Abstract
Bluetongue (BT), caused by Bluetongue virus (BTV), is an economically important disease affecting sheep, deer, cattle, and goats. Since 1998, a series of BT outbreaks have spread across much of southern and central Europe. To study why the epidemiology of the virus happens to change, it is important to fully know the mechanisms resulting in its genetic diversity. Gene mutation and segment reassortment have been considered as the key forces driving the evolution of BTV. However, it is still unknown whether intragenic recombination can occur and contribute to the process in the virus. We present here several BTV groups containing mosaic genes to reveal that intragenic recombination can take place between the virus strains and play a potential role in bringing novel BTV lineages.
Bluetongue (BT) is an economically significant disease that seriously threatens sheep, some species of deer, and to a lesser extent cattle and goats. As a vector-borne viral disease of ruminants, BT is endemic in tropical and subtropical countries (46). However, a series of BT outbreaks have spread across much of southern and central Europe since 1998 (29). Thus, it is of great importance to fully understand the molecular basis driving the change of its epidemiology so as to prevent or limit future BT pandemics.
Bluetongue virus (BTV), the pathogen of BT, belongs to the Orbivirus genus of the Reoviridae family (46). The virus has a segmented double-stranded RNA (dsRNA) genome that is packaged in a nonenveloped, icosahedral particle (46). Its 10 dsRNA segments encode 11 proteins, VP1 to VP7 (encoded by segments 1, 2, 3, 4, 6, 9, and 7, respectively), NS1 to SN3 (encoded by segments 5, 8, and 10, respectively), and NS3A (encoded by segment 10) (46). Two structural proteins, VP2 and VP5, form the outer layer of the virion particle and are responsible for cell attachment and virus entry (18, 31, 32), neutralizing epitope (14, 21), and virus virulence (36). Both of them are highly variable and generate 24 serotypes of the virus (44). The inner layers contain VP1, VP3, VP4, VP6, and VP7, and form the “core” of the BTV capsid. VP1 and VP6 are involved in RNA replication as the RNA-dependent RNA polymerase (54) and helicase/NTPase, respectively (49). VP7 forms the surface of the core and functions during the entry of the core into insect cells (44) and also can react with “core neutralizing” antibodies as a major serogroup-specific antigen (32, 44). These core proteins and two nonstructural proteins, NS1 and NS2, are thought to be relatively conservative, so that antigenic cross-reaction can take place between different BTV strains and serotypes, whereas NS3/N3a is more variable than the other nonstructural or core proteins (46).
The genetic diversity and variation in sequences of different BTV genome segments were initially identified by RNA oligonucleotide fingerprint analysis of BTV field samples (47). Until now, reassortment and dynamic gene mutation, regarded as the key factors responsible for the genetic diversity of BTV, have been studied in details (46). The two mechanisms can result in both genetic drift and genetic shift and contribute to BTV evolution (47). It has been revealed that high-frequency genome segment reassortment occurs readily between different BTV serotypes (16). Thus, segment reassortment is an important factor in generation of genetic diversity in orbivirus populations in nature (45). In addition, it has been shown that homologous recombination can also play a role in the genetic diversity and evolution of some RNA viruses (24, 33) and bring on virulent variants of these viruses at last (8, 56). Although homologous recombination has been observed in rotavirus, a member of the Reoviridae (39, 40), it is still unknown whether the intragenic recombination can occur and play a role in the generation of genetic diversity in orbivirus populations.
To determine whether homologous recombination shaped the evolution of BTV and to provide some insights into the recombination itself in the virus, we analyzed roughly 690 complete segments of BTV deposited in GenBank to see whether some of them underwent intragenic recombination event. Several BTV groups isolated at different time points and in different countries were found containing the same (or similar) mosaic segments, demonstrating that intragenic recombination had occurred in the field and that these viruses with mosaic segments had become prevailing strains. That is, intragenic recombination can play a potential role in generating genetic diversity of BTV and exert its influence on the change of BTV epidemiology.
MATERIALS AND METHODS
All 692 complete segments (Seg)—15 Seg-1, 160 Seg-2, 58 Seg-3, 17 Seg-4, 48 Seg-5, 123 Seg-6, 88 Seg-7, 33 Seg-8, 47 Seg-9, and 103 Seg-10—were respectively retrieved from GenBank. These sequences were aligned with CLUSTAL W (52). Phylogenetic neighbor-joining (NJ) trees were built by MEGA4 (51), using the maximum-composite-likelihood model. Maximum-likelihood (ML) trees were constructed by using Phyml with HKY85 nucleotide substitution model and estimated Ts/Tv (17) and are displayed as graphics by using MEGA4 to determine the topology of each NJ tree. Bootstrapping was used to assess the robustness of a tree with 1,000 replicates. Branches supported by an >70% bootstrap value are shown in each tree. The Shimodaira-Hasegawa test was used to prove whether phylogenetic trees estimated from different regions were significantly different (http://aix1.uottawa.ca/∼sarisbro/).
The aligned sequences were analyzed with the help of the RDP software package (v3.34) to seek potential recombination events (30). SimPlot software (28) was used to determine each recombinant as described in a previous study (19). Similarity plot analysis, bootscanning plot analysis, and the Findsite subprogram were performed to identify the potential recombination events. For each mosaic, the window sizes were differently set to avoid the overlook of small recombinant region due to large step size and the confused recombination information. Recombination breakpoints were analyzed by maximization of χ2 (3), using SimPlot combined with genetic algorithms using GARD (http://www.datamonkey.org/GARD/) (25). Using SplitTrees (v4.9) (22), we analyzed informative sites by phi test to differentiate recombination from convergent evolution through mutation. The structure of RNA sequence around breakpoint was analyzed using the RNAstructure 5.03 program (42).
