Abstract
Coxsackievirus B3 (CVB3) is a small RNA virus associated with diseases such as myocarditis, meningitis, and pancreatitis. We have previously demonstrated that proteasome inhibition reduces CVB3 replication and attenuates virus-induced myocarditis. However, the underlying mechanisms by which the ubiquitin/proteasome system regulates CVB replication remain unclear. In this study, we investigated the role of REGγ, a member of the 11S proteasome activator, in CVB3 replication. We showed that overexpression of REGγ promoted CVB3 replication but that knockdown of REGγ led to reduced CVB3 replication. We further demonstrated that REGγ-mediated p53 proteolysis contributes, as least in part, to the proviral function of REGγ. Although total protein levels of REGγ remained unaltered after CVB3 infection, virus infection induced a redistribution of REGγ from the nucleus to the cytoplasm, rendering an opportunity for a direct interaction of REGγ with viral proteins and/or host proteins (e.g., p53), which controls viral growth and thereby enhances viral infectivity. Further analyses suggested a potential modification of REGγ by SUMO following CVB3 infection, which was verified by both in vitro and in vivo sumoylation assays. Sumoylation of REGγ may play a role in its nuclear export during CVB3 infection. Taken together, our results present the first evidence that the host REGγ pathway is utilized and modified during CVB3 infection to promote efficient viral replication.
Viruses often adapt to the existing host cellular machinery to complete their own life cycle. The ubiquitin/proteasome system (UPS), a primary intracellular protein degradation system in eukaryotic cells, has emerged as a key modulator in viral infectivity and virus-mediated pathogenesis (6).
Coxsackievirus B3 (CVB3) is a small RNA virus associated with diseases such as myocarditis, meningitis, and pancreatitis (36). We have previously studied the function and regulation of the UPS in CVB3 infection and CVB3-induced myocarditis (7, 16, 17, 33). We demonstrated that CVB3 utilizes and manipulates the host UPS to achieve successful replication (17, 33). We provided evidence that proteasome inhibition reduces CVB3 replication and attenuates virus-induced myocarditis (7). However, we recognize the potential toxicity of general inhibition of proteasome function as a therapeutic means. Further investigation to identify specific targets within the UPS utilized during CVB3 infection is urgently needed and will allow for more-precise targeting in drug therapy.
The 20S proteasome is a multisubunit protease complex responsible for the degradation of misfolded proteins or short-lived regulatory proteins (16, 18). In the absence of proteasome activators, the 20S proteasome is latent and the protein substrates are barred from entering the 20S proteasome (16, 18). There are at least two families of proteasome activators, the 19S proteasome (also known as PA700) and the 11S proteasome (also known as REG or PA28) (16, 18). The 19S activator binds to proteasome to form the 26S proteasome, which primarily performs degradation of proteins in a ubiquitin-dependent manner.
The REG activator binds to and activates the proteasome in an ATP-independent manner to promote mainly ubiquitin-independent protein degradation. Three classes of REG have been identified, REGα, REGβ, and REGγ. REGα/β forms a heteroheptamer which is mainly localized to the cytosol (16, 18). The level of REGα/β is inducible by gamma interferon, and the main function of REGα/β has been implicated in major histocompatibility complex (MHC) class I antigen presentation (16, 18). REGγ exists in a homoheptamer and is primarily found in the nucleus (16, 18). Although the functional significance of REGγ has not been fully defined, studies of REGγ-deficient mice reveal a role for REGγ in the regulation of cell cycle progression and cell survival/apoptosis (1, 27). These effects appear to be related to REGγ-mediated degradation of several important intracellular proteins, such as cyclin-dependent kinase inhibitors p21, p16, and p19 (2, 14) and tumor suppressor p53 (43). Moreover, an interaction between the REGγ system and the viral proteins has recently been reported. It was shown that REGγ binds to and regulates the stability and nuclear retention of hepatitis C core protein (26), contributing to hepatitis C core protein-induced insulin resistance and hepatocarcinoma (24, 25).
We have previously reported that gene silencing of ubiquitin reduces viral protein synthesis and viral titers (33). However, such inhibitions are not as potent as by proteasome inhibition, suggesting that 11S proteasome-mediated proteasomal degradation may also play a role. In the present study, we seek to further understand the underlying mechanisms by which the UPS regulates CVB3 replication by investigating the interplay between REGγ and CVB3 infection and exploring the potential mechanisms of how REGγ controls CVB3 replication. Here, we provided the first evidence that the host REGγ pathway was utilized and modulated during CVB3 infection to promote efficient viral replication.
MATERIALS AND METHODS
Cells and cell culture.
HeLa cells obtained from the American Type Culture Collection were grown and maintained in complete medium (Dulbecco's modified Eagle's medium [DMEM]) supplemented with 10% heat-inactivated newborn calf serum.
The HEK293 stable cell line overexpressing REGγ under the control of a tet-on promoter was previously established (15), and overexpression of REGγ was induced by addition of doxycycline (1 μg/ml).
Antibodies.
The mouse monoclonal anti-VP1 antibody was purchased from DakoCytomation. The monoclonal mouse anti-p53 (DO-1), anti-caspase-3, and anti-p21 (F-5) antibodies as well as the horseradish peroxidase-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology. The rabbit polyclonal anti-REGγ antibody was purchased from the Zymed Laboratories. The mouse monoclonal anti-β-actin antibody was from Sigma.
Virus infection.
HeLa cells and REGγ stable cells were mock treated with phosphate-buffered saline or infected with the Kandolf strain of CVB3 (from Reinhard Kandolf, University of Tubingen, Germany) (12) for 1 h in serum-free DMEM at different multiplicities of infection (MOI) as specified in the figure legends. The cells were then washed with PBS and cultured in complete DMEM for various times. For proteasome or apoptosis inhibition, the cells were infected with CVB3 in the presence or absence of different concentrations of the proteasome inhibitor lactacystin (Calbiochem) or the general caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (zVAD.fmk) (BD Biosciences) as indicated in the figures.
Plaque assay.
The virus titer was measured on a monolayer of HeLa cells by an agar overlay plaque assay as previously described (38). In brief, cell supernatant was serially diluted and overlaid on a 90 to 95% confluent monolayer of HeLa cells. Following 1 h of incubation, medium was removed and complete DMEM containing 0.75% agar was overlaid. At 3 days postincubation, cells were fixed with Carnoy's fixative (25% acetic acid and 75% ethanol) and then stained with 1% crystal violet. Viral titer was determined as the number of PFU per milliliter.
