Abstract
Nonsyndromic cleft lip with or without cleft palate (nsCL/P, MIM 119530) is perhaps the most common major birth defect. Homozygous PVRL1 loss-of-function mutations result in an autosomal recessive CL/P syndrome, CLPED1, and a PVRL1 nonsense mutation is associated with sporadic nsCL/P in Northern Venezuela. To address the more general role of PVRL1 variation in risk of nsCL/P, we carried out mutation analysis of PVRL1 in North American and Australian nsCL/P cases and population-matched controls. We identified a total of 15 variants, 5 of which were seen in both populations and 1 of which, an in-frame insertion at Glu442, was more frequent in patients than in controls in both populations, though the difference was not statistically significant. Another variant, which is specific to the PVRL1 β (HIgR) isoform, S447L, was marginally associated with nsCL/P in North American Caucasian patients, but not in Australian patients, and overall variants that affect the β-isoform were significantly more frequent among North American patients. One Australian patient had a splice junction mutation of PVRL1. Our results suggest that PVRL1 may play a minor role in susceptibility to the occurrence of nsCL/P in some Caucasian populations, and that variation involving the β (HIgR) isoform might have particular importance for risk of orofacial clefts. Nevertheless, these results underscore the need for studies that involve very large numbers when assessing the possible role of rare variants in risk of complex traits such as nsCL/P.
Introduction
Cleft lip with or without cleft palate (CL/P) is one of the most common birth defects, occurring in approximately 1 per 800 North American Caucasian infants (Tolarova and Cervenka, 1998), and also with high frequency in other populations around the world. Approximately two-thirds of CL/P cases occur as an isolated, sporadic birth defect. Such nonsyndromic CL/P (nsCL/P) appears to be a multifactorial, polygenic disorder, each locus exerting a relatively modest effect against a complex outbred background (Mitchell and Risch, 1992; Mitchell, 1997).
Many candidate genes for nsCL/P have been assessed, with varying degrees of support for a large number (Schutte and Murray, 1999; Bender, 2000; Spritz, 2001; Cobourne, 2004; Stanier and Moore, 2004). Several lines of evidence support a possible role in nsCL/P for one or more genes of the nectin family, which encode a group of cell adhesion molecules. Homozygous loss-of-function mutations in the gene encoding nectin-1, PVRL1, result in a rare autosomal recessive CL/P syndrome, termed CLPED1 (Suzuki et al., 2000). A heterozygous nonsense mutation of PVRL1 has been associated with sporadic nsCL/P in Northern Venezuela (Sozen et al., 2001). Several subsequent studies have supported (Turhani et al., 2005; Avila et al., 2006; Neiswanger et al., 2006; Scapoli et al., 2006) or not supported (Scapoli et al., 2004; Ichikawa et al., 2006; Tseng et al., 2006) a role for PVRL1 in risk of nsCL/P. Two other genes of the nectin family, PVR and PVRL2, have also been considered as possible candidate genes for nsCL/P (Neiswanger et al., 2006; Warrington et al., 2006; Pezzetti et al., 2007), due to their paralogy with PVRL1 and their location in chromosome segment 19q13.2, which corresponds to a linkage region for nsCL/P, OFC3 (MIM 600757; Stein et al., 1995).
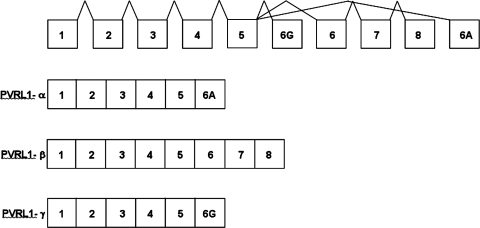
PVRL1 encodes three distinct proteins (Lopez et al., 2001), the result of alternative RNA splicing, each sharing an amino-terminal segment, consisting of three immunoglobulin domains, but with three completely different carboxyl-terminal segments encoded by different exons (Suzuki et al., 2000) (Fig. 1). The PVRL1 α-isoform encodes nectin-1 (PRR1), the cell-surface transmembrane receptor of a cell–cell adhesion system (Takahashi et al., 1999). The PVRL1 γ-isoform encodes a truncated PVRL1 protein that may regulate nectin-mediated cell adhesion by competitive inhibition (Lopez et al., 2001). The PVRL1 β-isoform encodes HIgR, an apparent transmembrane receptor with a carboxyl segment entirely different from nectin-1 and whose specific function is unknown. As shown in Figure 1, the PVRL1 β-isoform is encoded by exons 1–8, the PVRL1 α-isoform by exons 1–5 and exon 6A, and the PVRL1 γ-isoform by exons 1–5 and exon 6G.