RESULTS
Using different segment alignments, we performed analysis of available BTV gene sequences to find evidence of recombination between BTVs. Mosaics were found in almost all BTV genomic segments except for segments 5 and 6, including three vp1 (3/15), three vp2 (3/160), one vp3 (1/58), one vp4 (1/17), two vp7 (2/88), one ns1 (1/48), two ns2 (2/33), and nine ns3 (9/103) mosaic genes (Table 1) . Furthermore, the data also indicated that some mosaic RNAs represented single-crossover events (Fig. 1), while others represented double-crossover events (Fig. 2 and 3) or multirecombination events (Fig. 4D).
TABLE 1.
BTV strains with evidence for potential recombination
| GenBank no. | Mosaic virus strain | Source or reference | Gene | Putative parent lineage(s)a | Breakpoint position(s) | Country of origin |
|---|---|---|---|---|---|---|
| AM498051 | BTV8/NT | 29 | vp1 | BTV17/USA/L20447; BTV2/USA/L20508 | 1623 | Netherlands |
| FJ183374 | BTV8/NT | 40a | vp1 | BTV17/USA/L20447; BTV2/USA/L20508 | 1623 | Netherlands |
| AY154458 | BTV2/Frances (Corsican) | Unpublished | vp1 | BTV17/USA/L20447; BTV2/USA/L20508 | 1623 | France |
| AF481096 | BTV2/SA/vaccine | Unpublished | vp2 | BTV2/France/AF356601; BTV2/Spain/AM773697 | 165 | South Africa |
| AY839948 | BTV4/vaccine | 10a | vp2 | BTV4/Greece/AY839946; BTV4/Italy/DQ191276 | 2503 | South Africa |
| AY839945 | BTV4/France | 10a | vp2 | BTV4/Greece/DQ191277; BTV4/Italy/DQ191276 | 2540 | France |
| L19967 | BTV2/USA | 22b | vp3 | BTV2/SA/DQ186793; BTV17/USA/L19968 | 743, 906, 1203 | United States |
| L08637 | BTV2/USA/2M1 | 20a | vp4 | BTV11/USA/L08638; BTV8/NT/AM498054 | 444, 622 | United States |
| AY606206 | BTV18/India/Bangalore | 26 | vp7 | BTV23/India/AY643511; BTV1/Aus/M63417 | 460, 623 | India |
| AM261976 | India/Avikanagar | Unpublished | vp7 | BTV23/India/AY643511; BTV1/Aus/M63417 | 460, 623 | India |
| M97680 | BTV2/USA | 22a | ns1 | BTV2/Italy/AM773690; BTV13/USA/M97762 | 179 | United States |
| L08675 | BTV10/USA | 56a | ns2 | BTV11/USA/L08674; BTV2/USA/AY855287 | 485 | United States |
| AM900372 | BTV4/Greece/00 | 29 | ns2 | BTV4/Turkey/DQ825673; BTV4/Greece/AY857500 | 566 | Greece |
| AY775161 | BTV9/Italy/AY775161 | 32a | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Italy |
| EF554845 | BTV9/Turkey/EF554845 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554844 | BTV9/Turkey/EF554844 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554849 | BTV9/Turkey/EF554849 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554847 | BTV9/Turkey/EF554847 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554846 | BTV9/Turkey/EF554846 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554839 | BTV9/Turkey/EF554839 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554853 | BTV9/Turkey/EF554853 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
| EF554851 | BTV9/Turkey/EF554851 | 37 | ns3 | BTV9/Turkey/EF554850; BTV21/Aus/AF529054 | 158, 259 | Turkey |
Abbreviations: Aus, Australia; NT, Netherlands; SA, South Africa.
FIG. 1.
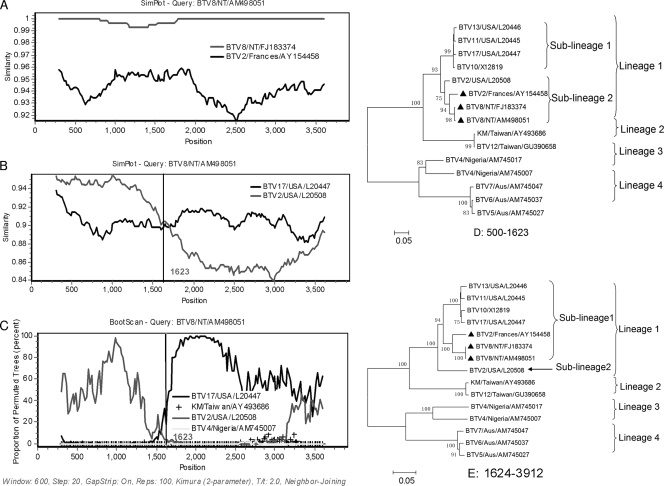
Evidence for recombination in segment 1 of three BTVs isolated from the Netherlands and France. (A) Comparison of segment 1 from three BTV isolates: BTV8/NT/AM498051, BTV8/NT/FJ183374, and BTV2/France/AY154468. BTV8/NT/AM498051 was used as the query strain. The y axis gives the percentage of identity within a sliding window 600 bp wide centered on the position plotted, with a step size between plots of 20 bp. (B) Comparison of segment 1 from BTV8/NT/AM498051 and its putative parents. (C) Bootscanning of the mosaic BTV8/NT/AM498051 and its parent sequences. The y axis gives the percentage of permutated trees using a sliding window. Two BTV isolates BTV4/Nigeria/AM745007 and KM/Taiwan/AY493686 are used as an outgroup. The vertical line shows the recombination breakpoint. The rest is the same as panels A and B. (D and E) Phylogenic trees of the segment in different regions from positions 500 to 1623 and positions 1623 to 3912, respectively. Mosaic isolates are marked with a “▴”.
FIG. 2.
