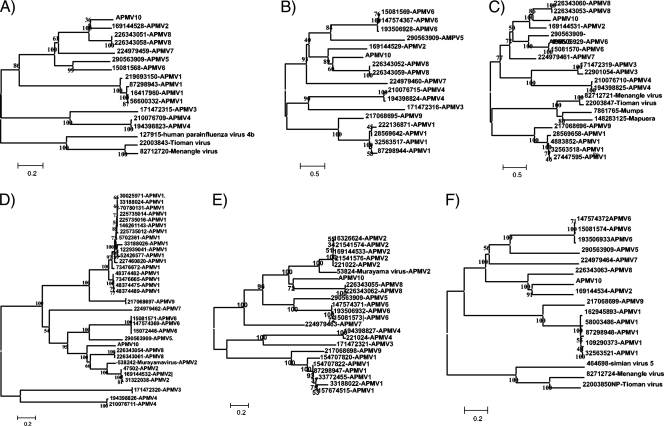
The virus, although highly divergent from other APMVs at the nucleotide level, was most closely related to APMV2 and APMV8 viruses at the amino acid sequence level (Fig. 2A to F). However, distances between APMV10 and either APMV2 or APMV8 varied depending on the gene analyzed. At the nucleotide level, the best alignment was obtained using the entire length (6,581 positions) of the highly conserved polymerase gene. Phylogenetic analysis of the polymerase gene at the nucleotide level suggest that APMV2 and APMV10 may be closer to each other than APMV8 and APMV10 (Table 2), and similar results were obtained when the 14,048 nucleotides of the entire coding regions of APMV2, −8 and −10, were aligned (data not shown). The analysis of the distances and the standard error (shown in italics) among all of these viruses indicates that differences in distances among the three serotypes may be very similar (Table 2). The calculated distance between APMV10 and APMV2 (GI 169133527) is 0.843, while the distance between APMV10 and APMV8 (GI 226343057) is 0.892 (with standard errors of 0.0274 and 0.0271, respectively). This type of distance is similar to the distances observed between APMV2 (GI 169133527) and APMV8 (GI 226343057) (e.g., 0.941 with a standard error of 0.029) (Table 2).
FIG. 2.
Phylogenetic analysis of the six proteins of the penguin virus in comparison to serologically identified APMVs in genomic order: (A) nucleocapsid protein (NP), (B) phosphoprotein (P), (C) matrix (M), (D) fusion (F), (E) hemagglutinin-neuraminidase (HN), and (F) large polymerase (L).
The close relationship between APMV10, APMV2, and APMV8 is reflected throughout the genome in the phylogenetic trees of individual open reading frames at the amino acid level (Fig. 2A to F). A detailed analysis of the distances separating the HN protein in individual viruses is also provided (Table 3). The HN is a surface protein believed to be key in determining the serological classification of viruses currently based on the serological HI assay. Here, the analysis of distances among individual viruses indicates that the shortest distance to APMV10 (1.0268) corresponds to an APMV2 virus (GI 21541576).
To determine distances among serotypes, rather than distances among individual viruses, available sequences were first grouped in serotypes, and the average distances between serotypes was calculated (Table 5). In this experiment, a total of 254 sequences of the HN protein were aligned and separated into the nine groups corresponding to APMV1 to APMV9 to determine the average distances between groups (see the supplemental material). Data suggest that APMV2 (1.027) is the group most closely related to APMV10, with APMV8 being the next closest (1.122) (Table 3). Regardless of the phylogenetic origin of APMV10, it is clear from the analysis that the distances that separate APMV10 and APMV2 and APMV10 and APMV8 (1.027 and 1.122, respectively) are similar to the distances existing among other distinct serotypes (the distance of APMV2 to APMV8 is 1.160) and in some cases are larger than the distances reported for different serotypes (APMV9 to APMV1 is 0.624).
TABLE 5.
