Abstract
Small interfering RNAs (siRNAs) direct RNA interference (RNAi) in eukaryotes. In flies, somatic cells produce siRNAs from exogenous double-stranded RNA (dsRNA) as a defense against viral infection. We identified endogenous siRNAs (endo-siRNAs), 21 nucleotides in length, that correspond to transposons and heterochromatic sequences in the somatic cells of Drosophila melanogaster. We also detected endo-siRNAs complementary to messenger RNAs (mRNAs); these siRNAs disproportionately mapped to the complementary regions of overlapping mRNAs predicted to form double-stranded RNA in vivo. Normal accumulation of somatic endo-siRNAs requires the siRNA-generating ribonuclease Dicer-2 and the RNAi effector protein Argonaute2 (Ago2). We propose that endo-siRNAs generated by the fly RNAi pathway silence selfish genetic elements in the soma, much as Piwi-interacting RNAs do in the germ line.
Three RNA-silencing pathways have been identified in flies and mammals: RNA interference (RNAi), guided by small interfering RNAs (siRNAs) derived from exogenous double-stranded RNA (dsRNA); the microRNA (miRNA) pathway, in which endogenous small RNAs repress partially complementary mRNAs; and the Piwi-interacting RNA (piRNA) pathway, whose small RNAs repress transposons in the germ line (1-3) and can activate transcription in heterochromatin (4).
Endogenous siRNAs (endo-siRNAs) silence retrotransposons in plants (5, 6), and siRNAs corresponding to the L1 retrotransposon have been detected in cultured mammalian cells (7). Genetic and molecular evidence suggests that in addition to suppressing viral infection, the RNAi pathway silences selfish genetic elements in the fly soma: Mutations in the RNAi gene rm62 (8) suppress mutations caused by retroelement insertion (9); depletion of the Argonaute proteins Ago1 or Ago2 increases transposon expression in cultured Drosophila Schneider 2 (S2) cells (10); small RNAs have been detected in Drosophila Kc cells for the 1360 transposon (11) and are produced during transgene silencing in flies (12); and siRNAs have been proposed to repress germline expression of suffix, a short interspersed nuclear element (SINE) (13).
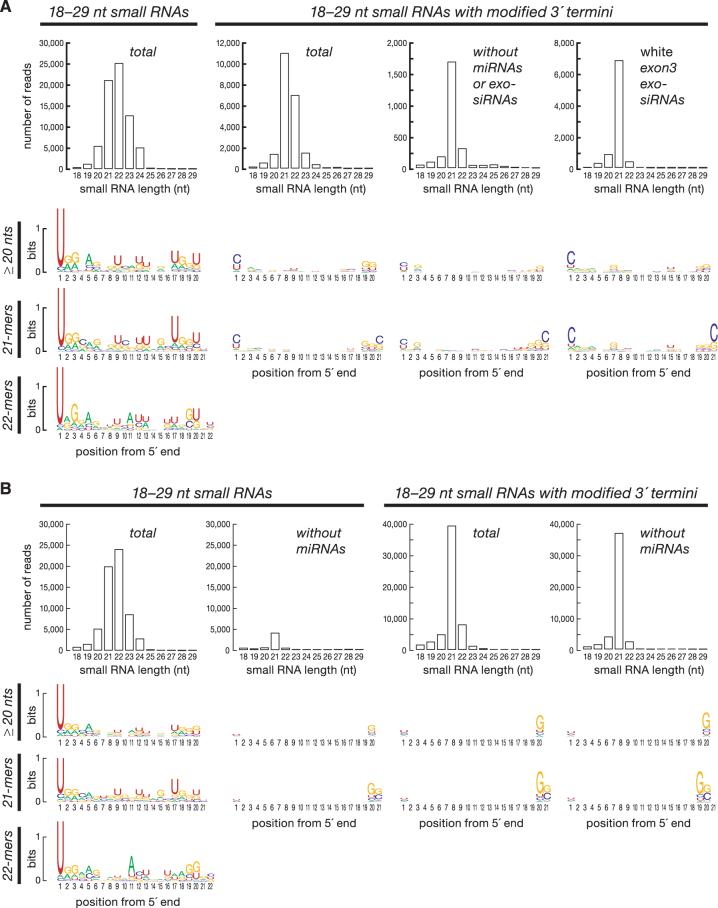
The defining properties of Drosophila siRNAs are their production from long dsRNA by Dicer-2 (Dcr-2), which generates 5′-monophosphate termini; their loading into Argonaute2 (Ago2); and their Ago2-dependent, 3′-terminal, 2′-O-methylation by the methyltransferase Hen1 (14-16), unlike most miRNAs (17). In vivo (Fig. 1A, rightmost panel) and in vitro (18), nearly all siRNAs produced by Dcr-2 from exogenous dsRNA are 21 nucleotides (nt) in length.