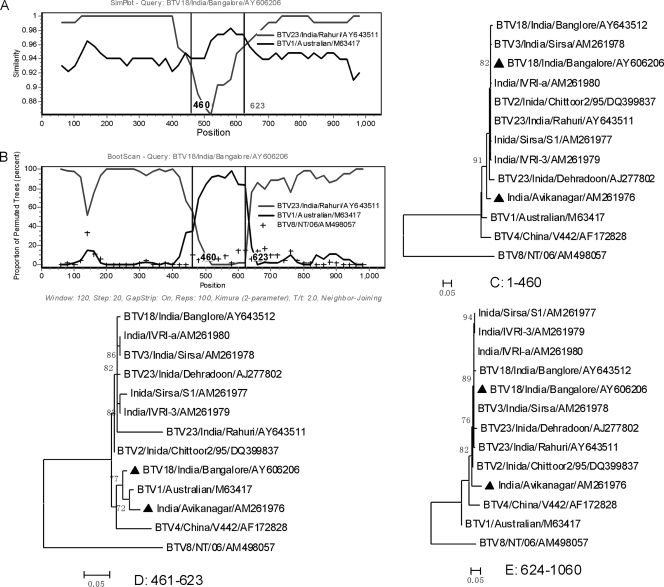
Evidence for recombination in segment 7 of two BTV strains isolated from India: BTV18/India/Bangalore/AY606206 and India/Avikanagar/AM261976. (A) Comparison of segment 7 of BTV18/India/Bangalore/AY606206 and its putative parent lineages. BTV18/India/Bangalore/AY606206 was used as the query. The y axis gives the percentage of identity within a sliding window 120 bp wide centered on the position plotted, with a step size between plots of 20 bp. (B) Bootscanning of BTV18/India/Bangalore/AY606206 mosaic gene and its parent sequences. The y axis gives the percentage of permutated trees using a sliding window. Two BTV isolates BTV8/NT/06/AM498057 and BTV4/China/V442/AF172828 are used as outgroups. The width of the sliding window is 120 bp, with a step size of 20 bp. The vertical line shows the recombination breakpoint. The rest is the same as panel A. (C to E) Phylogenic trees of the segment in different regions from positions 1 to 460, positions 461 to 623, and positions 624 to 1060, respectively. Mosaic isolates are marked with a “▴”.
FIG. 3.
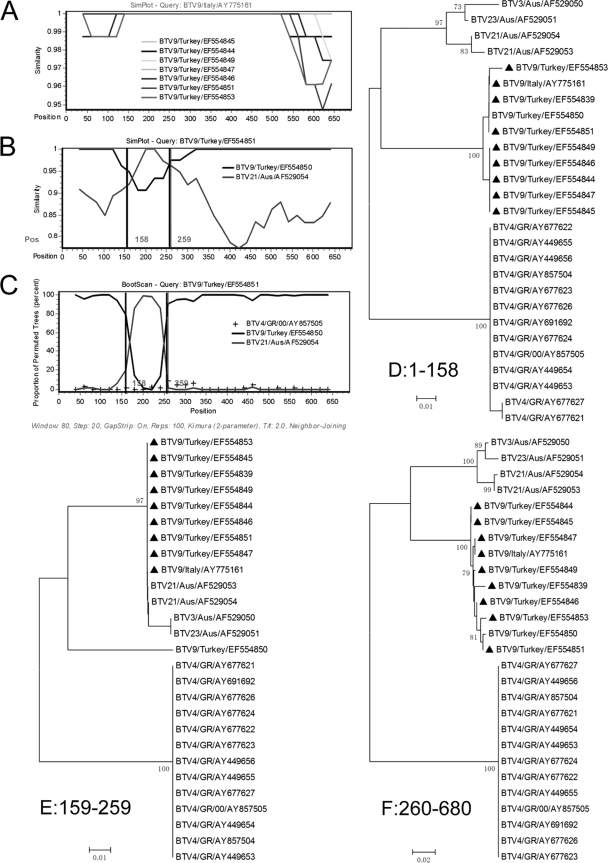
Evidence for recombination in segment 10 of a BTV group from Turkey and Italy. (A) Comparison of segment 10 from BTV isolates: BTV9/Turkey/EF554845, BTV9/Turkey/EF554844, BTV9/Italy/AY775161, BTV9/Turkey/EF554849, BTV9/Turkey/EF554847, BTV9/Turkey/EF554846, BTV9/Turkey/EF554851, and BTV9/Turkey/EF554853. BTV9/Italy/AY775161 was used as the query. The y axis gives the percentage of identity within a sliding window 80 bp wide centered on the position plotted, with a step size between plots of 20 bp. (B) Comparison of segment 1 from BTV9/Italy/AY775161 and its putative parents. (C) Bootscanning of the mosaic BTV9/Italy/AY775161 and its parent sequences. The y axis gives the percentage of permutated trees using a sliding window. A BTV isolate BTV21/Aus/AF529054 was used as an outgroup. The vertical line shows the recombination breakpoint. The rest is the same as panels A and). (D to F) Phylogenic trees of the segment in different regions from positions 1 to 158, positions 159 to 259, and positions 260 to 690, respectively. Mosaic isolates are marked with a “▴”. Greece and Australia are abbreviated as GR and Aus, respectively.
FIG. 4.
Bootscanning analysis of several single mosaics, their parent sequences, and different outgroups. The y axis gives the percentage of permutated trees using a sliding window. The vertical line shows the recombination breakpoints. (A) Segment 2 of BTV2/SA/vaccine/AF481096; (B) segment 2 of BTV4/vaccine/AY839948; (C) segment 2 of BTV4/France/03AY839945; (D) segment 3 of BTV2/USA/L19967; (E) segment 4 of BTV2/USA/2M1-L08637; (F) segment 5 of BTV2/USA/M97680; (G) segment 8 of BTV10/USA/L08675; (H) segment 8 of BTV4/Greece/00/AM900372.