Estimates of inter- and intraserotype average distances (D) among the grouped viruses for each serotype at the HN protein
| APMV strainc | Interserotype D or SEa |
Intraserotypeb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APMV10 | APMV1 | APMV2 | APMV3 | APMV4 | APMV5 | APMV6 | APMV7 | APMV8 | APMV9 | D | SE | |
| APMV10 | 0.255 | 0.092 | 0.280 | 0.335 | 0.155 | 0.151 | 0.163 | 0.102 | 0.253 | n/c | n/c | |
| APMV1 | 2.537 | 0.253 | 0.219 | 0.231 | 0.258 | 0.254 | 0.208 | 0.233 | 0.055 | 0.092 | 0.008 | |
| APMV2 | 1.027 | 2.570 | 0.277 | 0.276 | 0.136 | 0.130 | 0.150 | 0.101 | 0.258 | 0.006 | 0.002 | |
| APMV3 | 2.782 | 2.312 | 2.762 | 0.153 | 0.267 | 0.271 | 0.244 | 0.287 | 0.216 | n/c | n/c | |
| APMV4 | 3.180 | 2.469 | 2.768 | 1.732 | 0.327 | 0.314 | 0.237 | 0.282 | 0.225 | 0.020 | 0.006 | |
| APMV5 | 1.666 | 2.562 | 1.535 | 2.742 | 3.249 | 0.074 | 0.139 | 0.148 | 0.270 | n/c | n/c | |
| APMV6 | 1.748 | 2.671 | 1.564 | 2.829 | 3.051 | 0.851 | 0.137 | 0.157 | 0.274 | 0.020 | 0.005 | |
| APMV7 | 1.762 | 2.280 | 1.678 | 2.504 | 2.410 | 1.617 | 1.562 | 0.166 | 0.212 | n/c | n/c | |
| APMV8 | 1.122 | 2.444 | 1.160 | 2.971 | 2.827 | 1.671 | 1.820 | 1.765 | 0.244 | 0.014 | 0.005 | |
| APMV9 | 2.554 | 0.624 | 2.619 | 2.234 | 2.466 | 2.762 | 2.868 | 2.270 | 2.593 | n/c | n/c | |
Standard errors are shown in italics.
n/c, not calculated since only one sequence available.
GI numbers for viruses can be found in the supplemental material. Numbers of unique sequences used for APMV10 and APMV1 through APMV9 are 1, 238, 5 (not including Murayama virus), 1, 2, 3, 1, 2 and 1 respectively.
The distances for the HN protein of viruses that have been previously considered to be of one serotype have been analyzed (intraserotype distances) and are reported in the two furthest right columns of Table 5. Values range from 0.092 for the many viruses of the APMV1 group to 0.006 for APMV2 viruses. The number of isolates used to calculate the distances is dependent on the number of isolates available in GenBank. The large distance in APMV1, the most well studied serotype, is likely to be a reflection of the larger number of isolates and the large period of time covered. The 238 HN sequences isolated over 61 years for APMV1 were compared to 5 available sequences of APMV2, 4 of which were isolated in China over 4 years (67).
The difficulties associated with a sequence-based classification system are illustrated by the fact that the interserotype distances between some viruses, such as APMV1 and APMV9 (0.624) and APMV5 and APMV6 (0.851), are less than the distances of the other AMPV serotypes whose values are all greater than 1.0 (Table 5). In addition, a large range of genetic diversity can be seen in one serotype alone. When 119 nonredundant HN sequences (571 amino acids each) of APMV1 were compared using a different program, PROTDIST (http://evolution.genetics.washington.edu/phylip/progs.data.prot.html), to again calculate the intraserotype distance among the APMV1 isolates, the average was 0.092288256, with the maximum distance (of all 119 versus 118 combinations) equaling 0.195126 (see the supplemental material). The larger distance observed, 0.195126, among APMV1, collected over decades, illustrates that a large range of genetic diversity can be found in viruses of one serotype.
DISCUSSION
The combined serological and genomic data presented here characterize a novel virus that likely represents a new APMV, APMV10. The ability to classify new APMVs using computer-based programs that provide increased resolution by comparative analysis of genomic sequences is demonstrated. The APMV10 described here was isolated from three sampling sites within a 10-km area from four juvenile birds of one penguin species (Table 6). Serological evidence of APMV10 was found in adults in this same area (Table 4). This newly isolated hemagglutinating paramyxovirus, which is significantly different from APMV1 to APMV9 in sequence, is unable to grow in VERO cells and DF1 cells without the addition of trypsin and has a fusion cleavage site with an isoleucine at position 117 similar to that reported in APMV9 (Table 1) (47, 48). The electron microscopy confirmed paramyxovirus morphology with projections (glycoproteins) protruding from the envelope (Fig. 1B) and the occasional herringbone nucleocapsid extruded from some virions (data not shown). However, not surprisingly, it was not possible to detect large morphological differences between APMV8 and APMV10.
TABLE 6.
