Abstract
Ferryl species are important catalytic intermediates in heme enzymes. A recent experimental investigation of a diheme protein MauG reported the first case of using two Fe(IV) species as an alternative to compound I in catalysis. Both Fe(IV) species have unusual Mössbauer properties, which was found to originate from novel structural features based on a quantum chemical investigation. With comparison to the previously reported FeIV=O and FeIV–OH species, results here provide the first evidence of a couple of new mechanisms by which proteins influence the properties of ferryl species by directly providing the O via Tyr, or stabilizing exogenous O via hydrogen bonding interaction. These results expand our ability to identify and evaluate high-valent heme proteins and models.
Keywords: Fe(IV) species, heme, Mössbauer, DFT, hydrogen bond
High-valent Fe(IV) species are important intermediates in the catalytic cycles of many heme enzymes.1–9 57Fe Mössbauer spectroscopy is an invaluable tool to probe iron sites and determine quadrupole splitting (ΔEQ) and isomer shift (δFe) parameters, which are related to the electric field gradient and charge density at the iron nucleus, respectively.4 Fe(IV) species in heme proteins4–8 are characterized by small δFe values ranging from 0.03–0.14 mm/s (Figure 1). In contrast, ΔEQ values4–8 span a larger range of 1.02–2.29 mm/s. Thus, ΔEQ may be a more sensitive structural probe. Recent studies suggest that ΔEQ is an indicator of the protonation state of the oxo group,5,6,8 with large and small ΔEQ values proposed for the protonated and unprotonated ferryl species, respectively. A model compound with an unprotonated FeIV=O moiety9 defined crystallographically has indeed a ΔEQ value (1.24 mm/s) at the lower end of this range.
Figure 1.
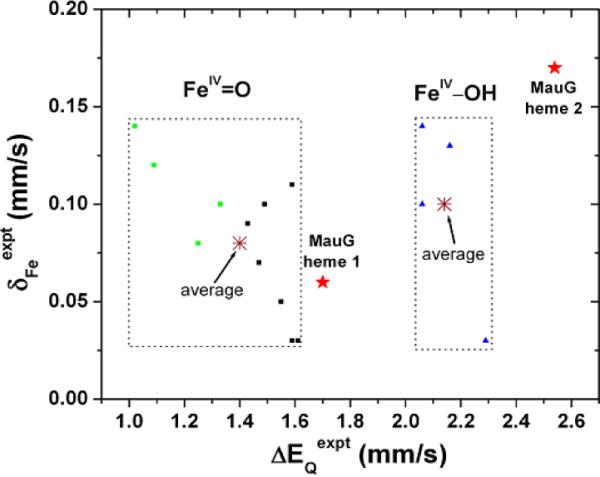
Experimental Mössbauer properties for Fe(IV) heme proteins.4–8,12 Green squares, black squares, and blue triangle points are for compounds I FeIV=O, compound II/ES FeIV=O, and compound II FeIV–OH species, respectively.
MauG10 contains two c-type hemes and catalyzes a six-electron oxidation to complete the biosynthesis of the tryptophan tryptophylquinone cofactor of methylamine dehydrogenase.11 Mössbauer spectroscopy revealed that MauG stabilizes a bis-FeIV intermediate with unusual ΔEQ values for each FeIV heme.12 Heme 1 was regarded as an FeIV=O species, but its ΔEQ value of 1.70 mm/s lies between the average experimental ΔEQ values in heme proteins for protonated and unprotonated forms4–8 (Figure 1). For heme 2, both the ΔEQ and δFe values are larger than any previous known data for Fe(IV) species in heme proteins.12 These results suggest that they may possess structural features that have not been described before. MauG is also the first known protein using two FeIV centers as an alternative to compound I in biological oxidation reactions.12
Quantum chemical investigations of Mössbauer parameters have been useful in elucidating structural features of iron sites in proteins and models.13–22 Here, we present a quantum chemical investigation of these two novel FeIV species in heme proteins, using a recently determined MauG X-ray crystal structure23 as a starting point. The DFT method used here has predicted δEQ and δFe values in iron proteins and model systems covering all iron spin states and coordination states and almost all the iron oxidation states. The theory-versus-experiment correlation coefficient for ΔEQ prediction is R2=0.98 in 48 systems covering an experimental range of 8.80 mm/s and that for δFe prediction is R2=0.97 in 49 systems covering an experimental range of 2.34 mm/s (see Supporting Information for computational details). The standard deviation of these δFe calculations is 0.07 mm/s. It should be noted that our method was calibrated using the small molecules' X-ray structures and the residual errors were found to generally decrease upon using better quality X-ray structures.20 For instance, the error in ΔEQ prediction for the ferryl model compound9 that has a high resolution X-ray structure is 0.01 mm/s.21 Therefore, this type of calculations has assisted in structure refinement for iron-containing proteins.20–22 In this work, this approach was used to evaluate different Fe(IV) models in MauG.