Fig. 1.
High-throughput pyrosequencing revealed 3′-terminally modified 21-nt RNAs in the fly soma. (A) Length and sequence composition of the small RNA sequences from a library of total small RNA from the heads of flies expressing an inverted repeat (IR) silencing the white gene and for a parallel library enriched for RNAs modified at their 3′ ends. (B) Similar analysis for small RNA sequences from Drosophila S2 cells. For data labeled “without miRNAs,” pre-miRNA–matching sequences were removed computationally.
We characterized the somatic small RNA content of S2 cells (19) and of heads expressing an RNA hairpin silencing the white gene by RNAi (20). To identify endo-siRNA candidates, we analyzed two types of RNA libraries. For total 18- to 29-nt RNA libraries, 89% (S2 cells) and 96% (heads) mapped to annotated miRNA loci. In contrast, libraries enriched for small RNAs bearing a 3′-terminal, 2′-O-methyl modification (21) were depleted of miRNAs: Only 19% (S2 cells) and 49% (heads) of reads and 2.4% (S2 cells; 58,681 reads; 12,036 sequences) and 12% (heads; 22,685 reads; 2929 sequences) of unique sequences mapped to miRNA loci.
Figure 1 shows the length distribution and sequence composition of the four libraries. The total RNA samples were predominantly miRNAs, a bias reflected in their modal length (22 nt) and pronounced tendency to begin with uracil. Exclusion of miRNAs revealed a class of small RNAs with a narrow length distribution and no tendency to begin with uracil. Except for an unusual cluster of X-chromosome small RNAs (fig. S1) and a miRNA-like sequence with an unusual putative precursor on chromosome 2 (fig. S2), few of these small RNAs are likely to correspond to novel miRNAs: None lie in the arms of hairpins predicted to be as thermodynamically stable as most pre-miRNAs (i.e., < −15 kcal/mol).
After excluding known miRNAs, 64% (heads) (Fig. 1A) and 78% (S2 cells) (Fig. 1B) of sequences in the libraries enriched for 3′-terminally modified small RNAs—that is, those likely to be Ago2-associated—were 21 nt long. For fly heads, 37% (8404 reads) derived from the white dsRNA hairpin. The abundance of these exo-siRNAs can be estimated by comparing them to the number of reads for individual miRNAs in the total small RNA library, where 1.6% (660 antisense and 491 sense reads) were 21-nt oligomers (21-mers) and matched the white sequences in the dsRNA-expressing transgene. The collective abundance of all white exo-siRNAs was less than the individual abundance of the 10 most abundant miRNAs in this sample; the median abundance of any one exo-siRNA species was two reads. The white–inverted repeat (IR) transgene phenocopies a nearly null mutation in white, yet the sequence of the most abundant exo-siRNA was read just 37 times.
In heads, the sequence composition of the 21-nt, 3′-terminally modified small RNAs closely resembled that of exo-siRNAs, which tended to begin and end with cytosine. In heads and S2 cells, the 21-mers lacked the sequence features of piRNAs, which either begin with uracil (Auband Piwi-bound) or contain an adenine at position 10 (Ago3-bound) and are 23 to 29 nt long (1, 2). These data suggest that the 21-nt small RNAs are somatic endo-siRNAs.
In S2 cells, endo-siRNAs mapped largely to transposons (86%); in fly heads, they mapped about equally to transposons, intergenic and unannotated sequences, and mRNAs. The finding that 41% of endo-siRNAs mapped to mRNAs without mapping to transposons suggests that endo-siRNAs may regulate mRNA expression. Endo-siRNAs mapping to mRNAs were likelier by a factor of >10 than expected by chance (5.22 × 10−161 < P < 8 × 10−151) to derive from genomic regions annotated to produce overlapping, complementary transcripts (Table 1 and table S1). These data suggest that such overlapping, complementary transcripts anneal in vivo to form dsRNA that is diced into endo-siRNAs. We note that among the mRNAs for which we detected complementary 21-mers was ago2 itself.
Table 1.