Identification and characterization of swab samples from juvenile penguins using hemagglutination inhibition assay, PCR and VI
| Bird no.a | Resultb |
Location | ||
|---|---|---|---|---|
| APMV10 serumc | PCR | VI | ||
| 324OP | 32 | + | + | Eagle Hill 3 |
| 323CL | 32 | + | + | Eagle Hill 3 |
| 437CL | 32 | + | + | Rugged Hill 1 |
| 539CL | 128 | + | + | Rugged Hill 2 |
| 528OP | n/a | + | − | Rugged Hill 2 |
Letters indicate sample type: OP, oropharyngeal; CL, cloacal.
n/a, not applicable since no virus was isolated. −, negative; +, positive.
Antibody titer calculated from the highest dilution of serum able to inhibit hemagglutinin, using B HA units of the APMV10 viruses.
Closer phylogenetic distances between APMV10, APMV8, and APMV2 were observed throughout the genome, at both the amino acid and nucleotide levels, suggesting the existence of a common ancestor to these three viruses. Not only are the calculated distances between APMV10 and APMV2 and between APMV10 and APMV8 similar to each other, but they are also similar to the distances observed between APMV2 and APMV8 (Table 2). The phylogenetic analysis by the methods of neighbor joining (Fig. 2) and maximum likelihood (see the supplemental material) demonstrate that APMV10 is closer to both AMPV2 and APMV8 than to other serotypes; however, it was clearly separated from both of those serotypes with variable distances depending on the gene used for analysis.
The apparent positioning of different APMV10 genes in the same phylogenetic branch with either APMV2 or APMV8, and the occasional lack of bootstrap support of the nodes that separate APMV10 virus in some of the trees, is evidence of the genetic equidistance of APMV10 from serotypes APMV2 and APMV8. However, this lack of bootstrap support is not likely to reflect the possibility of recombination among these three serotypes (data not shown). In fact, an ungapped alignment of the noncoding regions from the genomes of APMV10, APMV2, and APMV8 evaluating the sequences using seven algorithms found no events compatible with a recombination (data not shown).
Historically, APMVs have been grouped or separated by serological tests, primarily the HI test, which has been used to classify APMV into nine serotypes (3). This system has worked remarkably well to distinguish viruses that are distantly related. However, as more and more isolates become available and new intermediate isolates are discovered, it is possible that a serology-based classification may not provide enough resolution for exact classification. This may be the case with APMV10, which clearly does not belong to serotype APMV2 or APMV8 but may cross-react antigenically with viruses from those serotypes depending on the source of the serum used in the assay. For example in this instance, serum harvested from week-old survivors of an ICPI assay or hyperimmune serum made after multiple inoculations inhibited both the APMV8 antigen and the homologous APMV10 antigen, with HI titers of 32. These antisera failed to inhibit hemagglutination against antigens of the other APMV serotypes. This cross-reactivity with the APMV8 antigen was not observed when the serum used was produced with only one intravenous injection (data not shown). Here, inhibition was seen only with the homologous APMV10 antigen. While a correlation between the HN amino acid phylogenetic trees and the serological classification exists, the reliance of an entire classification system on the antigenic response to the single HN protein is not ideal. The possibility of the HN gene losing the ability to hemagglutinate red blood cells and undergo antigenic drift and even the unlikely possibility of recombination suggest that changes in the HN may not reflect the type of changes observed in the rest of the genome.
Antibodies to prototype viruses representative of the nine currently defined APMV serotypes are already known to cross-react with antigens of other serotypes and with mammalian paramyxoviruses (4, 5, 14, 25, 33, 41, 51, 52, 60). In addition, one-way antigenicity suggests that the immunogenic potential of the HN protein of some viruses may not be comparable in all serotypes. Currently there are at least eight other avian paramyxoviruses isolated from different penguin species from various geographical locations that are not inhibited by known APMV sera and likely representing at least three serotypes different from each other (2, 8) and from APMV10 (data not shown). In addition, the difficulties associated with easily procuring antiserum to each of the known APMVs (except for reference laboratories) highlight the need for alternative tools to classify paramyxoviruses.