Heme 1 is five-coordinate with a His residue as the axial ligand23 and a vacant site to bind O2 or H2O2. Five FeIV-oxo models (1a–1e in Table 1) were investigated to examine the difference between the unprotonated FeIV=O and protonated FeIV–OH species, and possible hydrogen bonding effects from nearby amino acid residues. As found with the experimental studies,4–9,12 the predicted δFe values in these models are similar and within the expected region, while ΔEQ values are much more sensitive to the structural variations.
Table 1.
Geometric, Electronic, and Mössbauer Properties of MauG Fe(IV) Models (S=1)
| MauG modelsa | RFeO (Å) | RFeN-His (Å) | RFeO-Tyr (Å) | RFeN-Por (Å) | ∠O-Fe-NHis (degrees) | ραβFe (e) | ραβO (e) | ΔEq (mm/s) | δFe (mm/s) | |
|---|---|---|---|---|---|---|---|---|---|---|
| heme 1 | Exptlb | 1.70 | 0.06 | |||||||
| 1a: FeIV(Por)2−(His)0(O)2− | Calcd | 1.653 | 2.333 | / | 2.042 | 176.6 | 1.18 | 0.89 | 1.45 | 0.15 |
| 1b: FeIV(Por)2−(His)0(OH)1− | Calcd | 1.797 | 2.202 | / | 2.032 | 175.3 | 1.99 | 0.09 | 2.95 | 0.10 |
| 1c: FeIV(Por)2−(His)0(O…HB1)2− | Calcd | 1.658 | 2.328 | / | 2.042 | 176.0 | 1.25 | 0.83 | 1.59 | 0.14 |
| 1d: FeIV(Por)2−(His)0(O…HB1')2− | Calcd | 1.658 | 2.330 | / | 2.043 | 176.2 | 1.27 | 0.80 | 1.67 | 0.13 |
| 1e: FeIV(Por)2−(His)0(O…HB2)2− | Calcd | 1.660 | 2.290 | / | 2.044 | 177.2 | 1.28 | 0.79 | 1.55 | 0.13 |
| heme 2 | Exptlb | 2.54 | 0.17 | |||||||
| 2a: FeIV(Por)2−(His)0(O)2− | Calcd | 1.661 | 2.125 | / | 2.031 | 177.8 | 1.13 | 0.94 | 0.84 | 0.13 |
| 2b: FeIV(Por)2−(His)0(OH)1− | Calcd | 1.796 | 2.042 | / | 2.021 | 177.6 | 1.85 | 0.16 | 2.58 | 0.03 |
| 2c: FeIV(Por)2−(Tyr)1−(O)2− | Calcd | 1.677 | / | 2.027 | 2.037 | 175.5 | 1.18 | 0.88 | 0.51 | 0.17 |
| 2d: FeIV(Por)2−(Tyr)1−(OH)1− | Calcd | 1.815 | / | 1.882 | 2.033 | 176.3 | 1.65 | 0.15 | 1.79 | 0.11 |
| 2e: FeIV(Por)2−(His)0(Tyr)1− | Calcd | / | 2.041 | 1.839 | 2.010 | 177.3 | 1.25 | 0.30 | 2.48 | 0.24 |
| 2f: FeIV(Por')2−(His')0(Tyr')1− | Calcd | / | 2.016 | 1.839 | 2.005 | 174.9 | 1.18 | 0.37 | 2.75 | 0.24 |
Por, HB1, and HB2 stand for the porphyrin structure, Gln103, and Gln103/Pro107 residues.