Western blot analysis.
Cell lysates were harvested with NETN lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40) containing proteasome inhibitor cocktail (Roche) as previously described (15). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a nitrocellulose membrane. The membrane was blocked with a 5% nonfat dry milk solution containing 0.1% Tween 20 for 1 h. The membrane was then incubated for 1 h with the primary antibody, followed by incubation with a horseradish peroxidase-conjugated secondary antibody for another 1 h. Immunoreactive bands were visualized with an enhanced chemiluminescence detection system (GE Healthcare).
Plasmid DNA and small interfering RNA (siRNA) transfection.
HeLa cells at ∼90% confluence were transiently transfected with a plasmid expressing REGγ (pcDNA-Flag-REGγ) (15) or a construct encoding wild-type p53 (pCMV-p53) (Clontech) using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions. The empty vectors were transfected as a control.
Knockdown of REGγ in HeLa cells was achieved through siRNA. HeLa cells were grown to ∼50% confluence and then transiently transfected with a pool of four REGγ siRNA duplexes (Dharmacon) at a concentration of 30 nM using Oligofectamine 2000 (Invitrogen). A scrambled siRNA duplex was used as a negative control. The silencing efficiency was measured by Western blot analysis using anti-REGγ antibody. Twenty-four hours after transfection, cells were infected with CVB3 as described above.
Caspase-3 activity assay.
HeLa cells transfected with REGγ or control siRNAs were infected with CVB3 for 18 h, cell lysates were collected, and caspase-3 activities were measured using a synthetic fluorogenic substrate (R&D Systems) in accordance with the manufacturer's instructions.
Cell viability assay.
The cell viability assay was performed using 3,4-(5-di- methylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS; Promega) as previously described (41). Following treatment, cells were incubated with MTS solution for 2 h and absorbance was measured at a wavelength of 490 nm using an enzyme-linked immunosorbent assay (ELISA) plate reader. Morphological changes of cells were visualized by phase-contrast microscopy.
Immunofluorescence.
Cells grown on glass slides were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and double labeled by indirect immunofluorescence with anti-REGγ or anti-VP1, followed by incubation with Alexa Fluor 488-conjugated anti-rabbit or Alexa Fluor 594 anti-mouse IgG (Molecular Probes), respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and cells were imaged and analyzed with a Leica SP2 AOBS confocal fluorescence microscope.
In vitro and in vivo sumoylation assay.
The in vitro sumoylation assay was carried out with a sumoylation assay kit according to the manufacturer's protocol (Biomol). In brief, 200 nM purified recombinant REGγ (BML-PW9875-0100; Biomol) was mixed with SUMO E1 (Aos1/Uba2), SUMO E2 (Ubc9), and SUMO in a reaction buffer in the presence or absence of Mg2+-ATP. The reaction mixture was incubated at 30°C for 1 h and then quenched with SDS-PAGE loading buffer. The samples were separated by SDS-PAGE, and Western blot analysis was performed with anti-SUMO antibody.
For in vivo sumoylation ELISA, HEK293-REGγ inducible cells were treated with or without doxycycline for 48 h, followed by transient transfection with a construct expressing SUMO-1 (pCMV3T-HA-SUMO-1, a generous gift from Louis Flamand at the Laval University) for another 48 h. After 20 h of mock or CVB3 infection (MOI = 1), cell extracts were collected in NETN buffer and sumoylated REGγ was detected by an ELISA using an EpiQuik in vivo universal protein sumoylation assay kit in accordance with the manufacturer's instructions (Epigentek). Briefly, equal amounts of proteins from the cell extracts were added to the strip wells, which were precoated with either anti-REGγ antibody or IgG, and incubated in SUMO assay buffer for 1 h at room temperature. After three washes, SUMO antibody was added and incubated for 15 min at room temperature. Following color development by a SUMO detection system, absorbance was measured at 450 nm using an ELISA plate reader.
Statistical analysis.
The results shown are the means ± standard deviations (SD), and statistical analysis was performed using the paired Student t test. A P value of <0.05 was considered significant. At least three replicates were performed for each experiment.
RESULTS
Effect of REGγ knockdown or overexpression on coxsackieviral replication and virus-induced apoptosis.
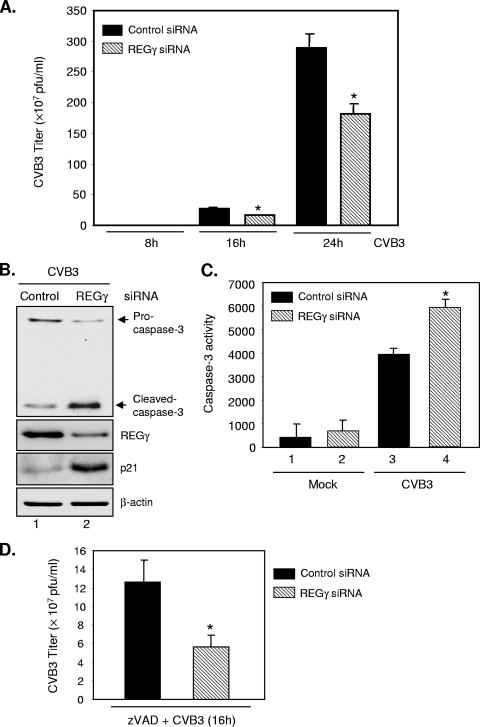
To determine whether REGγ-mediated proteolysis plays a role in the regulation of UPS-mediated coxsackieviral replication, we employed a pool of REGγ siRNA duplexes to silence the gene expression of REGγ and assessed the role of REGγ knockdown in coxsackieviral replication by a plaque assay. As shown in Fig. 1B, gene-silencing of REGγ in HeLa cells was able to induce an accumulation of p21 protein, which was previously demonstrated to be a target of REGγ (2, 14). The plaque assay results in Fig. 1A demonstrate that the virus titers for REGγ-silenced cells were significantly lower than those for control siRNA-transfected cells, suggesting that knockdown of REGγ attenuates CVB3 replication.
FIG. 1.