Because the APMV10 antigen did not react with antiserum from the other nine APMVs and antigens from the prototype viruses of the other serotypes did not react with the antiserum prepared against APMV10, this virus should be considered a new serotype. However, when the serum was prepared in young birds or under conditions that induced a hyperimmune response with a broader antibody spectrum, cross-reactivity with viruses from a closely related serotype (APMV8) did occur. This reaction has been documented for many years with paramyxoviruses and is the reason that reference serum is prepared with one inoculation (4, 34, 60). The APMVs so far recognized, for the most part, have been classified because they are viruses that impact domestic poultry species and the poultry industry. While it is unlikely that APMV10 will significantly impact the poultry industry and it is true that there have been only four viruses of this new serotype identified, there are other APMVs that are not yet classified because conventional methods have proved unreliable or ambiguous and no alternative has been put forward. Here, we demonstrate that using sequence data that is widely available in public databases (GenBank), a random sequencing approach that does not require the use of specifically designed primers and the use of computer programs to compare and relate viruses using distance provides a possible system to obtain a good understanding of the phylogenetic relationship among other hitherto unclassified AMPVs. After large numbers of complete sequences of each serotype representing viruses isolated in different parts of the world are deposited into GenBank, distance data along with negative serology can be used to determine definitive thresholds to assign new serotypes.
Supplementary Material
Acknowledgments
We thank the Falkland Islands government for issuing research permits and Nic Huin of Falkland Conservation, the Falkland Government Veterinary Service, Vic Epstein, Joe Hollins, and Lyn Dent for logistical support and field assistance. We thank Joan Beck, Scott Lee, Tim Olivier, and Dawn Williams-Coplin for their excellent technical assistance and Roger Brock for animal care. We thank Ruth Manvell for providing the APMV5 antigen and antisera. We also thank Mary Ard at the College of Veterinary Medicine Electron Microscopy Laboratory and Joyce Bennett at the South Atlantic Area Sequencing Facility for their assistance.
The mention of trade names does not imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 11 August 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Afonso, C. L. 2007. Sequencing of avian influenza virus genomes following random amplification. Biotechniques 43:188, 190, 192. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J. 1980. Avian paramyxoviruses. Vet. Bull. 50:737-752. [Google Scholar]
- 3.Alexander, D. J. (ed.). 1988. NDV—an avian paramyxovirus structure. Kluwer Academic Publishers, Boston, MA.
- 4.Alexander, D. J., and N. J. Chettle. 1978. Relationship of parakeet/Netherlands/449/75 virus to other avian paramyxoviruses. Res. Vet. Sci. 25:105-106. [PubMed] [Google Scholar]
- 5.Alexander, D. J., and M. S. Collins. 1981. The structural polypeptides of avian paramyxoviruses. Arch. Virol. 67:309-323. [DOI] [PubMed] [Google Scholar]
- 6.Alexander, D. J., V. S. Hinshaw, and M. S. Collins. 1981. Characterization of viruses from doves representing a new serotype of avian paramyxoviruses. Arch. Virol. 68:265-269. [DOI] [PubMed] [Google Scholar]
- 7.Alexander, D. J., V. S. Hinshaw, M. S. Collins, and N. Yamane. 1983. Characterization of viruses which represent further distinct serotypes (PMV-8 and PMV-9) of avian paramyxoviruses. Arch. Virol. 78:29-36. [DOI] [PubMed] [Google Scholar]
- 8.Alexander, D. J., R. J. Manvell, M. S. Collins, S. J. Brockman, H. A. Westbury, I. Morgan, and F. J. Austin. 1989. Characterization of paramyxoviruses isolated from penguins in Antarctica and sub-Antarctica during 1976-1979. Arch. Virol. 109:135-143. [DOI] [PubMed] [Google Scholar]
- 9.Alexander, D. J., and D. A. Senne. 2008. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, p. 75-116. In Y. M. Saif, A. M. Fadly, J. R. Glisson, L. R. McDougald, L. K. Nolan, and D. E. Swayne (ed.), Diseases of poultry, 12th ed. Iowa State University Press, Ames, IA.
- 10.Alexander, D. J., and D. E. Swayne. 1998. Newcastle disease virus and other avian paramyxoviruses, p. 156-163. In D. E. Swayne, J. R. Glisson, M. W. Jackwood, J. E. Pearson, and W. M. Reed (ed.), A laboratory manual for the isolation and identification of avian pathogens, 4th ed. American Association of Avian Pathologists, Kennett Square, PA.