See Text for HB1', Por', His', and Tyr'.
As seen from Table 1, for the unprotonated FeIV=O model 1a, the predicted Fe-oxo distance and O-Fe-NHis angle are similar to those seen in the X-ray structure of an FeIV=O model compound (1.646 Å and 178.9°)9 with a neutral N-coordination ligand similar to His investigated here. The predicted spin densities in Fe and O are also similar.21 Its ΔEQ value of 1.45 mm/s is close to the average value of 1.4 mm/s seen for unprotonated FeIV=O species in heme proteins (Figure 1). For the protonated FeIV–OH model 1b, the Fe-O distance and Fe spin density are similar to those of the FeIV–OH species in heme proteins.5,6,8 The 103% increase in ΔEQ caused by protonation of the oxo group (see Table 1 for results of 1b vs. 1a) is comparable to an average increase of 112% in other heme proteins.5,6,8
It can be seen from Table 1 that the experimental ΔEQ value of MauG heme 1 lies between the ΔEQ values of the protonated FeIV–OH model 1b and the unprotonated FeIV=O model 1a, and is closer to the latter one. This suggests that a secondary effect from nearby residue(s) may operate on the unprotonated FeIV=O species in MauG heme 1. Therefore, models 1c–1e were built on the basis of the unprotonated FeIV=O model which includes residues Gln103 and Pro107 that reside near heme 1 in the crystal structure to investigate such effects. Model 1c includes only Gln103 with its terminal N-H hydrogen bonded to the oxo group of the ferryl moiety (see Supporting Information for computational details). Interestingly, as shown in Table 1, this hydrogen bond reduces the error in the δEQ prediction from 0.25 mm/s in 1a to 0.11 mm/s in 1c. The error can be further reduced to be 0.03 mm/s in 1d (see Figure 2A), if Gln103 is allowed to move with no constraints from the resting state X-ray structure to further optimize its interaction with the FeIV=O group. This kind of hydrogen bond effect is similar to the ΔEQ changes of 0.1–0.2 mm/s reported previously in other heme protein systems.8,24 Results here suggest for the first time that an Fe(IV)=O species may be stabilized by an active site residue, which in MauG was experimentally found to be remarkably stable.12 It is also intriguing that the alignment of the amino acid sequences of MauG proteins indicates that Gln103 is absolutely conserved (see Figure S2 in reference 23). These results suggest that Gln103 may play a functional role in this site, which will be further investigated by mutation studies. In contrast to Gln103, the presence of Pro107 in 1e compared to 1c has minimal effects on the geometries, spin densities, and Mössbauer parameters. Thus, the role of Pro107 is likely structural and perhaps related to the fact that MauG does not require substrate binding to prime it for reactions with oxygen.23 These results suggest that quantum chemical studies of characteristic spectroscopic properties for proteins may help identify the roles of active site residues.
Figure 2.
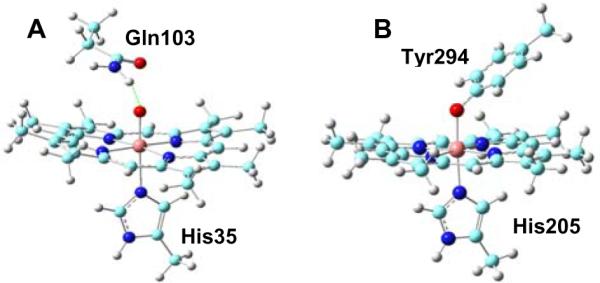
Active site models of MauG FeIV heme sites: (A) 1d; (B) 2e. The green dotted line in (A) represents a hydrogen bond.