Knockdown of REGγ reduces coxsackieviral progeny titers. HeLa cells were transiently transfected with the REGγ siRNA (30 nM) or a scramble control siRNA for 24 h, followed by mock infection or CVB3 infection (MOI = 1) for various times as indicated (A) or for 16 h (B, C, D). (A) Supernatants of infected cells were harvested, and progeny virion titers were measured by a plaque assay. The results are presented as means ± SD (n = 3). *, P < 0.01 for comparison to the scramble siRNA control. Similar results were observed with HEK293 cells. (B) Western blot analysis was performed to detect caspase-3, p21, REGγ, and β-actin (loading control). (C) Caspase-3 activities were measured using a synthetic fluorogenic substrate, and the results are expressed as means ± SD (n = 3). *, P < 0.01 for comparison to the siRNA control. (D) HeLa cells were transfected with REGγ or control siRNAs for 24 h and then infected with CVB3 for 16 h in the presence of zVAD (50 μM). Plaque assay results are shown as means ± SD (n = 3). *, P < 0.01 for comparison to the scramble siRNA control.
The function of REGγ in the control of cell survival/apoptosis has been increasingly recognized (14, 27). It has been shown that REGγ-deficient murine embryonic fibroblasts (MEFs) have markedly enhanced apoptosis compared to the wild-type cells. To assess the effect of REGγ depletion on CVB3-induced apoptosis, we performed experiments to measure caspase-3 cleavage and activity. As shown in Fig. 1B and C, CVB3 infection resulted in the cleavage (Fig. 1B, lane 1) and increased activity of caspase-3 (Fig. 1C, lane 3 versus lane 1). Following CVB3 infection, cells with REGγ depletion had enhanced caspase-3 cleavage (Fig. 1B, lane 2 versus lane 1) and increased caspase-3 activity (Fig. 1C, lane 4 versus lane 3) compared with cells transfected with scramble siRNAs. It was noted that gene silencing of REGγ did not promote increased apoptosis in mock-infected cells (Fig. 1C, lane 2 versus lane 1). These results suggest that inhibition of REGγ sensitizes HeLa cells to CVB3-induced apoptosis. Similar results were observed with HEK293 cells.
CVB3-induced apoptosis plays a key role in facilitating virus progeny release during late stages of viral infection. However, untimely cell death could be harmful to virus by creating an unpleasant environment for viral replication (40). To determine whether inhibition of CVB3 by REGγ knockdown is through induction of early apoptosis, we examined the influence of inhibition of apoptosis on CVB3 infection in both control and REGγ-depleting cells. We found that general caspase inhibition by zVAD did not eliminate the antiviral properties of REGγ depletion (Fig. 1D). These results suggest that the effect of REGγ knockdown on CVB3 infectivity is unlikely due to enhanced apoptosis.
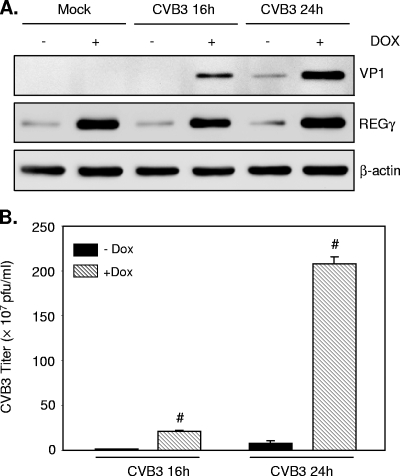
The role of REGγ in viral replication was also assessed by REGγ overexpression. REGγ was induced in the HEK293 “tet-on” stable cell line with the addition of doxycycline. Figure 2 demonstrates that overexpression of REGγ increased viral protein synthesis (Fig. 2A) and viral replication (Fig. 2B) in a time-dependent manner. To exclude the possible influence of doxycycline on viral replication, we also performed an experiment to examine the effect of doxycycline incubation on virus protein expression. We found that VP1 protein expression was unchanged (data not shown), suggesting that the increased viral production in HEK293-REGγ cells is not due to the treatment of doxycycline. Collectively, these results demonstrate an important role for the proteasome activator REGγ in controlling CVB3 infectivity.
FIG. 2.
Overexpression of REGγ promotes CVB3 replication. HEK293 tet-inducible REGγ cells were grown in the presence or absence of doxycycline (Dox) for 24 h, followed by mock or CVB3 infection (MOI = 1) for various times as indicated. (A) Cell lysates were collected, and expression of viral capsid protein VP1, REGγ, and β-actin (loading control) was detected by Western blot analysis. (B) Supernatants of infected cells were harvested, and progeny virion titers were measured by a plaque assay. The results are presented as means ± SD (n = 3). #, P < 0.001 for comparison to control cells without Dox induction.
Overexpression of REGγ decreases p53 levels.
To further explore the molecular mechanisms by which REGγ regulates CVB3 infection, we decided to focus on determining the role of identified REGγ substrate in CVB3 growth. We speculate that REGγ-mediated proteolysis of certain host intracellular proteins may target specific aspects of the viral replication process and thus control its replication.
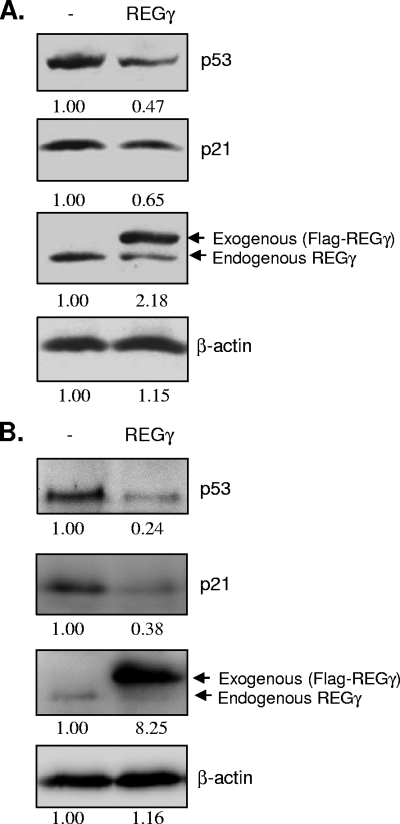
The tumor suppressor protein p53 has been suggested to be a common barrier to viral replication by directly inhibiting virus transcription and through promoting premature apoptosis (3, 4, 19, 29, 32, 35). Recent study demonstrates a role for the REGγ-proteasome pathway in regulating the stability of p53 (43). It was reported that REGγ facilitates p53 degradation by promoting MDM2-mediated p53 ubiquitination (43). Consistent with this report, Fig. 3 shows that transient transfection of HeLa cells (Fig. 3A) or HEK293 cells (Fig. 3B) with Flag-tagged REGγ expression vector reduced p53 and p21 levels.