In particular, nine BTV strains isolated from Turkey and Italy were observed to contain the same mosaic ns3/ns3a gene, and three strains isolated from the Netherlands and France were also found to have the similar mosaic vp1 gene. These findings suggested that homologous recombination could drive BTV evolution as a molecular mechanism of genetic diversity and might result in the potential change of its biologic character.
One segment 1 mosaic group was found.
As the largest protein in BTV, it has been known that VP1 is encoded by segment 1 and plays a role as RADP (54). According to the available vp1 complete sequences, three vp1 mosaic isolates constituted a separate sublineage in lineage I of four BTV lineages (data not shown). One mosaic was isolated from Corsican of France in 2000 and 2001 (10). The other two were isolated from the Netherlands in 2006 (29) and had nearly 100% sequence identity (Fig. 1A). The three mosaics shared similar recombination events (Fig. 1). According to the time frame of virus isolation, the segment 1 of the two Netherlands mosaics might be descended from the France mosaic sublineage, which is supported by the phylogenetic relationship of the three mosaics (Fig. 1D and E).
According to the results from RDP, two U.S. strains, BTV2/USA/L20508 and BTV17/USA/L20447, might act as the putative parent lineages of the mosaics. Therefore, they were used to compare with the Netherlands mosaic BTV8/NT/AM498051 (BTV-8NT) for recombination analysis. By using the Findsites subprogram of Simplot, we located a potential breakpoint in a parsimonious region with the maximization of χ2, from positions 1560 to 1657 (χmax2 = 20.1). Using GARD, a breakpoint (position 1623) was also found falling into this region. Combining the results from the two methods, we located the most probable breakpoint at position 1623. A similarity plot (Fig. 1B), constructed by using all sites, revealed that the Seg-1 sequence of BTV-8NT exhibited greater affinity with the lineage of BTV2/USA/L20508 before the breakpoint. On the contrary, the mosaic shared higher sequence similarity with BTV17/USA/L20477 after the breakpoint. Similar results were observed in bootscanning analysis (Fig. 1C). The recombination event was also supported by statistical evaluations from the RDP software (SIScan gave a Z-score of 5.69 with P = 2.16E-12, and 3SEQ gave a P value of 4.002E-9). The phi test gave significantly statistical evidence of recombination as well (P < 0.05). The most compelling phylogenetic evidence for recombination was the occurrence of incongruent phylogenetic trees (34). When phylogenetic trees inferred from nucleotide sequences of different regions were constructed (Fig. 1D and E), all of the three mosaics were clustered into the sublineage of BTV2/USA/L20508 before the breakpoint (with a 75% bootstrap value) but fell into the same sublineage with BTV17/USA/L20477 after the breakpoint (with 95% bootstrap value) (Fig. 1D and E). Moreover, a statistically significant discrepancy between the topologies of the two phylogenetic trees (Fig. 1D and E) also provided a powerful evidence for recombination (Shimodaira-Hasegawa test, P < 0.05).
Similar results can be found when vp1 of BTV-2/Corsican/AY154458 and BTV-8/FJ183374 were considered as queries in recombination analysis (data not shown).
Two similar segment 7 intragenic recombinants were identified.
VP7 is encoded by Seg-7 and plays some key roles in virus replication, such as forming the surface of the core and being responsible for core entry into insect cells (46). As the outer core protein, VP7 is the major serogroup-specific antigen and shows variations that are considered to be associated with the insect populations that act as vectors for different virus strains in different geographic areas (9, 55). All available 88 VP7 sequences constituted five lineages (data not shown). Moreover, two India isolates (BTV18/India/Bangalore/AY606206 and India/Avikanagar/AM261976) of lineage III were isolated in Bangalore and Avikanagar, respectively (26). They had similar mosaic vp7 genes.
When vp7 of BTV18/India/Bangalore/AY606206 was used as a query in bootscanning analysis, two breakpoints were located at positions 460 and 623 (Fig. 2 A and B). A similarity plot revealed that its sequence exhibited greater affinity with the BTV1/Australian/M63417 sublineage in the region between the two breakpoints (Fig. 2A). In contrast, it shared higher sequence similarity with BTV23/India/Rahuri/AY643511 before position 460 and after position 623. In the three phylogenetic trees inferred from different regions, the two mosaics were clustered into different parent lineages with high bootstrap values (>70%) (Fig. 2C to E). When vp7 sequences of the mosaic, its putative parents, and an outgroup BTV8/NT/06/AM498057 were submitted to conduct the phi test, statistically significant evidence for recombination was found (P < 0.001). Running 3SEQ and SIScan subprogram in the RDP software, the P value of delta distributions and Z-score was 4.5E-9 (P ≪ 0.001) and 6.36 (with P = 6.36E-12), respectively, which likewise strongly supported the recombination event. In addition, India/Avikanagar/AM261976 was also identified as a recombinant sharing similar mosaic vp7 gene with BTV18/India/Bangalore/AY606206 (Fig. 2C to E).
BTVs containing mosaic segment 10 could become prevailing strains.
To clarify whether intragenic recombination could exert its influence on the BTV evolutionary process, it would be important to find a group sharing similar recombination events. A good case in point was a BTV lineage isolated from 1999 to 2001 in Turkey and Italy which had the same mosaic Seg-10. According to Seg-10, BTV could be divided into four groups: Western 1 and 2 and Eastern 1 and 2 (29). Two subgroups of Eastern 1 were found in the global BTV phylogenetic tree (data not shown). BTVs of subgroup 1 were mainly isolated from Australia, and BTVs of subgroup 2 were mainly isolated from several Asian and European countries. It was the nine BTV9 isolates (eight from Turkey and one from Italy) of subgroup 1 that had the same mosaic segment 10.