Endo-siRNAs preferentially map to overlapping, complementary mRNAs.
| Sample | Enrichment | Enrichment after randomization |
Z score | P | |
|---|---|---|---|---|---|
| Mean | SD | ||||
| Fly heads | 10.9 | 1.0 | 0.38 | 26.1 | 7.9 × 10−151 |
| S2 cells | 12.3 | 1.1 | 0.42 | 27.0 | 5.2 × 10−161 |
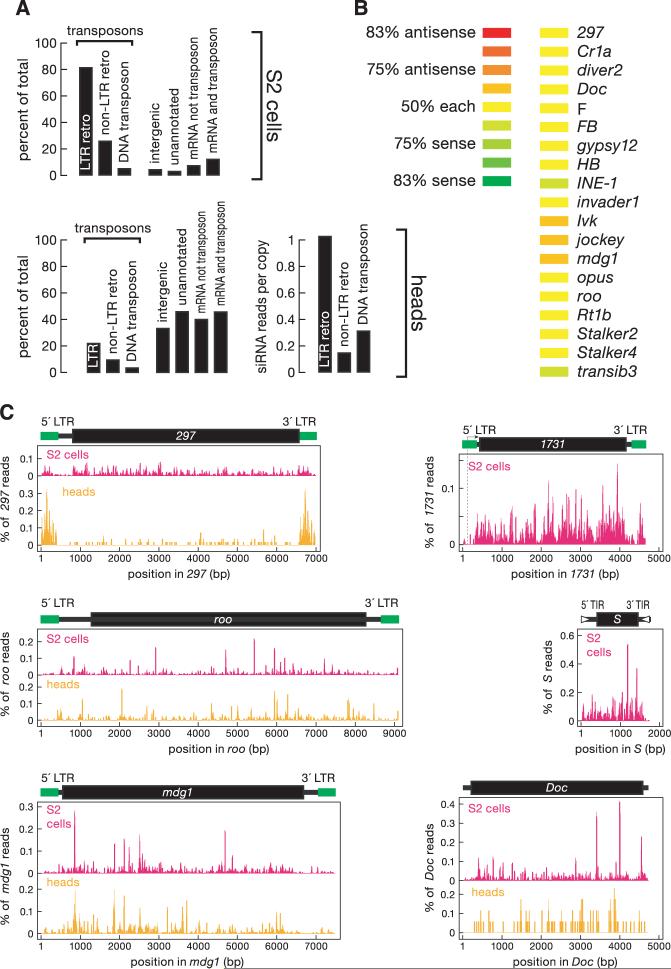
Endo-siRNAs mapped to all three large chromosomes (figs. S3 to S5). siRNAs corresponding to the three transposon types in Drosophila were detected, but long terminal repeat (LTR) retrotransposons, the dominant class of selfish genetic elements in flies, were overrepresented even after accounting for their abundance in the genome (Fig. 2A and table S2). Unlike piRNAs, which are disproportionately antisense to transposons, but like siRNAs derived from exogenous dsRNA, about equal numbers of sense and anti-sense transposon-matching endo-siRNAs were detected (Fig. 2B and fig. S6) (1-3, 22). Like piRNAs, endo-siRNAs map to large genomic clusters (table S3). Of 172 endo-siRNA clusters in S2 cells, four coincided with previously identified piRNA clusters (cluster 1, at 42A of chromosome 2R; clusters 7 and 10 in unassembled genomic sequence; and cluster 15 in the chromosome 3L heterochromatin). In heads, we detected 17 clusters; five corresponded to clusters found in S2 cells, but only one was shared with the germline piRNAs: the flamenco locus, consistent with recent genetic evidence that a Piwi-independent but flamenco-dependent pathway represses the Idefix and ZAM transposons in the soma (23). That both endo-siRNAs and piRNAs can arise from the same region suggests either that a single transcript can be a substrate for both piRNA and siRNA production or that distinct classes of transcripts arise from a single locus. The abundance and distribution of endo-siRNAs across the sequences of individual transposon species reflected the natural history of when the elements entered the fly genome, but not their mechanism of transposition (Fig. 2C) (24).
Fig. 2.
Endo-siRNAs correspond to transposons. (A) Distribution of annotations for the genomic matches of endo-siRNA sequences. Bars total more than 100% because some siRNAs match both LTR and non-LTR retrotransposons or match both mRNA and transposons. (B) Transposon-derived siRNAs with more than 50 21-nt reads mapped about equally to sense and antisense orientations. (C) Alignment of endo-siRNA sequences to Drosophila transposons. The abundance of each sequence is shown as a percentage of all transposon-matching siRNA sequences. LTR, long terminal repeat; TIR, terminal inverted repeat. Here and in subsequent figures, data from high-throughput pyrosequencing and sequencing-by-synthesis were pooled for wild-type heads.