FIG. 3.
Overexpression of REGγ decreases p21 and p53 levels. HeLa cells (A) or HEK293 cells (B) were transiently transfected with Flag-tagged REGγ expression vector or control plasmid pcDNA for 48 h. Cell lysates were collected, and expression of p53, p21, REGγ, and β-actin (loading control) was detected by Western blot analysis. Protein expression was quantitated by densitometric analysis using National Institutes of Health ImageJ 1.41o and normalized to the level for the control vector-transfected cells, which was arbitrarily set to a value of 1.0. Relative protein levels are listed below each panel. −, empty-vector control.
Overexpression of p53 reduces CVB3 replication and attenuates the proviral function of REGγ.
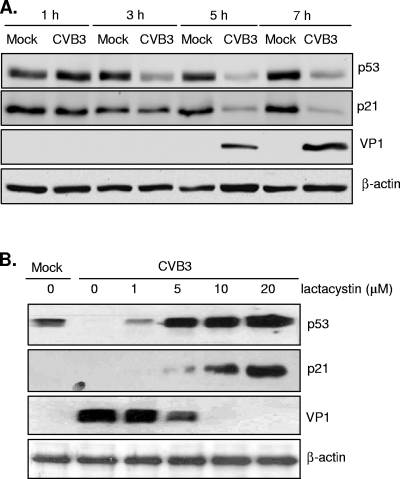
Several viruses have been shown to inactivate p53 during early viral infection for their own benefits via different mechanisms (3, 29, 32). The results in Fig. 4A show that CVB3 infection resulted in increased viral protein synthesis, accompanied by decreased levels of p53 and p21. We have previously reported that proteasome inhibition reduces CVB3 replication (17, 33). Figure 4B demonstrates that inhibition of p53 and p21 degradation during CVB3 infection by the proteasome inhibitor lactacystin was associated with decreased viral infectivity, suggesting a potential link between the p53 pathway and viral replication.
FIG. 4.
Blockage of p21 and p53 degradation by proteasome inhibitor lactacystin is associated with decreased virus protein expression. (A) HeLa cells were mock infected or infected with CVB3 (MOI = 10) for different time courses as indicated. Western blot analysis was carried out for detection of p53, p21, VP1, and β-actin. (B) HeLa cells were infected with CVB3 (MOI = 10) in the presence of increasing concentrations of lactacystin as indicated. Cell lysates were collected at 7 h postinfection for Western blot analysis of p21, p53, VP1, and β-actin.
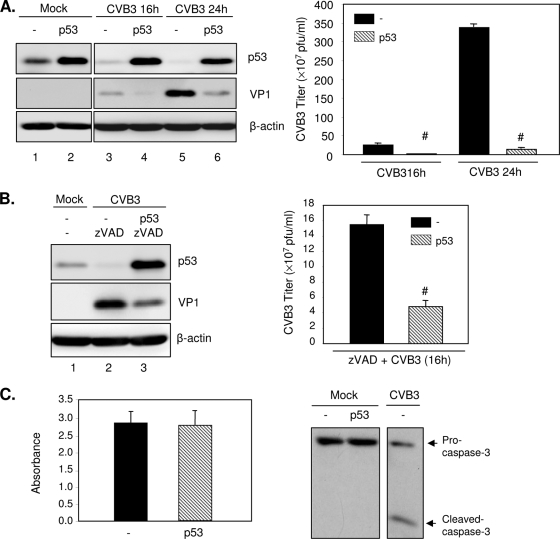
To delineate the potential functions of p53 degradation on CVB3 replication, we overexpressed p53 in HeLa cells, which were then exposed to CVB3 for various times. As shown in Fig. 5, CVB3 infection led to a reduction of p53 protein expression in a time-dependent manner (Fig. 5A, left panel, lanes 3 and 5 versus lane 1, and B, left panel, lane 2 versus lane 1), which is consistent with the observation in Fig. 4. Overexpression of p53 overcame the suppression induced by CVB3 infection (Fig. 5A, left panel, lanes 4 and 6 versus lanes 3 and 5, respectively), resulting in significant reduction of viral protein expression (Fig. 5A, left panel, lanes 4 and 6 versus lanes 3 and 5, respectively) and virus titers (Fig. 5A, right panel). These results indicate a mechanism by which REGγ regulates viral replication via regulation of p53 levels.
FIG. 5.
Overexpression of p53 inhibits CVB3 infection. (A) HeLa cells were transiently transfected with a plasmid expressing p53 or empty vector pCMV for 24 h, followed by infection with CVB3 (MOI = 1) for 16 or 24 h. Cell lysates were analyzed by Western blotting for protein expression of p53, VP1, and protein loading control β-actin (left panel). Supernatants of infected cells were harvested, and progeny virion titers were measured by a plaque assay (right panel). The results are presented as means ± SD (n = 3). #, P < 0.001 for comparison to vector control cells. (B) HeLa cells were transfected with p53 construct or empty vector for 24 h and then infected with CVB3 for 16 h in the presence of zVAD (50 μM). Western blot analysis (left panel) and a plaque assay (right panel) were performed to examine VP1 expression and virus titers, respectively. The results are shown as means ± SD (n = 3). #, P < 0.001 for comparison with the vector control. (C) HeLa cells were transiently transfected with p53 or empty vector for 48 h. (Left panel) Cell viability was determined by the MTS assay (mean ± SD; n = 3). (Right panel) Western blot analysis for the cleavage of caspase-3. HeLa cells infected with CVB3 were used as a positive control. −, empty-vector control.
To further determine whether p53 inhibits viral replication by promoting early apoptosis, we treated the cells with zVAD and examined the effect of apoptosis inhibition on viral infection. As shown in Fig. 5B, inhibition of apoptosis did not prevent the inhibitory effects of p53 on viral protein expression (left panel) and virus titers (right panel). Furthermore, we performed a cell viability assay and Western blot analysis to examine cell death and apoptosis following overexpression of p53. Figure 5C shows that overexpression of p53 for 48 h did not induce increased cell death (left panel) and apoptosis (right panel) compared to the level for the vector control. Our results suggest that p53 appears to inhibit CVB3 replication by a direct mechanism independent of its regulatory role of initiating apoptosis.