We compared the potential mosaics and found that they shared more than 99% sequence similarity (Fig. 3A). For recombination analysis, BTV9/Turkey/EF554851 was used as a representative strain. Two isolates, BTV9/Turkey/EF554850 and BTV21/Aus/AF529054, could be supposed as putative parents of the mosaics since they shared more than 99.8% identity with the mosaic in different regions delimited by the breakpoints (Fig. 3B). Combined with the results of the maximization of χ2 and GARD, two breakpoints were located at the positions 158 and 259 with maximum χ2 (5.8 and 14.3, respectively). We compared Seg-10 of BTV9/Turkey/EF554851 with its potential parent lineages (Fig. 3B). Between the two breakpoints, BTV9/Turkey/EF554851 shared 100% homology with the putative parent strain BTV21/Australasia/AF529054 (versus 92.8%, 93/101 with BTV9/Turkey/EF554850). However, before and after the two breakpoints, the mosaic only shared 90.5 and 87.3% sequence similarity, respectively, with BTV21/Australasia/AF529054 (versus 100 and 99.8%, respectively, with BTV9/Turkey/EF554850). The bootscanning result also supported the hypothesis that the mosaic was descended from lineages BTV21/Australasia/AF529054 and BTV9/Turkey/EF554850 lineages (Fig. 3C). Phylogenetics analysis showed that the mosaic fell into the Australia group between the two breakpoints but was clustered into the BTV9/Turkey/EF554850 lineage before and after the breakpoints (Fig. 3D to F). Moreover, the statistical tests did find significant evidence for recombination (P < 0.05 [phi test], P < 0.0005 [Siscan], and P < 0.0001 [3SEQ]).
As well, other eight Seg-10 mosaics had the same recombination events. These mosaics were isolated from different countries but shared the same recombination events, suggesting that they might be descended from the same mosaic ancestor, which could become prevailing strain with good adaptability.
DISCUSSION
In the study, 22 mosaic genes composed up to 3.2% (22/692) of the analyzed sequences. Given that similar mosaics might be descended from the same mosaic ancestor, there are only 11 unique recombinants among these mosaics; that is, only ca. 1.6% (11/681) of the BTV genes have undergone recombination events. Interestingly, it seems that vp1 has a higher recombination rate (20%, 3/15). However, considering that the three vp1 mosaics are inherited from the same mosaic ancestor, the recombination rate should be corrected to 8.3% (1/12). Moreover, the recombination rate of vp1 might also be overestimated because there are only small sample sequences available in the public database. Therefore, intragenic recombination might not be a usual event but may have selective advantage, leading to gene persistence in the population.
In our analysis, three vp1 mosaics—BTV2/France/AY154458, BTV8/NT/FJ183374, and BTV8/NT/AM498051—were found to share the similar recombination event (Fig. 1). However, they were isolated from different countries by different research groups. Similar observations were also obtained with vp7 in two India isolates (Fig. 2) and with ns3/ns3A from a mosaic group from Turkey and Italy (Fig. 3). The fact that different isolates, originating from the same mosaic ancestor, could be found in different locations and at different time points provides robust evidence that intragenic recombination lineages results in novel BTV strains. Moreover, these mosaic viruses can circulate in the field. Therefore, intragenic recombination does act as an evolutionary force and plays an important role in shaping the genetic diversity of BTV.
In addition, we also found some single mosaic segments (listed in Table 1 and Fig. 4). For these mosaics, we also considered the possibility that the detected recombinants had resulted from laboratory artifacts. With the exception of two mosaics, BTV4/Greece/00/AM900372 and BTV4/vaccine/AY839948, the presence of unique nucleotide substitutions in the recombinant regions compared to the comparable region of the predicted parental strains suggested that these regions did not arise due to contamination with BTV strains deposited in the sequence database. For the mosaics BTV4/Greece/00/AM900372 and BTV4/vaccine/AY839948, resequencing might still be necessary to objectively determine the two recombination events, although the mosaics and their putative parents were isolated by different laboratories in different countries.
In an insect vector, BTV coinfection has been considered to be more universal, since greater frequency reassortment often takes place (48), which also provides the basis for intragenic recombination. Hence, it is not surprising that intragenic recombination can occur between BTVs in insect host. Similar to reassortment, recombination processes will allow some viruses to acquire many of the key adaptive mutations in a single step and thus make a major leap in fitness, which might result in a change in host tropism (27, 50). Therefore, it should be necessary to pay attention to the potential influence of intragenic recombination on its biology and epidemiology. Among the viral proteins, NS3 is proposed to be associated with the transmission of the virus in different insect populations and species (4, 5, 35), and VP7 is also thought to be related to the infection of insect populations (9, 55). In the present study, stable and genetic vp7 and ns3 mosaics were found, suggesting that insects might be more permissive for the events leading to intragenic recombination as well, making insects an important diversity-generating host.
Interestingly, there was no amino acid difference between the putative parents of the Seg-10 mosaics in the exchanged region, although recombination was found to bring about amino acid changes in other mosaic segments (e.g., 2 in the vp7 mosaics and 13 in the vp1 mosaics). Also, the Seg-10 mosaics constituted the biggest mosaic group in the study. An explanation might be that intragenic recombination can occur readily between some BTV strains because it has little or no effect on the protein encoded by the viral gene. Thus, there would be no selective pressure against recombination which results in a stable mosaic gene. On the other hand, the rapid change of functional proteins, such as VP1, due to recombination might play a potential role in the rapid evolution of BTV.
Homologous recombination is important process driving genotype diversity for some RNA viruses (11, 20, 38). Recombination, especially between different serotypes, may speed up the evolution of BTV and result in a rapid change in its epidemiology. In the two quickly evolving genes vp2 and vp5, only intragenic vp2 recombinants are found within the same serotype. It is possible that selection disfavors interserotype recombinants, partly due to incompatibilities between interacting viral proteins or between viral proteins and their cis-acting sequences. After all, they have the highest variability among all genes of the virus and show great differences between serotypes. For relatively conservative genes, such as vp7 and ns3, intragenic recombination can occur between different serotypes (Fig. 2 and 3).