For heme 2, the MauG X-ray structure reveals an unusual His/Tyr ligation.23 It has never been observed for c-type hemes or any other heme proteins where function requires the formation of an FeIV oxidation state. In principle, three major types of FeIV hemes may be formed upon the binding of O2 or H2O2 to MauG: 1) His-FeIV-O(H) (2a and 2b); 2) (H)O-FeIV-Tyr (2c and 2d); 3) His-FeIV-Tyr (2e). However, as shown in Table 1, for type 1 and 2 models, either ΔEQ or δFe predictions have much large errors compared to the experimental data, which supports the proposal in the original experimental investigation of MauG that this FeIV heme site has two protein residues as axial ligands.12 To examine the consequence of an FeIV heme with the unique His/Tyr ligand set (type 3), model 2e of FeIV(Por)2−(His)0(Tyr)1− was investigated (Figure 2B). The average Fe and porphyrin nitrogen distance (RFeN-Por) in 2e is similar to the values of 2.01–2.03 Å seen in the isoelectronic FeIV–OH species in previously studied heme proteins6 and the FeIV–OH heme model (1b) here. The long Fe-O bond length of 1.839 Å in 2e is similar to the Fe–O bond (1.84 Å) in a model compound,8 FeIV(TMP)(OCH3)2 (TMP = tetramesitylporphyrin), with the same coordinate state and a similarly high ΔEQ value of 2.12 mm/s.25 It should be noted that the FeIV–OCH3 group is isoelectronic to a FeIV–OH species. So, the ΔEQ value of FeIV (TMP)(OCH3)2 is close to the ΔEQ value of 2.06–2.29 mm/s seen with the FeIV–OH species in other heme proteins.5,6,8
A notable difference from the previously investigated FeIV=O and FeIV–OH heme species is that the Mulliken spin densities of the oxo and iron atoms (ραβO and ραβFe) in 2e are ca. 0.5 e smaller than the two electrons expected for an S=1 state. However, the spin densities of the whole Tyr group and Fe of 1.92 e are indeed close to the expected value, suggesting a delocalization effect of the conjugated Tyr residue. Compared to the FeIV–OH/OCH3 species reported before5,6,8,25 that have dual anionic axial ligands, the unique ligand set of His/Tyr makes 2e isoelectronic to 1b, which also has one neutral His axial ligand and one anionic axial ligand. Note that 1b has a ΔEQ value larger than those of previously reported FeIV–OH/OCH3 species, which is the same for 2e. Since Tyr has only one formal negative charge, which is smaller than the two formal negative charges for an oxo group, 2e has a much longer Fe-O bond compared to those in other FeIV-oxo porphyrins investigated here (see Table 1) and previously.4–8 This decreases the electron charge density at the iron nucleus, which is negatively proportional to δFe and thus results in a much large δFevalue.4 To further examine the effect of this unique Tyr/His ligand set on Mössbauer parameters, a calculation of FeIV(Por')2−(His')0(Tyr')1− using a simple non-substituted porphyr with no protein structural restraints (2f) was performed. As seen from Table 1, both the geometric and Mössbauer results are not identical to those of 2f, indicating an effect of the protein environment. However, both ΔEQ and δFe values of 2f are again much larger than those reported previously, which further supports an important role of the His/Tyr ligand set in determining the unusually large Mössbauer parameters. These results support for a novel FeIVprotein state without an exogenous non-protein ligand.
Overall, the geometric, electronic, and Mössbauer properties from this work suggests new mechanisms by which proteins influence the properties of FeIV=O hemes by directly providing the O via Tyr, or stabilizing exogenous O via hydrogen bonding interaction. These results expand our ability to identify and evaluate high-valent heme proteins and models.
Supplementary Material
Acknowledgment
This work was supported by the NIH grants GM-085774 (YZ) and GM-41574 (VLD). We thank Mississippi Center for Supercomputing research and USM Vislab for using the computing facilities and Lyndal M. R. Jensen and Carrie M. Wilmot for providing the coordinates prior to publication and helpful discussions, and Aimin Liu for his assistance.