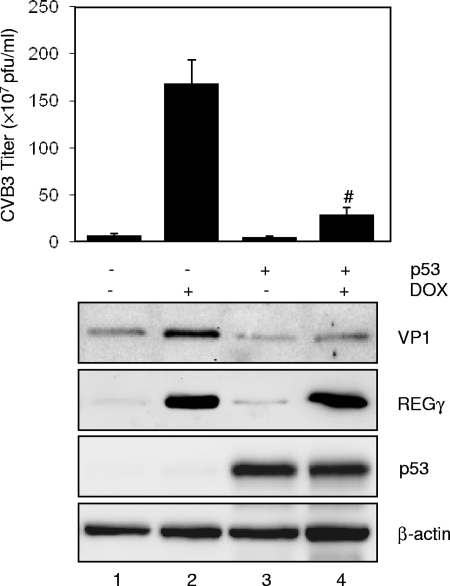
Finally, we examined whether overexpression of p53 can attenuate the effect of REGγ on viral replication. HEK293-REGγ inducible cells were transiently transfected with p53 or control vector for 24 h in the presence or absence of doxycycline and then infected with CVB3 for 16 h. Consistent with the results shown in Fig. 2, REGγ overexpression markedly enhanced CVB3 VP1 expression and virus titer (Fig. 6, lane 2 versus lane 1). However, the positive effect of REGγ on viral replication was significantly attenuated by p53 overexpression (lane 4 versus lane 2), supporting a role for REGγ in regulating CVB3 infection via destabilization of p53.
FIG. 6.
Overexpression of p53 attenuates the effect of REGγ on promotion of CVB3 replication. HEK293 tet-inducible REGγ cells were grown in the presence or absence of doxycycline (Dox) as indicated for 24 h, followed by transient transfection with either p53 (+) or empty vector (−). Twenty-four hours later, cells were infected with CVB3 (MOI = 1) for 16 h. (Upper panel) Supernatants of infected cells were harvested, and progeny virion titers were measured by a plaque assay. The results are presented as means ± SD (n = 3). #, P < 0.001 for comparison to the vector control with induction of Dox (lane 2). (Lower panel) Cell lysates were collected for detection of viral capsid protein VP1, REGγ, p53, and β-actin (loading control) by Western blot analysis.
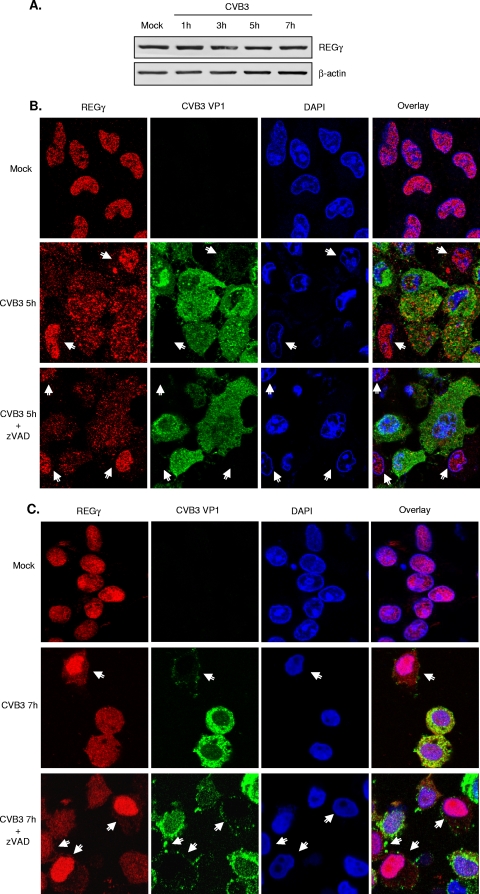
CVB3 infection leads to cytoplasmic relocalization of REGγ.
To understand the interaction between REGγ and CVB3 infection, we next assessed the impact of CVB3 infection on protein expression, subcellular localization, and activation of REGγ. Figure 7A shows that the protein expression levels of REGγ appeared to be unchanged throughout the course of virus infection. We further examined the cellular localization of REGγ after CVB3 infection. Double immunostaining for REGγ and viral protein VP1 was performed on mock- or CVB3-infected HeLa cells at 1 h, 3 h, 5 h, and 7 h postinfection. As shown in Fig. 7B and C, REGγ was exclusively localized in the nucleus in mock-infected cells (top panels). However, following 5 h (Fig. 7B) or 7 h (Fig. 7C) of viral infection, REGγ was largely redistributed to the cytoplasm in CVB3-infected cells (green-stained cells), whereas in noninfected cells (green-negative cells), REGγ remained in the nucleus (middle panels). The bottom panels in Fig. 7B and C show that inhibition of apoptosis by zVAD did not prevent REGγ redistribution, suggesting that cytoplasmic translocation of REGγ is unlikely a consequence of CVB3-induced apoptosis. It should be noted that detection of VP1 protein by immunostaining was not very sensitive when it was at a relatively low level. It was found that VP1 was undetectable until 5 h postinfection, consistent with the Western blot results shown in Fig. 4A. Thus, Fig. 7B and C only show cytoplasmic relocalization of REGγ at 5 h and 7 h postinfection in virus-infected cells. However, REGγ redistribution likely occurred earlier, similar to what was observed by Western blot analysis in Fig. 4A, that expression of p53 and p21 was downregulated at 3 h postinfection, prior to the detection of VP1 protein expression (i.e., 5 h postinfection).
FIG. 7.
CVB3 infection leads to redistribution of REGγ. (A) HeLa cells were either mock infected or infected with CVB3 (MOI = 10) for different times. Western blot analysis was performed for detecting REGγ and β-actin. (B, C) HeLa cells were infected with CVB3 (MOI = 10) for 5 h (B) or 7 h (C) in the presence or absence of zVAD (50 μM). Double-immunocytochemical staining was carried out for examination of the expression and localization of REGγ (red) and viral protein VP1 (green). The nucleus was stained with DAPI (blue). Arrows denote cells without or with low levels of viral protein expression. The yellow staining in the merged image indicates colocalization of these two proteins. It is noteworthy that REGγ is redistributed to the cytoplasm in CVB3-infected cells (green-positive cells) but that REGγ remains in the nuclei of noninfected cells (green-negative cells).