Recently, attenuated vaccine strains of BTV developed in South Africa (Onderstepoort Biological Products, Onderstepoort, South Africa) have been used in animal populations. In the early stages of the recent outbreaks in the eastern Mediterranean region, these live attenuated vaccine strains of BTV-2, BTV-4, and BTV-9 (western topotype) and BTV-16 (eastern topotype) were used in the Iberian peninsula, eastern Mediterranean islands, and Italy to reduce the circulation of BTV (7). There is clear evidence showing that the vaccine strains of BTV-2 and BTV-16 have been transmitted in the field (1, 6, 10). The release of BTV vaccine strains into the field has added further genetic diversity to the pool of field strains circulating in the field, generating an unprecedented mix of viruses that includes several eastern and western viruses, as well as both field and vaccine strains (7). Therefore, BTV vaccine strains cannot avoid exchanging their genetic materials with the field strains through reassortment or intragenic recombination. In fact, it has been found that a European field BTV strain derived from two parental vaccine strains by genome segment reassortment is circulating in the field (7). In the present study, two BTV2 vaccine strains were also found to bear mosaic VP2 partly descended (about 170 and 360 bp, respectively) from European field BTV lineages (Fig. 4A and B). This highlighted existing concerns over the use and transmission of BTV vaccine strains in the field. It also suggested that a live avirulent vaccine could play a role in speeding up the evolution of BTV via intragenic recombination. Indeed, polio epidemics in Hispaniola that arose from similar recombinants involving vaccine virus and other endogenous enteroviruses have been threatening the entire World Health Organization polio eradication program (23). Therefore, it might be necessary to evaluate the effect of recombination associated with attenuated vaccine on BTV epidemiology. From an environmental perspective, inactivated BTV vaccine should be safer.
Accumulated data have shown that homologous recombination can occur between double- or negative-stranded RNA viruses that undergo “caged replication” (12, 13, 15, 19, 41, 57). However, the mechanism resulting in homologous recombination in these viruses is still unknown. The recombination rate of BTV is found to be much lower than that of the positive-sense RNA viruses, such as the foot-and-mouth disease virus, in which 10 to 20% of the viral genomes undergo recombination during a single replication cycle (2). In poliovirus, recombination occurred in RNA regions where RNA could potentially form a secondary structure and play a role in recombination as an activator (33). This allows two parental RNAs of different origins to form a complex and thereby forces recombination to occur (43, 53). Therefore, analyzing the characteristics of breakpoints could also provide clues to understand the recombination mechanism in BTVs. When the sequences around the breakpoints of recombinants were analyzed, potential secondary structures were also found in all mosaics (data not shown), which could offer a molecular basis for recombination to occur.
In conclusion, we provide here evidence demonstrating that recombination can occur in BTVs and plays a potential role in the evolution and genetic diversity of the virus. Studies will need to evaluate its influence on virulence and host tropism and to study the mechanisms underlying BTV recombination.
Acknowledgments
We thank the sequence submitters for their original work and the anonymous reviewers for helpful comments.
This study was supported by a Postdoctoral Innovative Project Foundation of Shandong Province in China (200702013), a Project of Shandong Province Higher Educational Science and Technology Program (J09LCD209-12), and a Shandong Province Young and Middle-Aged Scientists Research Awards Fund (BS2009NY011).
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Aguero, M., M. Arias, L. J. Romero, M. J. Zamora, and J. M. Sanchez-Vizcaino. 2002. Molecular differentiation between NS1 gene of a field strain Bluetongue virus serotype 2 (BTV-2) and NS1 gene of an attenuated BTV-2 vaccine. Vet. Microbiol. 86:337-341. [DOI] [PubMed] [Google Scholar]
- 2.Alejska, M., A. Kurzyniska-Kokorniak, M. Broda, R. Kierzek, and M. Figlerowicz. 2001. How RNA viruses exchange their genetic material. Acta Biochim. Pol. 48:391-407. [PubMed] [Google Scholar]
- 3.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. [DOI] [PubMed] [Google Scholar]
- 4.Balasuriya, U. B., S. A. Nadler, W. C. Wilson, L. I. Pritchard, A. B. Smythe, G. Savini, F. Monaco, P. De Santis, N. Zhang, W. J. Tabachnick, and N. J. Maclachlan. 2008. The NS3 proteins of global strains of bluetongue virus evolve into regional topotypes through negative (purifying) selection. Vet. Microbiol. 126:91-100. [DOI] [PubMed] [Google Scholar]
- 5.Bansal, O. B., A. Stokes, A. Bansal, D. Bishop, and P. Roy. 1998. Membrane organization of bluetongue virus nonstructural glycoprotein NS3. J. Virol. 72:3362-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barros, S. C., F. Ramos, T. M. Luis, A. Vaz, M. Duarte, M. Henriques, B. Cruz, and M. Fevereiro. 2007. Molecular epidemiology of bluetongue virus in Portugal during 2004-2006 outbreak. Vet. Microbiol. 124:25-34. [DOI] [PubMed] [Google Scholar]
- 7.Batten, C. A., S. Maan, A. E. Shaw, N. S. Maan, and P. P. Mertens. 2008. A European field strain of bluetongue virus derived from two parental vaccine strains by genome segment reassortment. Virus Res. 137:56-63. [DOI] [PubMed] [Google Scholar]