FIG. 1.
Schematic genomic organization of PVRL1, and alternative RNA splicing to generate PVRL1 α-, β-, and γ-mRNA isoforms.
The aim of this study was to investigate possible involvement of the PVRL1 gene in risk of nsCL/P in Caucasian populations. We carried out mutation analysis of both nsCL/P patients and population-matched controls, screening all coding exons of the PVRL1 gene encompassing all three PVRL1 gene isoforms, so as to determine whether variants of PVRL1 or any specific isoform may contribute to risk of nsCL/P in Caucasians.
Materials and Methods
Mutation screening, genotyping, and statistics
Genomic DNA samples were obtained with informed consent from patients with nsCL/P and controls from different populations in North America. We analyzed DNA samples from 104 nsCL/P patients and 105 controls from North America, including 44 from Texas, 20 from Maryland, 20 from Ohio, and 20 from Iowa, as well 112 nsCL/P patients and 118 controls from Australia. DNA was isolated from bloodspots (Polski et al., 1998) and used as template for polymerase chain reaction (PCR), using primers for amplicons spanning the 10 PVRL1 exons described previously (Suzuki et al., 2000), as well as additional primers specified in Table 1. We screened amplicons spanning the 10 PVRL1 exons, and adjacent intron and noncoding sequences, by simultaneous single-strand conformation polymorphism (SSCP)/heteroduplex analysis for the North American samples, and by denaturing high-performance liquid chromatography (dHPLC) for the Australian samples. Variants were defined by purifying the amplified products by electrophoresis in 0.5 × MDE gels (Biowhittaker Molecular Applications, Rockland, ME) containing 10% glycerol (Lee et al., 1995) and sequencing PCR products showing novel aberrant SSCP/heteroduplex patterns. Allele frequencies were analyzed using Fisher's exact test, one-tailed.
Table 1.
PVRL1 Polymerase Chain Reaction Primers
| Amplicon | Primer sequence | Isoform |
|---|---|---|
| Exon 1F | 5′-CTG GTTTCTGCTGCGCGAGGA-3′ | All |
| Exon 1R | 5′-AGCAGAGGTGAGCGCCTCTTA-3′ | All |
| Exon 2Fa | 5′-CAGAGCACAGGAACTGTGTGGGT-3′ | All |
| Exon 2R | Suzuki et al. (2000) | All |
| Exon 3F | Suzuki et al. (2000) | All |
| Exon 3R | Suzuki et al. (2000) | All |
| Exon 4F | Suzuki et al. (2000) | All |
| Exon 4R | Suzuki et al. (2000) | All |
| Exon 5F | Suzuki et al. (2000) | All |
| Exon 5R | Suzuki et al. (2000) | All |
| Exon 6 gamma F | 5′-AGCCCAAAGCCATAGTGC-3′ | γ |
| Exon 6 gamma R | 5′-CAGCTGTCTGACATCAAGC-3′ | γ |
| Exon 6 gamma R2 | 5′-GTCCCAGGTGAAGTCTCTC-3′ | γ |
| Exon 6A F | Suzuki et al. (2000) | α (PRR1) |
| Exon 6A R | Suzuki et al. (2000) | α (PRR1) |
| Exon 6A F 1. half | Suzuki et al. (2000) | α (PRR1) |
| Exon 6A R2 1. half | 5′-GTCCTCGTCATATTTGGGGT-3′ | α (PRR1) |
| Exon 6A F 2 2. half | 5′-TCAGACGACGAGAAGAAGGC-3′ | α (PRR1) |
| Exon 6A R 2. half | Suzuki et al. (2000) | α (PRR1) |
| Exon 6b F3 | 5′-CAGGGTCTGACTCTCTCACA-3′ | β (HIgR) |
| Exon 6b R2 | 5′-ACCAGTGGGAGTTTAGTGGGCA-3′ | β (HIgR) |
| Exon 6b R3 | 5′-CTGAGCTTTCACAAGTTTAG-3′ | β (HIgR) |
| Exon 7b F | Suzuki et al. (2000) | β (HIgR) |
| Exon 7b R | Suzuki et al. (2000) | β (HIgR) |
| Exon 8b F | Suzuki et al. (2000) | β (HIgR) |
| Exon 8b R | Suzuki et al. (2000) | β (HIgR) |
| Exon 8b F2 | 5′-GAAAGGTCTGTGCAGCACTC-3′ | β (HIgR) |
| Exon 8b R2 | 5′-CTAAGGCAGCTGGGCTCAT-3′ | β (HIgR) |
| Exon 8b F3 | 5′-ATGGAAAGGTCTGTGCAGCA-3′ | β (HIgR) |
| Exon 8b R3 | 5′-CTCTAAGGCAGCTGGGCTCA-3′ | β (HIgR) |
Slightly modified from Suzuki et al. (2000).