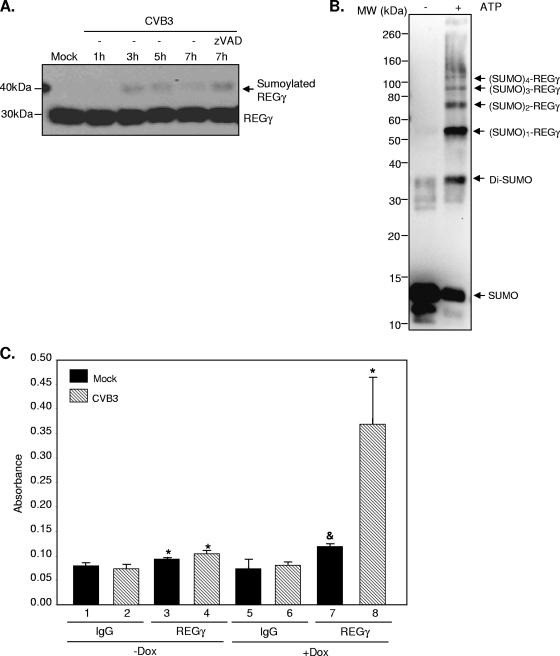
CVB3 infection promotes REGγ sumoylation.
REGγ activity may also be regulated by posttranslational modifications, such as ubiquitination, phosphorylation, and sumoylation. By Western blot analysis, a higher-molecular-mass protein band above the regular REGγ band was detected starting at 3 h postinfection (Fig. 8A). This result suggests a potential posttranslational modification of REGγ after CVB3 infection. The molecular mass of the higher band (∼45 kDa) seemed to correspond in size to the sumoylated REGγ. We further showed that general caspase inhibition did not block the appearance of this band, suggesting that this modification of REGγ is not due to virus-induced apoptosis.
FIG. 8.
CVB3 infection promotes REGγ sumoylation. (A) HeLa cells were infected with CVB3 (MOI = 10) for different times in the presence or absence of zVAD (50 μM). Western blot analysis was performed to detect the expression of REGγ and β-actin. (B) An in vitro sumoylation assay was performed on purified REGγ protein according to the manufacturer's instructions (Biomol). Following in vitro reaction, sumoylated proteins were detected by Western blot analysis using anti-SUMO antibody. (C) HEK293-REGγ stable cells were treated with Dox or without Dox as indicated for 48 h, followed by transient transfection with SUMO-1 for an additional 48 h. After 20 h of mock or CVB3 infection (MOI = 1), cell extracts were harvested for an in vivo sumoylation ELISA as described in Materials and Methods. The data displayed are means ± SD (n = 3). *, P < 0.01 for comparison to mock or CVB3 IgG controls (lane 3 versus lane 1, lane 4 versus lane 2, and lane 8 versus lane 6). &, P < 0.05 for comparison to the mock IgG control (lane 7 versus lane 5).
Small ubiquitin-related modifier proteins (SUMO) are a family of small proteins that are structurally similar to ubiquitin (11). Protein modification by sumoylation is directed by an enzymatic cascade catalyzed by three enzymes (11). The SUMO E1-activating enzyme is a heterodimeric complex consisting of Aos1 and Uba2. Ubc9 is the only identified SUMO E2-conjugating enzyme. At least three classes of SUMO E3-ligases have been reported, RanBP2, PIAS, and the polycomb protein Pc2. A recent report shows that CVB5 infection results in cytoplasmic redistribution of SUMO-1 and UBC9 (8).
To verify the potential of REGγ sumoylation, we performed in vitro and in vivo sumoylation assays. As shown in Fig. 8B, in the presence of SUMO E1 (Aos/UBa2), SUMO E2 (UBC9), and ATP, REGγ can be sumoylated to form multiple sumoylated REGγ. REGγ sumoylation was further confirmed by an in vivo sumoylation ELISA via overexpression of SUMO-1 in HEK293-REGγ stable cells either in the absence or in the presence of doxycycline induction (Fig. 8C, lane 3 versus lane 1 [−Dox] and lane 7 versus lane 5 [+Dox]). Importantly, we showed that CVB3 infection significantly enhanced REGγ sumoylation (Fig. 8C, lane 4 versus lane 2 [−Dox] and lane 8 versus lane 6 [+Dox]).
DISCUSSION
The major findings of this study are as follows: (i) p53 negatively regulates CVB3 replication, (ii) REGγ promotes CVB3 replication, at least in part, through facilitation of p53 degradation, and (iii) CVB3 infection enhances REGγ sumoylation, which may result in nuclear export of REGγ and subsequent p53 degradation in the cytoplasm. Our earlier work has demonstrated that general proteasome inhibition blocks CVB3 infection (17, 33); however, the mechanisms and the downstream targets of this pathway responsible for such effect have not been identified. In the present paper, we provide evidence that p53, a target of the UPS, plays a critical role in the control of CVB3 replication. Although REGγ was originally characterized as a proteasome activator (18), recent study has shown that REGγ promotes the turnover of p53 by acting as a coactivator to facilitate MDM2-mediated p53 polyubiquitination independent of its function in directly activating the 20S proteasome (43). Most recently, we also provide evidence that REGγ can enhance monoubiquitination of p53, resulting in nuclear export of REGγ and subsequent cytoplasmic degradation, which serves as an additional mechanism for p53 inactivation by REGγ (data not shown). Thus, the regulatory effect of REGγ on CVB3 infection via p53 appears to be independent of the proteasome-activating function of REGγ.
The tumor suppressor protein p53 is a transcription factor which plays a central role in regulating cell growth arrest and apoptosis in response to DNA damage (21). Many viruses have evolved different strategies to inactivate p53 to prevent early apoptosis, allowing for effective viral replication (3, 29, 32). In addition, it was reported that p53 can directly interfere with the propagation of several viruses, such as human immunodeficiency virus type 1 (4), simian virus (35), and encephalomyocarditis virus (19). CVB3 infection leads to a dramatic downregulation of p53 protein, suggesting that CVB3 may have evolved strategies to regulate p53 function and stability to permit efficient viral replication. We have previously found that cardiomyocytes and HEK293 cells, which express higher levels of p53, produce much lower yields of virus than HeLa cells, in which the level of p53 expression is relatively low (data not shown). This observation implicates a potential inhibitory function of p53 in the control of coxsackievirus replication. Furthermore, in the study of poliovirus, a close family member of coxsackievirus, knockdown of endogenous p53 by siRNA in U2OS cells has been shown to result in a higher level of viral replication (28). In this study, we investigate the role of p53 in coxsackievirus infection by overexpressing p53 in HeLa cells. Our results provide direct evidence that p53 functions as an antiviral protein against CVB3 replication. Although the mechanism of the antiviral action of p53 has not yet been fully defined, our results in the present study suggest that p53 appears to inhibit virus replication by a direct mechanism, independent of its regulatory role of initiating apoptosis.