- 8.Becher, P., M. Orlich, and H. J. Thiel. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J. Virol. 75:6256-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonneau, K. R., N. Z. Zhang, W. C. Wilson, J. B. Zhu, F. Q. Zhang, Z. H. Li, K. L. Zhang, L. Xiao, W. B. Xiang, and N. J. MacLchlan. 2000. Phylogenetic analysis of the S7 gene does not segregate Chinese strains of bluetongue virus into a single topotype. Arch. Virol. 145:1163-1171. [DOI] [PubMed] [Google Scholar]
- 10.Breard, E., C. Hamblin, S. Hammoumi, C. Sailleau, G. Dauphin, and S. Zientara. 2004. The epidemiology and diagnosis of bluetongue with particular reference to Corsica. Res. Vet. Sci. 77:1-8. [DOI] [PubMed] [Google Scholar]
- 10a.Breard, E., C. Sailleau, K. Nomikou, et al. 2007. Molecular epidemiology of bluetongue virus serotype 4 isolated in the Mediterranean Basin between 1979 and 2004. Virus Res. 125:191-197. [DOI] [PubMed] [Google Scholar]
- 11.Bruen, T. C., and M. Poss. 2007. Recombination in feline immunodeficiency virus genomes from naturally infected cougars. Virology 364:362-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao, D., M. Barro, and Y. Hoshino. 2008. Porcine rotavirus bearing an aberrant gene stemming from an intergenic recombination of the NSP2 and NSP5 genes is defective and interfering. J. Virol. 82:6073-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong, Y. L., A. Padhi, P. J. Hudson, and M. Poss. 2010. The effect of vaccination on the evolution and population dynamics of avian paramyxovirus-1. PLoS Pathog. 6:e1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowley, J. A., and B. M. Gorman. 1989. Cross-neutralization of genetic reassortants of bluetongue virus serotypes 20 and 21. Vet. Microbiol. 19:37-51. [DOI] [PubMed] [Google Scholar]
- 15.Geue, L., S. Schares, C. Schnick, J. Kliemt, A. Beckert, C. Freuling, F. J. Conraths, B. Hoffmann, R. Zanoni, D. Marston, L. McElhinney, N. Johnson, A. R. Fooks, N. Tordo, and T. Muller. 2008. Genetic characterisation of attenuated SAD rabies virus strains used for oral vaccination of wildlife. Vaccine 26:3227-3235. [DOI] [PubMed] [Google Scholar]
- 16.Gorman, B. M. 1990. The bluetongue viruses. Curr. Top. Microbiol. Immunol. 162:1-19. [DOI] [PubMed] [Google Scholar]
- 17.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 18.Hassan, S. H., C. Wirblich, M. Forzan, and P. Roy. 2001. Expression and functional characterization of bluetongue virus VP5 protein: role in cellular permeabilization. J. Virol. 75:8356-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, C. Q., Z. X. Xie, G. Z. Han, J. B. Dong, D. Wang, J. B. Liu, L. Y. Ma, X. F. Tang, X. P. Liu, Y. S. Pang, and G. R. Li. 2009. Homologous recombination as an evolutionary force in the avian influenza A virus. Mol. Biol. Evol. 26:177-187. [DOI] [PubMed] [Google Scholar]
- 20.Herrewegh, A. A., I. Smeenk, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 72:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Huang, I. J., E. Hayama, Y. J. Jeong, and J. K. Li. 1993. Conservation of the segment 4 gene sequence and of a leucine zipper motif in VP4 among five US bluetongue viruses. Virology 195:772-779. [DOI] [PubMed] [Google Scholar]
- 21.Huismans, H., N. T. van der Walt, M. Cloete, and B. J. Erasmus. 1987. Isolation of a capsid protein of bluetongue virus that induces a protective immune response in sheep. Virology 157:172-179. [DOI] [PubMed] [Google Scholar]
- 22.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 22a.Hwang, G. Y., J. F. Chiou, Y. Y. Yang, and J. K. Li. 1993. High-sequence conservation among the United States bluetongue viruses cognate M2 genes which encode the nonstructural NS1 tubule protein. Virology 192:321-327. [DOI] [PubMed] [Google Scholar]
- 22b.Hwang, G. Y., M. Xiang, and J. K. Li. 1994. Analyses and conservation of sequences among the cognate L3 segments of the five United States bluetongue viruses. Virus Res. 32:381-389. [DOI] [PubMed] [Google Scholar]
- 23.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 24.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosakovsky Pond, S. L., D. Posada, M. B. Gravenor, C. H. Woelk, and S. D. Frost. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891-1901. [DOI] [PubMed] [Google Scholar]
- 26.Kovi, R. C., S. Dahiya, G. Prasad, and Minakshi. 2006. Nucleotide sequence analysis of VP7 gene of Indian isolates of bluetongue virus vis-a-vis other serotypes from different parts of the world. DNA Seq. 17:187-198. [DOI] [PubMed] [Google Scholar]
- 27.Kuiken, T., E. C. Holmes, J. McCauley, G. F. Rimmelzwaan, C. S. Williams, and B. T. Grenfell. 2006. Host species barriers to influenza virus infections. Science 312:394-397. [DOI] [PubMed] [Google Scholar]
- 28.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maan, S., N. S. Maan, N. Ross-smith, C. A. Batten, A. E. Shaw, S. J. Anthony, A. R. Samuel, K. E. Darpel, E. Veronesi, C. A. Oura, K. P. Singh, K. Nomikou, A. C. Potgieter, H. Attoui, E. van Rooij, P. van Rijn, K. De Clercq, F. Vandenbussche, S. Zientara, E. Breard, C. Sailleau, M. Beer, B. Hoffman, P. S. Mellor, and P. P. Mertens. 2008. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 377:308-318. [DOI] [PubMed] [Google Scholar]
- 30.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 31.Mertens, P. P., J. N. Burroughs, and J. Anderson. 1987. Purification and properties of virus particles, infectious subviral particles, and cores of bluetongue virus serotypes 1 and 4. Virology 157:375-386. [DOI] [PubMed] [Google Scholar]