- 11.Austin, F. J., and R. G. Webster. 1993. Evidence of ortho- and paramyxoviruses in fauna from Antarctica. J. Wildl. Dis. 29:568-571. [DOI] [PubMed] [Google Scholar]
- 12.Bankowski, R. A., R. E. Corstvet, and G. T. Clark. 1960. Isolation of an unidentified agent from the respiratory tract of chickens. Science 132:292-293. [DOI] [PubMed] [Google Scholar]
- 13.Beard, C. W. 1970. Avian influenza antibody detection by immunodiffusion. Avian Dis. 14:337-341. [PubMed] [Google Scholar]
- 14.Brostrom, M. A., G. Bruening, and R. A. Bankowski. 1971. Comparison of neuraminidases of paramyxoviruses with immunologically dissimilar hemagglutinins. Virology 46:856-865. [DOI] [PubMed] [Google Scholar]
- 15.Chang, C. M., C. Lebarbenchon, M. Gauthier-Clerc, C. Le Bohec, D. Beaune, Y. Le Maho, and S. van der Werf. 2009. Molecular surveillance for avian influenza A virus in king penguins (Aptenodytes patagonicus). Polar Biol. 32:663-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chua, K. B., L. F. Wang, S. K. Lam, G. Crameri, M. Yu, T. Wise, D. Boyle, A. D. Hyatt, and B. T. Eaton. 2001. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 283:215-229. [DOI] [PubMed] [Google Scholar]
- 17.Clark, H. F., F. S. Lief, P. D. Lunger, D. Waters, P. Leloup, D. W. Foelsch, and R. W. Wyler. 1979. Fer de Lance virus (FDLV): a probable paramyxovirus isolated from a reptile. J. Gen. Virol. 44:405-418. [DOI] [PubMed] [Google Scholar]
- 18.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins; matrices for detecting distant relationships, p. 345-358. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. V. National Biomedical Research Foundation, Washington, DC. [Google Scholar]
- 19.Djikeng, A., R. Halpin, R. Kuzmickas, J. Depasse, J. Feldblyum, N. Sengamalay, C. Afonso, X. Zhang, N. G. Anderson, E. Ghedin, and D. J. Spiro. 2008. Viral genome sequencing by random priming methods. BMC Genomics 9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauthier-Clerc, M., N. Eterradossi, D. Toquin, M. Guiettet, G. Kuntz, and Y. L. Maho. 2002. Serological survey of the king penguin, Aptenodytes patagonicus, in Crozet Archipelago for antibodies to infectious bursal disease, influenza A and Newcastle disease viruses. Polar Biol. 25:316-319. [Google Scholar]
- 22.Jack, P. J., D. B. Boyle, B. T. Eaton, and L. F. Wang. 2005. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J. Virol. 79:10690-10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 24.Karesh, W. B., M. M. Uhart, E. Frere, P. Gandini, W. E. Braselton, H. Puche, and R. A. Cook. 1999. Health evaluation of free-ranging rockhopper penguins (Eudyptes chrysocomes) in Argentina. J. Zoo Wildl. Med. 30:25-31. [PubMed] [Google Scholar]
- 25.Kessler, N., M. Aymard, and A. Calvet. 1979. Study of a new strain of paramyxoviruses isolated from wild ducks: antigenic and biological properties. J. Gen. Virol. 43:273-282. [DOI] [PubMed] [Google Scholar]
- 26.Kim, L. M., D. J. King, P. E. Curry, D. L. Suarez, D. E. Swayne, D. E. Stallknecht, R. D. Slemons, J. C. Pedersen, D. A. Senne, K. Winker, and C. L. Afonso. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81:12641-12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, L. M., D. L. Suarez, and C. L. Afonso. 2008. Detection of a broad range of class I and II Newcastle disease viruses using multiplex real-time reverse transcription polymerase chain reaction assay. J. Vet. Diagn. Invest. 20:414-425. [DOI] [PubMed] [Google Scholar]
- 28.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 29.Kusagawa, S., H. Komada, X. Mao, M. Kawano, F. Nishikawa, M. Tsurudome, H. Matsumura, H. Ohta, T. Yuasa, and M. Nishio. 1993. Antigenic and molecular properties of Murayama virus isolated from cynomolgus monkeys: the virus is closely related to avian paramyxovirus type 2. Virology 194:828-832. [DOI] [PubMed] [Google Scholar]