The current study provides evidence showing that REGγ-mediated p53 proteolysis may contribute, as least in part, to the proviral function of REGγ. However, we cannot exclude the potential roles of other host protein targets of REGγ in regulating coxsackieviral infectivity. In addition, future investigation will explore the possibility that REGγ may directly regulate CVB3 protein processing.
Coxsackievirus or poliovirus infection has been shown to induce cytosolic translocation of several host nuclear proteins, such as the nuclear protein La (22), Sam68 (20), and polypyrimidine tract binding protein (10). In this study, we have added REGγ to this list. Regarding the mechanism by which CVB3 infection triggers the subcellular redistribution of REGγ, one possibility may be related to the disruption of the nucleocytoplasmic trafficking pathways. It has been reported that poliovirus infection leads to the degradation of the nuclear pore complex protein Nup153 and p62, resulting in the blockage of nuclear import of nucleocytoplasmic shuttle proteins (9). The other possible explanation may be due to increased sumoylation modification of REGγ during CVB3 infection, which leads to nuclear export of REGγ. Although sumoylation has most often been implicated in promoting nuclear import (11), there is evidence that sumoylation can also serve as a signal for cytoplasmic translocation of some protein substrates, such as TEL (39), Smad3 (13), Med (23), MEK1 (34), and heterogeneous nuclear ribonucleoproteins M and C (37). REGγ has been suggested to be a potential nucleocytoplasmic shuttling protein (42). However, the mechanisms regulating REGγ nuclear import/export have not yet been fully characterized. The role of sumoylation modification in such regulation warrants further investigation.
REGγ is not a previously recognized substrate for SUMO modification. Here, we provide both in vitro and in vivo evidence that REGγ can be sumoylated and that such posttranslational modification is enhanced during CVB3 infection. However, the exact sumoylation site(s) and the SUMO E3 responsible for REGγ sumoylation have not been identified in the current study. Interestingly, the proteins RanBPM and PIAS, two known SUMO E3s, have been reported to physically associate with REGγ (30), suggesting a possible function for them as SUMO E3 ligases for REGγ sumoylation. By use of the SUMOplot analysis program (Abgent), six putative sumoylation sites are predicted. Further studies will be necessary to verify these predictions and to understand the exact functions of REGγ sumoylation.
The significance of cytosolic REGγ in relation to coxsackievirus replication is still unclear. Given that coxsackievirus replication takes place exclusively in the cytoplasm, cytoplasmic localization of REGγ may allow for easier access to virus proteins and closer interaction with host proteins which control viral replication. For example, it has been shown that efficient degradation of p53 by proteasome occurs in the cytoplasm (5, 31). Thus, nuclear export of REGγ may provide a more effective way to interact with and promote p53 degradation in the cytoplasm. REGγ has been reported to be required for nuclear retention and degradation of hepatitis C virus core protein (26). We expect that cytosolic translocation of REGγ may also render an opportunity for its direct interaction with viral proteins and thus enhance viral protein processing.
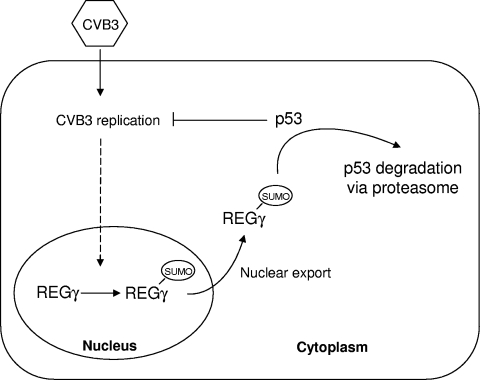
In summary, our results demonstrate that the proteasome activator REGγ plays an important role in facilitating coxsackieviral replication. As illustrated in Fig. 9, CVB3 infection results in increased sumoylation and cytoplasmic translocation of REGγ, which is associated with augmented p53 degradation. We also demonstrate an antiviral activity of p53 against CVB3 replication. Our results suggest that CVB3 infection promotes REGγ-mediated proteolysis of p53, which may enhance its own replication by reducing the inhibitory influence of p53 on viral replication.
FIG. 9.
Proposed mechanism by which REGγ enhances CVB3 infectivity. Following CVB3 infection, REGγ is sumoylated and exported from the nucleus. Cytoplasmic translocation of REGγ facilitates proteasomal degradation of tumor suppressor protein p53, which subsequently enhances CVB3 infection by suppressing the inhibitory effect of p53 on viral replication.
Acknowledgments
We thank Louis Flamand (Laval University, Quebec, Canada) for his generous gift of the pCMV3T-HA-SUMO-1 construct.
This work was supported by a joint grant from the Canadian Institutes of Health Research (CIHR)/the National Natural Science Foundation of China (30811120435 and 30811120435) (H.L./X.L.). This article was also partially supported by the Heart and Stroke Foundation of British Columbia and Yukon (H.L.) and the National Institutes of Health (1R01CA131914). J.W. is the recipient of studentships from the CIHR and the Heart and Stroke Foundation of Canada.
Footnotes
Published ahead of print on 18 August 2010.