F, forward primer; R, reverse primer.
Results
We carried out a case–control survey of PVRL1 variants among 104 unrelated Caucasian nsCL/P patients from North America versus 105 unrelated North American Caucasian controls, and 112 unrelated Australian Caucasian nsCL/P patients versus 118 unrelated Australian Caucasian controls. We screened the 10 exons of the PVRL1 gene, and adjacent intron and noncoding sequences, by simultaneous SSCP/heteroduplex or dHPLC analysis, followed by DNA sequencing of PCR products that contained apparent variants.
As shown in Table 2, among North American Caucasian nsCL/P patients and controls we identified a total of eight variants, two of which, 442insE and G361V, were relatively common and slightly more frequent in patients than in controls. Two variants affect all three PVRL1 isoforms, three affect only the α (PRR1) isoform, and three affect only the β (HIgR) isoform. When we considered each variant individually, only one, S447L, was significantly more prevalent among cases than among controls (p = 0.033). When we considered all variants affecting each isoform together, while there was no significant difference between nsCL/P cases and controls for variants affecting the α (PRR1) isoform or γ-isoform variants affecting the β (HIgR) isoform were significantly more frequent in nsCL/P cases than in controls (p = 0.006).
Table 2.
PVRL1 Variants in North American Caucasian Nonsyndromic Cleft Lip Patients with or without Cleft Palate, and Controls
| Variant | Exon | PVRL1 isoform affected | Allele frequency among nsCL/P patients (n = 104) | Allele frequency among controls (n = 105) | p-Valuea |
|---|---|---|---|---|---|
| V89M (GTG > ATG) | 2 | α,β,γ | 2/208 (0.010) | 0 | ns |
| R199Q (CGG > CAG) | 3 | α,β,γ | 0 | 2/210 (0.010) | ns |
| 442insE (insGAG) | 6A | α only | 22/208 (0.106) | 16/210 (0.080) | ns |
| 442insEE (insGAGGAG) | 6A | α only | 1/208 (0.005) | 2/210 (0.010) | ns |
| G507E (GGG > GAG) | 6A | α only | 0 | 1/210 (0.005) | ns |
| G361V (GGT > GTT) | 6 | β only | 20/208 (0.096) | 13/210 (0.062) | ns |
| P393P (CCG > CCA) | 7 | β only | 7/208 (0.034) | 2/210 (0.010) | ns |
| S447L (TCG > CCT) | 8 | β only | 7/208 (0.034) | 1/210 (0.005) | 0.033 |
| Total affecting α (PRR) isoform | 25 | 21 | ns | ||
| Total affecting β (HIgR) isoform | 36 | 18 | 0.006 | ||
| Total affecting γ isoform | 2 | 2 | ns |
Fisher's exact test, one-tailed.
ns, nonsignificant; nsCL/P, nonsyndromic cleft lip with or without cleft palate.
However, this finding was not replicated in Australian cases and controls, in whom we identified a total of 12 variants, of which 442insE and G361V again were relatively common. As shown in Table 3, no individual variant occurred significantly more frequently among cases than among controls. Further, when we considered variants affecting each isoform together, we likewise found no significant difference. Nevertheless, in one Australian nsCL/P patient we observed a splice junction mutation, IVS4 + 1G > A, that constitutes a clear loss-of-function mutation that would abolish expression of all three PVRL1 mRNA isoforms from this allele.
Table 3.