- 30.Lamb, R., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In B. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 31.Lee, K. E., T. Umapathi, C. B. Tan, H. T. Tjia, T. S. Chua, H. M. Oh, K. M. Fock, A. Kurup, A. Das, A. K. Tan, and W. L. Lee. 1999. The neurological manifestations of Nipah virus encephalitis, a novel paramyxovirus. Ann. Neurol. 46:428-432. [PubMed] [Google Scholar]
- 32.Li, Z., M. Yu, H. Zhang, D. E. Magoffin, P. J. Jack, A. Hyatt, H. Y. Wang, and L. F. Wang. 2006. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 346:219-228. [DOI] [PubMed] [Google Scholar]
- 33.Lipkind, M., Y. Weisman, E. Shihmanter, and D. Shoham. 1982. Isolation of yucaipa-like avian paramyxovirus from a wild mallard duck (Anas platyrhinchos) wintering in Israel. Vet Rec. 110:15-16. [DOI] [PubMed] [Google Scholar]
- 34.Mao, X., S. Kusagawa, M. Tsurudome, H. Komada, M. Kawano, M. Nishio, and Y. Ito. 1996. Characterization of Bangor virus proteins by using monoclonal antibodies. Avian Dis. 40:150-157. [PubMed] [Google Scholar]
- 35.Martin, D. P. 2009. Recombination detection and analysis using RDP3, p. 185-205. In J. M. Walker (ed.), Methods in molecular biology: bioinformatics of DNA sequence analysis, vol. 537. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 36.Miller, P. J., D. B. Boyle, B. T. Eaton, and L. F. Wang. 2003. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology 317:330-344. [DOI] [PubMed] [Google Scholar]
- 37.Miller, P. J., L. M. Kim, H. S. Ip, and C. L. Afonso. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391:64-72. [DOI] [PubMed] [Google Scholar]
- 38.Morgan, I. R., and H. A. Westbury. 1981. Virological studies of Adelie penguins (Pygoscelis adeliae) in Antarctica. Avian Dis. 25:1019-1026. [PubMed] [Google Scholar]
- 39.Morgan, I. R., H. A. Westbury, and J. Campbell. 1985. Viral infections of little blue penguins (Eudyptula minor) along the southern coast of Australia. J. Wildl. Dis. 21:193-198. [DOI] [PubMed] [Google Scholar]
- 40.Morgan, I. R., H. A. Westbury, I. W. Caple, and J. Campbell. 1981. A survey of virus infection in sub-antarctic penguins on Macquarie Island, Southern Ocean. Aust. Vet. J. 57:333-335. [DOI] [PubMed] [Google Scholar]
- 41.Nerome, K., M. Nakayama, M. Ishida, and H. Fukumi. 1978. Isolation of a new avian paramyxovirus from budgerigar (Melopsittacus undulatus). J. Gen. Virol. 38:293-301. [DOI] [PubMed] [Google Scholar]
- 42.Osterhaus, A. D., R. L. de Swart, H. W. Vos, P. S. Ross, M. J. Kenter, and T. Barrett. 1995. Morbillivirus infections of aquatic mammals: newly identified members of the genus. Vet. Microbiol. 44:219-227. [DOI] [PubMed] [Google Scholar]
- 43.Perozo, F., R. Merino, C. L. Afonso, P. Villegas, and N. Calderon. 2008. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 52:472-479. [DOI] [PubMed] [Google Scholar]
- 44.Philbey, A. W., P. D. Kirkland, A. D. Ross, R. J. Davis, A. B. Gleeson, R. J. Love, P. W. Daniels, A. R. Gould, and A. D. Hyatt. 1998. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 4:269-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierson, G. P., and C. J. Pfow. 1975. Newcastle disease surveillance in the United States. J. Am. Vet. Med. Assoc. 167:801-803. [PubMed] [Google Scholar]
- 46.Renshaw, R. W., A. L. Glaser, H. Van Campen, F. Weiland, and E. J. Dubovi. 2000. Identification and phylogenetic comparison of Salem virus, a novel paramyxovirus of horses. Virology 270:417-429. [DOI] [PubMed] [Google Scholar]
- 47.Samuel, A. S., S. Kumar, S. Madhuri, P. L. Collins, and S. K. Samal. 2009. Complete sequence of the genome of avian paramyxovirus type 9 and comparison with other paramyxoviruses. Virus Res. 142:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuel, A. S., A. Paldurai, S. Kumar, P. L. Collins, and S. K. Samal. Complete genome sequence of avian paramyxovirus (APMV) serotype 5 completes the analysis of nine APMV serotypes and reveals the longest APMV genome. PLoS One 5:e9269. [DOI] [PMC free article] [PubMed]
