Abstract
Identifying the nucleotides that cause gene expression variation is a critical step in dissecting the genetic basis of complex traits. Here, we focus on polymorphisms that are predicted to alter transcription factor binding sites (TFBSs) in the yeast, Saccharomyces cerevisiae. We assembled a confident set of transcription factor motifs using recent protein binding microarray and ChIP-chip data and used our collection of motifs to predict a comprehensive set of TFBSs across the S. cerevisiae genome. We used a population genomics analysis to show that our predictions are accurate and significantly improve on our previous annotation. Although predicting gene expression from sequence is thought to be difficult in general, we identified a subset of genes for which changes in predicted TFBSs correlate well with expression divergence between yeast strains. Our analysis thus demonstrates both the accuracy of our new TFBS predictions and the feasibility of using simple models of gene regulation to causally link differences in gene expression to variation at individual nucleotides.
Keywords: Saccharomyces cerevisiae, transcription factors, transcription factor binding sites, population genetics, gene expression, SNP, eQTL
Natural variation in gene expression underlies many diseases (Knight 2004; Cookson et al. 2009) and plays an important role in evolution (Wray 2007; Carroll 2008). A number of studies have demonstrated that gene expression variation is widespread and heritable across a wide range of species, including human, rat, mouse, yeast and Drosophila (Rockman and Kruglyak 2006). Identifying the specific genomic changes that cause gene expression variation is a vital step in understanding phenotypic diversity and the genetic architecture of complex traits. Loci that cause expression variation can be classified as cis- or trans-acting. Cis-acting variation is often thought to be prevalent in evolution because it is believed to cause fewer pleiotropic effects than trans-acting variation (Chen and Rajewsky 2007; Ronald and Akey 2007; Carroll 2008). Despite the presumed importance of cis-acting variation, only a few polymorphisms that cause gene expression differences have been identified, largely because of the difficulty of fine-mapping phenotypic traits in most organisms. We thus explored the feasibility of using genome-wide computational predictions of transcription factor binding sites (TFBSs) to predict nucleotides causing variation in transcript levels, using the yeast, Saccharomyces cerevisiae, as a model system. Although it is clear that gene expression can be regulated posttranscriptionally, our expectation was that transcriptional control would likely play a major role in determining transcript abundance.
In this study, we present two major results. The first result is a major reannotation of the yeast transcription regulatory network. A number of groups have produced TFBS predictions for S. cerevisiae based on motifs inferred from a large set of ChIP-chip experiments (Harbison et al. 2004; Erb and van Nimwegen 2006; Macisaac et al. 2006). We previously published algorithms for predicting transcription factor (TF) motifs (Siddharthan et al. 2005) and predicting TFBSs (van Nimwegen 2007) and also demonstrated the accuracy of the algorithms in the case of S. cerevisiae (Erb and van Nimwegen 2006). The current study builds on our previous work by incorporating a large set of new TF motifs from recent protein binding microarray experiments (Badis et al. 2008; Zhu et al. 2009), allowing us to significantly increase the scope of the network while maintaining a high degree of specificity. We expect that our annotations are likely to be of independent interest to the community, and they are freely available online (http://www.swissregulon.unibas.ch/).
Our second major result is the identification of a subset of genes for which we can significantly correlate changes in the predicted TFBSs with gene expression divergence. The problem of predicting gene expression from sequence alone is well known to be difficult because of the complexity of cis-regulatory regions, even in a relatively simple eukaryote, such as S. cerevisiae (Yuan et al. 2007). For example, the effects of a mutation at a given TFBS may depend on the constellation of other TFBSs in the promoter. Several authors examined the correlation between differences in TFBSs and gene expression divergence between different yeast species (Doniger and Fay 2007; Tirosh et al. 2008) or duplicated genes within S. cerevisiae (Zhang et al. 2004; Leach et al. 2007). These studies had only limited success in correlating expression with sequence that we hypothesize is partly because of the large evolutionary distances used in the comparisons. For example, the sequence divergence between two commonly studied S. cerevisiae strains, S288c and RM11-1a, is ∼0.5%, whereas the divergence between S. cerevisiae and S. paradoxus is as much as ∼12% for coding sequence and ∼18% for noncoding sequence (Cliften et al. 2001). Likewise, most gene duplications in yeast are ancient, with the majority of the duplication events occurring around the time of the eukaryote–prokaryote split (Gu et al. 2005). Therefore, at these larger evolutionary distances promoters typically differ at multiple positions. We reasoned that fewer complex changes in cis-regulatory region organization are likely to have occurred over the timescales separating S. cerevisiae strains, allowing us to more readily correlate sequence and expression divergence.