Footnotes
Supporting Information Available: Computational details, optimized coordinates, and additional results (Tables S1–S12) are available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Dawson JH. Probing Structure-function Relations in Heme-containing Oxyganses and Peroxidases. Science. 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- (2).Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet BM, Ringe D, Petsko GA, Sligar SG. The Catalytic Pathway of Cytochrome P450cam at Atomic Resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- (3).Shaik S, de Visser SP, Kumar D. One Oxidant, Many Pathways: A Theoretical Perspective of Monooxygenation Mechanisms by Cytochrome P450 enzymes. J. Biol. Inorg. Chem. 2004;9:661–668. doi: 10.1007/s00775-004-0576-6. [DOI] [PubMed] [Google Scholar]
- (4).Debrunner PG. In: Iron Porphyrins. Lever ABP, Gray HB, editors. Vol. 3. VCH Publishers; New York: 1989. pp. 139–234. [Google Scholar]
- (5).Stone KL, Hoffart LM, Behan RK, Krebs C, Green MT. Evidence for Two Ferryl Species in Chloroperoxidase Compound II. J. Am. Chem. Soc. 2006;128:6147–6153. doi: 10.1021/ja057876w. [DOI] [PubMed] [Google Scholar]
- (6).Behan RK, Hoffart LM, Stone KL, Krebs C, Green MT. Evidence for Basic Ferryls in Cytochromes P450. J. Am. Chem. Soc. 2006;128:11471–11474. doi: 10.1021/ja062428p. [DOI] [PubMed] [Google Scholar]
- (7).Garcia-Serres R, Davydov RM, Matsui T, Ikeda-Saito M, Hoffman BM, Huynh BH. Distinct Reaction Pathways Followed upon Reduction of Oxy-heme Oxygenase and Oxymyoglobin as Characterized by Mossbauer Spectroscopy. J. Am. Chem. Soc. 2007;129:1402–1412. doi: 10.1021/ja067209i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Horner O, Mouesca JM, Solari PL, Orio M, Oddou JL, Bonville P, Jouve HM. Spectroscopic Description of an Unusual Protonated Ferryl Species in the Catalase from Proteus mirabilis and Density Functional Theory Calculations on Related Models. Consequences for the Ferryl Protonation State in Catalase, Peroxidase and Chloroperoxidase. J. Biol. Inorg. Chem. 2007;12:509–525. doi: 10.1007/s00775-006-0203-9. [DOI] [PubMed] [Google Scholar]
- (9).Rohde JU, In JH, Lim MH, Brennessel WW, Bukowski MR, Stubna A, Munck E, Nam W, Que L. Science. Vol. 299. 2003. Crystallographic and Spectroscopic Characterization of a Nonheme Fe(IV)=O Complex; pp. 1037–1039. [DOI] [PubMed] [Google Scholar]
- (10).Wang YT, Graichen ME, Liu AM, Pearson AR, Wilmot CM, Davidson VL. MauG, a Novel Diheme Protein Required for Tryptophan Tryptophylquinone Biogenesis. Biochemistry. 2003;42:7318–7325. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- (11).Wilmot CM, Davidson VL. Uncovering Novel Biochemistry in the Mechanism of Tryptophan Tryptophylquinone Cofactor Biosynthesis. Curr. Opin. Chem. Biol. 2009;13:469–474. doi: 10.1016/j.cbpa.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Li XH, Fu R, Lee SY, Krebs C, Davidson VL, Liu AM. A Catalytic Di-heme Bis-Fe(IV) Intermediate, Alternative to an Fe(IV)=O Porphyrin Radical. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8597–8600. doi: 10.1073/pnas.0801643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Han WG, Liu TQ, Lovell T, Noodleman L. Active Site Structure of Class I Ribonucleotide Reductase Intermediate X: A Density Functional Theory Analysis of Structure, Energetics, and Spectroscopy. J. Am. Chem. Soc. 2005;127:15778–15790. doi: 10.1021/ja050904q. [DOI] [PubMed] [Google Scholar]