PVRL1 Variants in Australian Caucasian Nonsyndromic Cleft Lip Patients with or without Cleft Palate, and Controls
| Variant | Exon | PVRL1 isoform affected | Allele frequency among nsCL/P patients (n = 112) | Allele frequency among controls (n = 118) | p-Valuea |
|---|---|---|---|---|---|
| V28I (GTC > ATC) | 2 | α,β,γ | 0 | 1/230 (0.005) | ns |
| L114L (CTG > CTA) | 2 | α,β,γ | 0 | 1/230 (0.005) | ns |
| E125E (GAG > GAA) | 2 | α,β,γ | 0 | 1/230 (0.005) | ns |
| T187T (ACT > ACA) | 3 | α,β,γ | 1/220 (0.005) | 0 | ns |
| R199Q (CGG > CAG) | 3 | α,β,γ | 4/216 (0.019) | 3/230 (0.013) | ns |
| T206T (ACG > ACA) | 3 | α,β,γ | 1/220 (0.005) | 1/230 (0.005) | ns |
| IVS4 + 1G > A | 4 | α,β,γ | 1/220 (0.005) | 0 | ns |
| 442insE (insGAG) | 6A | α only | 30/220 (0.136) | 23/230 (0.100) | ns |
| 442insEE (insGAGGAG) | 6A | α only | 0 | 1/230 (0.005) | ns |
| E335D (GAA > GAC) | 6 | β only | 20/220 (0.091) | 19/236 (0.081) | ns |
| G361V (GGT > GTT) | 6 | β only | 0 | 1/234 (0.005) | ns |
| P393P (CCG > CCA) | 7 | β only | 1/222 (0.005) | 0 | ns |
| Total affecting α (PRR1) isoform | 37 | 31 | ns | ||
| Total affecting β (HIgR) isoform | 28 | 28 | ns | ||
| Total affecting γ isoform | 7 | 7 | ns |
Fisher's exact test, 1-tailed.
ns, nonsignificant; nsCL/P, nonsyndromic cleft lip with or without cleft palate.
Analysis of rare variants in a complex disease is problematic. In any given study, the observed frequencies of any specific variant may be too low to permit reliable conclusions to be drawn. This problem is compounded if the gene in question contributes only a small fraction of total liability. In the present study, we observed a higher frequency of the PVRL1 β (HIgR) isoform-specific variant, S447L, in North American nsCL/P cases than in North American controls, as well as a higher frequency of variants affecting the β-isoform overall. However, we did not observe the S447L variant among Australians at all, either in cases or in controls, suggesting that these two populations may not be strictly comparable. Moreover, in the course of this study we found one splice junction mutation (IVS4 + 1G > A) in an Australian nsCL/P patient. While this variant is of obvious functional significance, and would abolish expression of all three PVRL1 isoforms from the variant allele, the general frequency of obviously deleterious alleles of PVRL1 in different populations is not yet known. It is clear that sequence analyses of far larger numbers of cases and controls will be necessary to assess the relevance of rare PVRL1 variants to the pathogenesis of nsCL/P.
Discussion
PVRL1 is expressed in the developing palatal epithelium, tooth buds, and skin keratinocytes, consistent with the phenotype of patients with CLPED1 syndrome who completely lack all three isoforms because of null-mutant alleles (Suzuki et al., 2001). The slightly increased frequency of PVRL1 variants that affect the β- (HIgR) and α-isoforms in patients with sporadic, nsCL/P suggests that the β-isoform might be specifically involved in craniofacial development. The γ-isoform may thus relate more to the skin-hair, tooth, and hand aspects of the CLPED1 syndromic phenotype. It will thus be of interest to assess the developmental expression of each of the three PVRL1 isoforms individually. Further, whereas the PVRL1 α (PRR1) isoform is a cell–cell adhesion molecule that interfaces with afadin via its intracellular carboxyl domain (Takahashi et al., 1999), the intracellular carboxyl domain of the β (HIgR) isoform is entirely different, suggesting that its function likewise is entirely different. It thus will be of great importance to determine the function and biology of the PVRL1 HIgR isoform and clarify its role in craniofacial development.
Acknowledgments
This work was supported by National Institutes of Health Grant DE13571 to R.A.S. We thank Dr. Jeff Murray, Dr. Georgia Trench, and Dr. Iain McIntosh for their contribution of DNA samples for this study.
Disclosure Statement
No competing financial interests exist.