Statistically significant reductions in siRNA abundance were observed in dcr-2L811fsX null mutant heads relative to heads from heterozygous siblings for 38 transposons (fig. S7 and table S4). Normalized for sequencing depth, sequencing results from homozygous dcr-2 mutant heads yielded fewer 21-mers overall (by a factor of 3.1) and fewer 21-mers corresponding to transposons (by a factor of 6.3) than did their heterozygous siblings (P < 2.2 × 10−16; χ2 test). In contrast, overall miRNA abundance—normalized to sequencing depth—was essentially unchanged between dcr-2 heterozygotes and homozygotes (fig. S7 and table S5). These data suggest that endo-siRNAs are produced by Dcr-2, but we do not yet know why some endo-siRNAs persist in dcr-2L811fsX mutants.
Transposon expression in the soma reflects both the silencing of transposons—potentially by either or both posttranscriptional and transcriptional mechanisms—and the tissue specificity of transposon promoters. Drosophila somatic cells may contain siRNAs targeting transposons that would not be highly expressed even in the absence of those siRNAs, because the promoters of those transposons are not active in some or all somatic tissues or because they are repressed by additional mechanisms. We analyzed the expression of a panel of transposons in heads from ago2 and dcr-2 mutants and in S2 cells depleted of Dcr-1, Dcr-2, or Ago2 by RNAi (Fig. 3 and fig. S8). We found that the steady-state abundance of RNA from the LTR retrotransposons 297 and 412 increased in heads from dcr-2L811fsX null mutants (Fig. 3A). Similarly, the steady-state abundance of RNA from the LTR retrotransposons 297, 412, mdg1, and roo, the non-LTR retrotransposon F-element, and the SINE-like element INE-1 increased in ago2414 mutant heads (Fig. 3B).
Fig. 3.
Transposon silencing requires Dcr-2 and Ago2, but not Dcr-1. (A and B) The change in mRNA expression (mean ± SD, N = 3) for each transposon between dcr-2L811fsX (A) or ago2414 (B) heterozygous and homozygous heads was measured by quantitative reverse transcription polymerase chain reaction. The data were corrected for differences in transposon copy number between the paired genotypes. (C) The change in transposon expression (mean ± SD, N = 3) in S2 cells was measured for the indicated RNAi depletion relative to a control dsRNA.
In S2 cells, RNA expression from the LTR retrotransposons 297, 1731, mdg1, blood, and gypsy and from the DNA transposon S-element all increased significantly (0.00001 < P < 0.002) when Dcr-2 was depleted or when both Dcr-2 and Dcr-1 were depleted, but not when Dcr-1 alone was depleted (Fig. 3C). Similarly, ago2(RNAi) in S2 cells desilenced transposons, including nine LTR and non-LTR retrotransposons and the DNA transposon S-element (fig. S8).
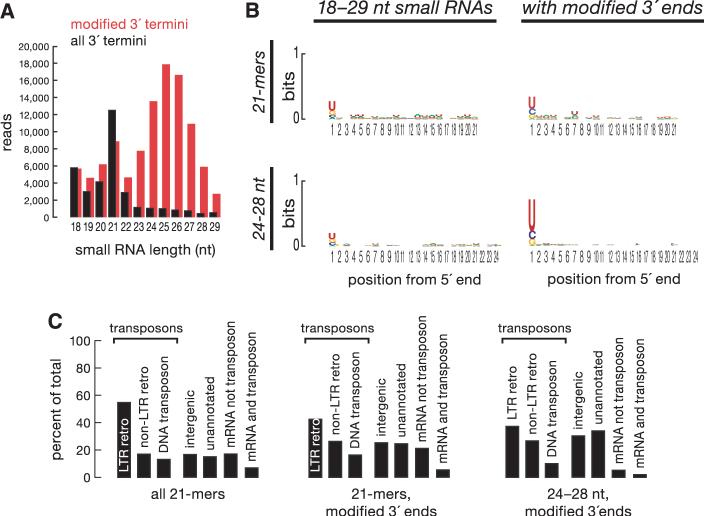
Is Ago2 required for the production or accumulation of endo-siRNAs? We sequenced 18- to 29-nt small RNAs from ago2414 homozygous fly heads and from the same small RNA sample treated to enrich for 3′-terminally modified RNAs. After computationally removing miRNAs, the sequences from the untreated library contained a prominent 21-nt peak (Fig. 4A) that predominantly began with uracil (Fig. 4B), much like miRNAs and unlike siRNAs in wild-type heads, which often began with cytosine (Fig. 1A). Perhaps in the absence of Ago2, only a subpopulation of endo-siRNAs that can bind Ago1 accumulates. The small RNAs from the ago2414 library enriched for 3′-terminally modified sequences were predominantly 24 to 27 nt long and often began with uracil—a length distribution and sequence bias characteristic of piRNAs, which, like siRNAs, are 2′-O-methylated at their 3′ ends. Both the 21-nt small RNAs and the piRNA-like RNAs in the ago2 mutant heads mapped to transposons, unannotated heterochromatic and unassembled sequences, but the piRNA-like sequences mapped to mRNAs far less frequently than did either the 21-mers or wild-type endo-siRNAs (Fig. 4C). How these piRNA-like small RNAs are generated and whether they contribute to transposon silencing in the fly soma remain unknown.