REFERENCES
- 1.Barton, L. F., H. A. Runnels, T. D. Schell, Y. Cho, R. Gibbons, S. S. Tevethia, G. S. Deepe, Jr., and J. J. Monaco. 2004. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J. Immunol. 172:3948-3954. [DOI] [PubMed] [Google Scholar]
- 2.Chen, X., L. F. Barton, Y. Chi, B. E. Clurman, and J. M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell 26:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 4.Duan, L., I. Ozaki, J. W. Oakes, J. P. Taylor, K. Khalili, and R. J. Pomerantz. 1994. The tumor suppressor protein p53 strongly alters human immunodeficiency virus type 1 replication. J. Virol. 68:4302-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 84:5-14. [DOI] [PubMed] [Google Scholar]
- 7.Gao, G., J. Zhang, X. Si, J. Wong, C. Cheung, B. McManus, and H. Luo. 2008. Proteasome inhibition attenuates coxsackievirus-induced myocardial damage in mice. Am. J. Physiol. Heart Circ. Physiol. 295:H401-H408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes, R., R. Guerra-Sa, and E. Arruda. 2010. Coxsackievirus B5 induced apoptosis of HeLa cells: effects on p53 and SUMO. Virology 396:256-263. [DOI] [PubMed] [Google Scholar]
- 9.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilgarth, R. S., L. A. Murphy, H. S. Skaggs, D. C. Wilkerson, H. Xing, and K. D. Sarge. 2004. Regulation and function of SUMO modification. J. Biol. Chem. 279:53899-53902. [DOI] [PubMed] [Google Scholar]
- 12.Huber, M., H. C. Selinka, and R. Kandolf. 1997. Tyrosine phosphorylation events during coxsackievirus B3 replication. J. Virol. 71:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imoto, S., N. Ohbayashi, O. Ikeda, S. Kamitani, R. Muromoto, Y. Sekine, and T. Matsuda. 2008. Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochem. Biophys. Res. Commun. 370:359-365. [DOI] [PubMed] [Google Scholar]
- 14.Li, X., L. Amazit, W. Long, D. M. Lonard, J. J. Monaco, and B. W. O'Malley. 2007. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell 26:831-842. [DOI] [PubMed] [Google Scholar]
- 15.Li, X., D. M. Lonard, S. Y. Jung, A. Malovannaya, Q. Feng, J. Qin, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2006. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 124:381-392. [DOI] [PubMed] [Google Scholar]
- 16.Luo, H., J. Wong, and B. Wong. 2010. Protein degradation systems in viral myocarditis leading to dilated cardiomyopathy. Cardiovasc. Res. 85:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, H., J. Zhang, C. Cheung, A. Suarez, B. M. McManus, and D. Yang. 2003. Proteasome inhibition reduces coxsackievirus B3 replication in murine cardiomyocytes. Am. J. Pathol. 163:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao, I., J. Liu, X. Li, and H. Luo. 2008. REGgamma, a proteasome activator and beyond? Cell. Mol. Life Sci. 65:3971-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques, J. T., D. Rebouillat, C. V. Ramana, J. Murakami, J. E. Hill, A. Gudkov, R. H. Silverman, G. R. Stark, and B. R. Williams. 2005. Down-regulation of p53 by double-stranded RNA modulates the antiviral response. J. Virol. 79:11105-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. U. S. A. 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meek, D. W. 2009. Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer 9:714-723. [DOI] [PubMed] [Google Scholar]
- 22.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miles, W. O., E. Jaffray, S. G. Campbell, S. Takeda, L. J. Bayston, S. P. Basu, M. Li, L. A. Raftery, M. P. Ashe, R. T. Hay, and H. L. Ashe. 2008. Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 22:2578-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyamoto, H., K. Moriishi, K. Moriya, S. Murata, K. Tanaka, T. Suzuki, T. Miyamura, K. Koike, and Y. Matsuura. 2007. Involvement of the PA28gamma-dependent pathway in insulin resistance induced by hepatitis C virus core protein. J. Virol. 81:1727-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moriishi, K., R. Mochizuki, K. Moriya, H. Miyamoto, Y. Mori, T. Abe, S. Murata, K. Tanaka, T. Miyamura, T. Suzuki, K. Koike, and Y. Matsuura. 2007. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 104:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriishi, K., T. Okabayashi, K. Nakai, K. Moriya, K. Koike, S. Murata, T. Chiba, K. Tanaka, R. Suzuki, T. Suzuki, T. Miyamura, and Y. Matsuura. 2003. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 77:10237-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata, S., H. Kawahara, S. Tohma, K. Yamamoto, M. Kasahara, Y. Nabeshima, K. Tanaka, and T. Chiba. 1999. Growth retardation in mice lacking the proteasome activator PA28gamma. J. Biol. Chem. 274:38211-38215. [DOI] [PubMed] [Google Scholar]
- 28.Pampin, M., Y. Simonin, B. Blondel, Y. Percherancier, and M. K. Chelbi-Alix. 2006. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J. Virol. 80:8582-8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao, L., M. Debbas, P. Sabbatini, D. Hockenbery, S. Korsmeyer, and E. White. 1992. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc. Natl. Acad. Sci. U. S. A. 89:7742-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechsteiner, M., and C. P. Hill. 2005. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 15:27-33. [DOI] [PubMed] [Google Scholar]
- 31.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17:554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 33.Si, X., G. Gao, J. Wong, Y. Wang, J. Zhang, and H. Luo. 2008. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One 3:e2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobko, A., H. Ma, and R. A. Firtel. 2002. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell 2:745-756. [DOI] [PubMed] [Google Scholar]
- 35.Tiemann, F., and W. Deppert. 1994. Stabilization of the tumor suppressor p53 during cellular transformation by simian virus 40: influence of viral and cellular factors and biological consequences. J. Virol. 68:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tracy, S., and C. Gauntt. 2008. Group B coxsackievirus virulence. Curr. Top. Microbiol. Immunol. 323:49-63. [DOI] [PubMed] [Google Scholar]
- 37.Vassileva, M. T., and M. J. Matunis. 2004. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 24:3623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong, J., J. Zhang, X. Si, G. Gao, I. Mao, B. M. McManus, and H. Luo. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood, L. D., B. J. Irvin, G. Nucifora, K. S. Luce, and S. W. Hiebert. 2003. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 100:3257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, J., P. K. Cheung, H. Zhang, D. Chau, B. Yanagawa, C. Cheung, H. Luo, Y. Wang, A. Suarez, B. M. McManus, and D. Yang. 2004. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab. Invest. 84:703-714. [DOI] [PubMed] [Google Scholar]
- 41.Yuan, J., J. Zhang, B. W. Wong, X. Si, J. Wong, D. Yang, and H. Luo. 2005. Inhibition of glycogen synthase kinase 3beta suppresses coxsackievirus-induced cytopathic effect and apoptosis via stabilization of beta-catenin. Cell Death Differ. 12:1097-1106. [DOI] [PubMed] [Google Scholar]
- 42.Zannini, L., D. Lecis, G. Buscemi, L. Carlessi, P. Gasparini, E. Fontanella, S. Lisanti, L. Barton, and D. Delia. 2008. REGgamma proteasome activator is involved in the maintenance of chromosomal stability. Cell Cycle 7:504-512. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Z., and R. Zhang. 2008. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 27:852-864. [DOI] [PMC free article] [PubMed] [Google Scholar]