References
- Avila JR. Jezewski PA. Vieira AR, et al. PVRL1 variants contribute to non-syndromic cleft lip and palate in multiple populations. Am J Med Genet A. 2006;140:2562–2570. doi: 10.1002/ajmg.a.31367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender PL. Genetics of cleft lip and palate. J Pediatr Nurs. 2000;15:242–249. doi: 10.1053/jpdn.2000.8148. [DOI] [PubMed] [Google Scholar]
- Cobourne MT. The complex genetics of cleft lip and palate. Eur J Orthod. 2004;26:7–16. doi: 10.1093/ejo/26.1.7. [DOI] [PubMed] [Google Scholar]
- Ichikawa E. Watanabe A. Nakano Y, et al. PAX9 and TGFB3 are linked to susceptibility to nonsyndromic cleft lip with or without cleft palate in the Japanese: population-based and family-based candidate gene analyses. J Hum Genet. 2006;51:38–46. doi: 10.1007/s10038-005-0319-8. [DOI] [PubMed] [Google Scholar]
- Lee ST. Park SK. Lee KH, et al. A non-radioactive method for simultaneous detection of single-strand conformation polymorphisms (SSCPs) and heteroduplexes. Mol Cells. 1995;5:668–672. [Google Scholar]
- Lopez M. Cocchi F. Avitabile E, et al. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J Virol. 2001;75:5684–5691. doi: 10.1128/JVI.75.12.5684-5691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE. Genetic epidemiology of birth defects: nonsyndromic cleft lip and neural tube defects. Epidemiol Rev. 1997;19:61–68. doi: 10.1093/oxfordjournals.epirev.a017947. [DOI] [PubMed] [Google Scholar]
- Mitchell LE. Risch N. Mode of inheritance of nonsyndromic cleft lip with or without cleft palate: a reanalysis. Am J Hum Genet. 1992;51:323–332. [PMC free article] [PubMed] [Google Scholar]
- Neiswanger K. Deleyiannis FW. Avila JR, et al. Candidate genes for oral-facial clefts in Guatemalan families. Ann Plast Surg. 2006;56:518–521. doi: 10.1097/01.sap.0000210261.65455.9d. [DOI] [PubMed] [Google Scholar]
- Pezzetti F. Palmieri A. Martinelli M, et al. Linkage disequilibrium analysis of two genes mapping on OFC3: PVR and PVRL2. Eur J Hum Genet. 2007;15:992–994. doi: 10.1038/sj.ejhg.5201868. [DOI] [PubMed] [Google Scholar]
- Polski JM. Kimzey S. Percival RW, et al. Rapid and effective processing of blood specimens for diagnostic PCR using filter paper and Chelex-100. Mol Pathol. 1998;51:215–217. doi: 10.1136/mp.51.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapoli L. Palmieri A. Martinelli M, et al. Study of the PVRL1 gene in Italian nonsyndromic cleft lip patients with or without cleft palate. Ann Hum Genet. 2006;70:410–413. doi: 10.1111/j.1529-8817.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- Scapoli L. Palmieri A. Pezzetti F, et al. Investigation of the W185 × nonsense mutation of PVRL1 gene in Italian nonsyndromic cleft lip and palate patients. Am J Med Genet A. 2004;127:211. doi: 10.1002/ajmg.a.20684. [DOI] [PubMed] [Google Scholar]
- Schutte BC. Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Sozen MA. Suzuki K. Tolarova MM, et al. Mutation of PVRL1 is associated with sporadic, non-syndromic cleft lip/palate in northern Venezuela. Nat Genet. 2001;29:141–142. doi: 10.1038/ng740. [DOI] [PubMed] [Google Scholar]
- Spritz RA. The genetics and epigenetics of orofacial clefts. Curr Opin Pediatr. 2001;13:556–560. doi: 10.1097/00008480-200112000-00011. [DOI] [PubMed] [Google Scholar]
- Stanier P. Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet. 2004;1:R73–R81. doi: 10.1093/hmg/ddh052. [DOI] [PubMed] [Google Scholar]
- Stein J. Mulliken JB. Stal S, et al. Nonsyndromic cleft lip with or without cleft palate: evidence of linkage to BCL3 in 17 multigenerational families. Am J Hum Genet. 1995;57:257–272. [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. Hu D. Bustos T, et al. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat Genet. 2000;25:427–430. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Nakanishi H. Miyahara M, et al. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolarova MM. Cervenka J. Classification and birth prevalence of orofacial clefts. Am J Med Genet. 1998;75:126–137. [PubMed] [Google Scholar]
- Tseng YT. Hsiao HH. Hsiao HP, et al. A study of PVRL1 mutations for non-syndromic cleft lip and/or palate among Taiwanese patients. Int J Oral Maxillofac Surg. 2006;35:453–455. doi: 10.1016/j.ijom.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Turhani D. Item CB. Watzinger E, et al. Mutation analysis of CLPTM 1 and PVRL 1 genes in patients with non-syndromic clefts of lip, alveolus and palate. J Craniomaxillofac Surg. 2005;33:301–306. doi: 10.1016/j.jcms.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Warrington A. Vieira AR. Christensen K, et al. Genetic evidence for the role of loci at 19q13 in cleft lip and palate. J Med Genet. 2006;43:e26. doi: 10.1136/jmg.2005.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]