- 49.Shi, L. Y., M. Li, L. J. Yuan, Q. Wang, and X. M. Li. 2008. A new paramyxovirus, Tianjin strain, isolated from common cotton-eared marmoset: genome characterization and structural protein sequence analysis. Arch. Virol. 153:1715-1723. [DOI] [PubMed] [Google Scholar]
- 50.Shortridge, K. F., and D. J. Alexander. 1978. Incidence and preliminary characterisation of a hitherto unreported, serologically distinct, avian paramyxovirus isolated in Hong Kong. Res. Vet. Sci. 25:128-130. [PubMed] [Google Scholar]
- 51.Shortridge, K. F., D. J. Alexander, and M. S. Collins. 1980. Isolation and properties of viruses from poultry in Hong Kong which represent a new (sixth) distinct group of avian paramyxoviruses. Res. Vet. Sci. 49:255-262. [DOI] [PubMed] [Google Scholar]
- 52.Smit, T., and P. R. Rondhuis. 1976. Studies on a virus isolated from the brain of a parakeet (Neophema sp). Avian Pathol. 5:21-30. [DOI] [PubMed] [Google Scholar]
- 53.Smith, K. M., W. B. Karesh, P. Majluf, R. Paredes, C. Zavalaga, A. H. Reul, M. Stetter, W. E. Braselton, H. Puche, and R. A. Cook. 2008. Health evaluation of free-ranging Humboldt penguins (Spheniscus humboldti) in Peru. Avian Dis. 52:130-135. [DOI] [PubMed] [Google Scholar]
- 54.Snoeck, C. J., M. F. Ducatez, A. A. Owoade, O. O. Faleke, B. R. Alkali, M. C. Tahita, Z. Tarnagda, J. B. Ouedraogo, I. Maikano, P. O. Mbah, J. R. Kremer, and C. P. Muller. 2009. Newcastle disease virus in West Africa: new virulent strains identified in non-commercial farms. Arch. Virol. 154:47-54. [DOI] [PubMed] [Google Scholar]
- 55.Spackman, E., D. A. Senne, T. J. Myers, L. L. Bulaga, L. P. Garber, M. L. Perdue, K. Lohman, L. T. Daum, and D. L. Suarez. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tidona, C. A., H. W. Kurz, H. R. Gelderblom, and G. Darai. 1999. Isolation and molecular characterization of a novel cytopathogenic paramyxovirus from tree shrews. Virology 258:425-434. [DOI] [PubMed] [Google Scholar]
- 59.Tikasingh, E. S., A. H. Jonkers, L. Spence, and T. H. Aitken. 1966. Nariva virus, a hitherto undescribed agent isolated from the Trinidadian rat, Zygodontomys b. brevicauda (J. A. Allen & Chapman). Am. J. Trop. Med. Hyg. 15:235-238. [DOI] [PubMed] [Google Scholar]
- 60.Tumova, B., J. H. Robinson, and B. C. Easterday. 1979. A hitherto unreported paramyxovirus of turkeys. Res. Vet. Sci. 27:135-140. [PubMed] [Google Scholar]
- 61.Tumova, B., A. Stumpa, V. Janout, M. Uvizl, and J. Chmela. 1979. A further member of the Yucaipa group isolated from the common wren (Troglodytes troglodytes). Acta Virol. 23:504-507. [PubMed] [Google Scholar]
- 62.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, L. F., W. P. Michalski, M. Yu, L. I. Pritchard, G. Crameri, B. Shiell, and B. T. Eaton. 1998. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J. Virol. 72:1482-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wild, T. F. 2009. Henipaviruses: a new family of emerging paramyxoviruses. Pathol. Biol. (Paris) 57:188-196. [DOI] [PubMed] [Google Scholar]
- 65.Wise, M. G., D. L. Suarez, B. S. Seal, J. C. Pedersen, D. A. Senne, D. J. King, D. R. Kapczynski, and E. Spackman. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39:306-314. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, G. Z., J. X. Zhao, H. W. Wang, A. M. Yang, C. Y. Bu, and M. Wang. 2006. Isolation, identification, and comparison of four isolates of avian paramyxovirus serotype 2 in China. Avian Dis. 50:386-390. [DOI] [PubMed] [Google Scholar]
- 68.Zwiebel, J. A. 2001. Cancer gene and oncolytic virus therapy. Semin. Oncol. 28:336-343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.