Fig. 4.
The composition of somatic small RNAs is altered in the absence of Ago2. (A and B) Size distribution (A) and sequence composition (B) of sequences from a library of total 18- to 29-nt RNA from the heads of ago2 null mutant flies or a library enriched for 3′-terminally modified RNAs. Reads matching pre-miRNA sequences were removed. (C) Distribution of annotations for the genomic matches of small RNA sequences from the two ago2 libraries.
Note added in proof: The loci described here in figs. S1 and S2 correspond to endo-siRNA–generating hairpins recently identified in (25-27).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1157396/DC1
Materials and Methods
Figs. S1 to S8
Tables S1 to S7
References
References and Notes
- 1.Gunawardane LS, et al. Science. 2007;315:1587. doi: 10.1126/science.1140494. published online 21 February 2007 (10.1126/science.1140494) [DOI] [PubMed] [Google Scholar]
- 2.Brennecke J, et al. Cell. 2007;128:1089. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Vagin VV, et al. Science. 2006;313:320. doi: 10.1126/science.1129333. published online 28 June 2006 (10.1126/science.1129333) [DOI] [PubMed] [Google Scholar]
- 4.Yin H, Lin H. Nature. 2007;450:304. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton A, Voinnet O, Chappell L, Baulcombe D. EMBO J. 2002;21:4671. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunkar R, Girke T, Zhu JK. Nucleic Acids Res. 2005;33:4443. doi: 10.1093/nar/gki758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang N, Kazazian HHJ. Nat. Struct. Mol. Biol. 2006;13:763. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 8.Ishizuka A, Siomi MC, Siomi H. Genes Dev. 2002;16:2497. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csink AK, Linsk R, Birchler JA. Genetics. 1994;138:153. doi: 10.1093/genetics/138.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehwinkel J, et al. Mol. Cell. Biol. 2006;26:2965. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes KA, Caudy AA, Collins L, Elgin SC. Curr. Biol. 2006;16:2222. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal-Bhadra M, Bhadra U, Birchler JA. Mol. Cell. 2002;9:315. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 13.Tchurikov NA, Kretova OV. PLoS ONE. 2007;2:e476. doi: 10.1371/journal.pone.0000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwich MD, et al. Curr. Biol. 2007;17:1265. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. J. Virol. 2007;81:1951. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito K, et al. Genes Dev. 2007;21:1603. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura K, Ishizuka A, Siomi H, Siomi MC. Genes Dev. 2004;18:1655. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nykanen A, Haley B, Zamore PD. Cell. 2001;107:309. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 19.Drosophila RNAi Screening Center at Harvard Medical School. ( http://flyrnai.org/cgi-bin/RNAi_FAQ_lines.pl)
- 20.Lee YS, Carthew RW. Methods. 2003;30:322. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 21.Seitz H, Ghildiyal M, Zamore PD. Curr. Biol. 2008;18:147. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamore PD, Tuschl T, Sharp PA, Bartel DP. Cell. 2000;101:25. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 23.Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. PLoS ONE. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.See supporting material on Science Online.
- 25.Czech B, et al. Nature. 2008 10.1038/nature07007. [Google Scholar]
- 26.Kawamura Y, et al. Nature. 2008 10.1038/nature06938. [Google Scholar]
- 27.Okamura K, et al. Nature. 2008 10.1038/nature07015. [Google Scholar]
- 28.We thank A. Boucher and S. Ma for technical assistance; G. Farley for encouragement, support, and technical assistance; and Roche Applied Science for high-throughput sequencing. P.D.Z. is a W. M. Keck Foundation Young Scholar in Medical Research. Supported by NIH grants GM62862 and GM65236 (P.D.Z.), GM080625 (J.X. and Z.W.), and HG003367 (S.L.); EMBO long-term (ALTF 910−2004) and Human Frontier Science Program (LT00575/2005-L) fellowships (H.S.); and a National Research Service Award predoctoral MD/PhD fellowship from the National Institute on Aging (F30AG030283) (M.D.H.). NCBI Gene Expression Omnibus accession numbers for sequence and abundance data are GSE9389 and GSE11019, respectively.