- 32.Mertens, P. P., J. N. Burroughs, A. Walton, M. P. Wellby, H. Fu, R. S. O'Hara, S. M. Brookes, and P. S. Mellor. 1996. Enhanced infectivity of modified bluetongue virus particles for two insect cell lines and for two Culicoides vector species. Virology 217:582-593. [DOI] [PubMed] [Google Scholar]
- 32a.Monacom, F., C. Cammà, S. Serini, and G. Savini. 2006. Differentiation between field and vaccine strain of bluetongue virus serotype 16. Vet. Microbiol. 116:45-52. [DOI] [PubMed] [Google Scholar]
- 33.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, M. I., and E. C. Holmes. 2007. The evolution of epidemic influenza. Nat. Rev. Genet. 8:196-205. [DOI] [PubMed] [Google Scholar]
- 35.Nikolakaki, S. V., K. Nomikou, M. Koumbati, O. Mangana, M. Papanastassopoulou, P. P. Mertens, and O. Papadopoulos. 2005. Molecular analysis of the NS3/NS3A gene of Bluetongue virus isolates from the 1979 and 1998-2001 epizootics in Greece and their segregation into two distinct groups. Virus Res. 114:6-14. [DOI] [PubMed] [Google Scholar]
- 36.O'Hara, R. S., A. J. Meyer, J. N. Burroughs, L. Pullen, L. A. Martin, and P. P. Mertens. 1998. Development of a mouse model system, coding assignments and identification of the genome segments controlling virulence of African horse sickness virus serotypes 3 and 8. Arch. Virol. Suppl. 14:259-279. [DOI] [PubMed] [Google Scholar]
- 37.Ozkul, A., A. Erturk, E. Caliskan, F. Sarac, C. Ceylan, P. Mertens, O. Kabakli, E. Dincer, and S. G. Cizmeci. 2009. Segment 10 based molecular epidemiology of bluetongue virus (BTV) isolates from Turkey: 1999-2001. Virus Res. 142:134-139. [DOI] [PubMed] [Google Scholar]
- 38.Palmenberg, A. C., D. Spiro, R. Kuzmickas, S. Wang, A. Djikeng, J. A. Rathe, C. M. Fraser-Liggett, and S. B. Liggett. 2009. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 324:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra, G. I., K. Bok, M. Martinez, and J. A. Gomez. 2004. Evidence of rotavirus intragenic recombination between two sublineages of the same genotype. J. Gen. Virol. 85:1713-1716. [DOI] [PubMed] [Google Scholar]
- 40.Phan, T. G., S. Okitsu, N. Maneekarn, and H. Ushijima. 2007. Evidence of intragenic recombination in G1 rotavirus VP7 genes. J. Virol. 81:10188-10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Potgeiter, A. C., N. A. Page, J. Liebenburg, I. M. Wright, O. Landt, and A. A. van Dijk. 2009. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J. Gen. Virol. 90:1423-1432. [DOI] [PubMed] [Google Scholar]
- 41.Qin, Z., L. Sun, B. Ma, Z. Cui, Y. Zhu, Y. Kitamura, and W. Liu. 2008. F gene recombination between genotype II and VII Newcastle disease virus. Virus Res. 131:299-303. [DOI] [PubMed] [Google Scholar]
- 42.Reuter, J. S., and D. H. Mathews. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform. 11:129. [DOI] [PMC free article] [PubMed]
- 43.Romanova, L. I., V. M. Blinov, E. A. Tolskaya, E. G. Viktorova, M. S. Kolesnikova, E. A. Guseva, and V. I. Agol. 1986. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology 155:202-213. [DOI] [PubMed] [Google Scholar]
- 44.Roy, P. 1992. Bluetongue virus proteins. J. Gen. Virol. 73(Pt. 12):3051-3064. [DOI] [PubMed] [Google Scholar]
- 45.Roy, P. 2008. Bluetongue virus: dissection of the polymerase complex. J. Gen. Virol. 89:1789-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy, P. 2006. Orbiviruses, p. 1975-1997. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 47.Roy, P., J. J. Marshall, and T. J. French. 1990. Structure of the bluetongue virus genome and its encoded proteins. Curr. Top. Microbiol. Immunol. 162:43-87. [DOI] [PubMed] [Google Scholar]
- 48.Samal, S. K., A. el-Hussein, F. R. Holbrook, B. J. Beaty, and R. F. Ramig. 1987. Mixed infection of Culicoides variipennis with bluetongue virus serotypes 10 and 17: evidence for high frequency reassortment in the vector. J. Gen. Virol. 68(Pt. 9):2319-2329. [DOI] [PubMed] [Google Scholar]
- 49.Stauber, N., J. Martinez-Costas, G. Sutton, K. Monastyrskaya, and P. Roy. 1997. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol. 71:7220-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinhauer, D. A., and J. J. Skehel. 2002. Genetics of influenza viruses. Annu. Rev. Genet. 36:305-332. [DOI] [PubMed] [Google Scholar]
- 51.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 54.Urakawa, T., D. G. Ritter, and P. Roy. 1989. Expression of largest RNA segment and synthesis of VP1 protein of bluetongue virus in insect cells by recombinant baculovirus: association of VP1 protein with RNA polymerase activity. Nucleic Acids Res. 17:7395-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, W. C., H. C. Ma, E. H. Venter, A. A. van Djik, B. S. Seal, and J. O. Mecham. 2000. Phylogenetic relationships of bluetongue viruses based on gene S7. Virus Res. 67:141-151. [DOI] [PubMed] [Google Scholar]
- 56.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80(Pt. 10):2535-2543. [DOI] [PubMed] [Google Scholar]
- 56a.Yang, Y. Y., J. F. Chiou, G. Y. Hwang, I. J. Huang, and J. K. Li. 1992. Evolutionary analyses of five US bluetongue viruses using the cognate S2 genes. Virus Res. 25:241-249. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, R., X. Wang, J. Su, J. Zhao, and G. Zhang. 2010. Isolation and analysis of two naturally occurring multi-recombination Newcastle disease viruses in China. Virus Res. 151:45-53. [DOI] [PubMed] [Google Scholar]