- (14).Lovell T, Han WG, Liu TQ, Noodleman L. A Structural Model for the High-valent Intermediate Q of Methane Monooxygenase from Broken-symmetry Density Functional and Electrostatics Calculations. J. Am. Chem. Soc. 2002;124:5890–5894. doi: 10.1021/ja0121282. [DOI] [PubMed] [Google Scholar]
- (15).Sinnecker S, Svensen N, Barr EW, Ye S, Bollinger JM, Neese F, Krebs C. Spectroscopic and Computational Evaluation of the Structure of the High-spin Fe(IV)-oxo Intermediates in Taurine: Alpha-Ketoglutarate Dioxygenase from Escherichia coli and its His99Ala Ligand Variant. J. Am. Chem. Soc. 2007;129:6168–6179. doi: 10.1021/ja067899q. [DOI] [PubMed] [Google Scholar]
- (16).Schoneboom JC, Neese F, Thiel W. Toward Identification of the Compound I Reactive Intermediate in Cytochrome P450 Chemistry: A QM/MM Study of its EPR and Mossbauer Parameters. J. Am. Chem. Soc. 2005;127:5840–5853. doi: 10.1021/ja0424732. [DOI] [PubMed] [Google Scholar]
- (17).Neese F. Prediction and Interpretation of the Fe-57 Isomer Shift in Mossbauer Spectra by Density Functional Theory. Inorg. Chim. Acta. 2002;337:181–192. [Google Scholar]
- (18).Grodzicki M, Flint H, Winkler H, Walker FA, Trautwein AX. Electronic Structure, Porphyrin Core Distortion, and Fluxional Behavior of Bis-ligated Low-spin Iron(II) Porphyrinates. J. Phys. Chem. A. 1997;101:4202–4207. [Google Scholar]
- (19).Chanda A, Shan XP, Chakrabarti M, Ellis WC, Popescu DL, de Oliveira FT, Wang D, Que L, Collins TJ, Munck E, Bominaar EL. (TAML)Fe-IV=O Complex in Aqueous Solution: Synthesis and Spectroscopic and Computational Characterization. Inorg. Chem. 2008;47:3669–3678. doi: 10.1021/ic7022902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang Y, Gossman W, Oldfield E. A Density Functional Theory Investigation of Fe-N-O Bonding in Heme Proteins and Model Systems. J. Am. Chem. Soc. 2003;125:16387–16396. doi: 10.1021/ja030340v. [DOI] [PubMed] [Google Scholar]
- (21).Zhang Y, Oldfield E. Cytochrome P450: An Investigation of the Mossbaluer Spectra of a Reaction Intermediate and an Fe(IV)=O Model System. J. Am. Chem. Soc. 2004;126:4470–4471. doi: 10.1021/ja030664j. [DOI] [PubMed] [Google Scholar]
- (22).Zhang Y, Oldfield E. On the Mossbauer Spectra of Isopenicillin N Synthase and a Model {FeNO}(7) (S=3/2) System. J. Am. Chem. Soc. 2004;126:9494–9495. doi: 10.1021/ja0401242. [DOI] [PubMed] [Google Scholar]
- (23).Jensen LMR, Sanishvili R, Davidson VL, Wilmot CM. In Crystallo Posttranslational Modification within a MauG/pre-methylamine Dehydrogenase Complex. Science. 2010;327:1392–1394. doi: 10.1126/science.1182492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ling Y, Zhang Y. In: Annual Reports in Computational Chemistry. Wheeler RA, editor. Vol. 6. Elsevier; New York: 2010. pp. 65–77. [Google Scholar]
- (25).Groves JT, Quinn RQ, Mc Murry TJ, Nakamura M, Lang G, Boso B. Preparation and Characterization of a Dialkoxyiron (IV) Porphyrin. J. Am. Chem. Soc. 1985;107:354–360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