Taken together, our evolutionary and gene expression analyses demonstrate that our new TFBS predictions significantly improve on the previous annotations and that for a subset of genes, changes in predicted TFBSs correlate significantly with changes in gene expression divergence. Our fine-scale sequence-based computational approach thus complements the classical phenotype-based approach in which quantitative trait locus (QTL) mapping methods are used to identify genomic loci associated with the phenotype. Ultimately, we expect that a combination of the two approaches will be necessary for elucidating the mapping of genotype to phenotype.
Materials and Methods
TF Binding Site Predictions
We combined 89 position-specific weight matrices (PWMs) from Zhu et al. (2009), 112 PWMs from Badis et al. (2008), and 72 PWMs from Erb and van Nimwegen (2006). Overall, visual inspection of TF motifs inferred by more than one method suggests that there is good agreement between the three data sets. Single PWMs for each TF were obtained using a Bayesian procedure that takes a set of PWMs as input and determines the relative alignment of the PWMs that maximizes the probability that the entire set derives from a single underlying PWM and also infers this underlying PWM (FANTOM Consortium 2009). This method also determines whether the data are consistent with all PWMs deriving from one common PWM. For 12 TFs, two of the methods agreed while the third was an outlier, so for each of these TFs, the outlier was manually removed and the two remaining PWMs were aligned. For two TFs, the protein binding microarray methods disagreed between a dimer and monomer motif so we resolved these cases manually. For the other TFs, we first aligned the two protein binding microarray PWMs and then aligned the resulting average protein binding microarray PWM with the ChIP-chip PWM. Finally, we manually trimmed the motif boundaries to exclude positions with little information content and discarded the motif for FHL, a forkhead-like TF that, based on in vitro assays, is suspected of not binding DNA directly (Rudra et al. 2005).
All analyses were performed on the April 2006 Saccharomyces Genome Database (SGD) version of the S. cerevisiae genome sequence to facilitate comparison with our previous TFBS predictions (Erb and van Nimwegen 2006). There have not been major changes in the S. cerevisiae genome since 2006. Intergenic regions were aligned to S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus using MLAGAN and used as input to the MOTEVO program as previously described (Erb and van Nimwegen 2006).
Sensitivity/Specificity Analysis
From the SCPD (Zhu and Zhang 1999) and TRANSFAC (Matys et al. 2003) databases, we curated a set of 452 binding for sites which a binding TF could be identified unambiguously (Erb and van Nimwegen 2006). For each TF for which both MotEvo predictions and at least one known site were available and for each intergenic region, we calculated the sum of the posteriors of all binding sites in the region. The predicted target TF/promoter–region combinations were then ordered by the sum of posteriors and at different cutoffs the fraction of all known targets that were among the predictions (sensitivity) and the fraction of all predictions that correspond to known targets (specificity) were calculated. For the ChIP-chip data of Harbison et al. (2004), TF/promoter–region target combinations were sorted by the P value of binding and the sensitivity and specificity were calculated at different P value cutoffs.
Population Genomics Analysis
In processing the raw single nucleotide polymorphism (SNP) data, we used a threshold of 40 on the Phred score and normalized the allele frequency by the sequencing coverage, following Liti et al. (2009). The results were similar for Phred cutoff of 20 or when excluding singleton polymorphisms (data not shown). For the derived allele frequency (DAF) distributions, we aligned the S. cerevisiae and S. paradoxus reference genomes with MAVID (Bray and Pachter 2004) and rooted the S. cerevisiae SNPs with the S. paradoxus reference sequence and vice versa. We defined conserved elements as 7-mers (possibly overlapping) conserved in the same five species used for TFBS prediction because the average information score of the PWMs was 14 bits. We varied the parameters by testing 6- to 12-mers and 4 or 5 species. The results were entirely consistent in that the inferred selective constraint increased with longer motifs or more species.
Analysis of PWM Score Changes
We collected all TFBSs that were predicted in the BY strain and contained exactly one SNP relative to the RM strain and determined the difference dl in log-likelihood of the BY and RM sequences for the corresponding PWM. From this, we determined the distribution of dl for all observed SNPs, weighing each SNP by the posterior probability of the TFBS in which it occurs. We compared the distribution of observed dl with two randomized distributions. First, we used the distribution of dl of all single point mutations of the TFBS containing SNPs, again weighing the mutations in a TFBS by the posterior probability of the TFBS. Second, we used the distribution of dl of all single point mutations at the same position as where the observed SNP occurred. To take into account the sequence composition of intergenic regions, different point mutations in the randomized sets were also weighed by the overall frequency of the corresponding nucleotide in intergenic regions.
Correlation of Sequence with Gene Expression
Unless specified otherwise, we used 600-nt upstream of the transcription start site to define the promoter region. Transcription starts were defined in Zhang and Dietrich (2005) and David et al. (2006). For divergently transcribed genes where the intergenic region was less than 1,200 nt, we divided the region into two equal-sized promoter regions, based on the result of Erb and van Nimwegen (2006) that most TFBSs likely regulate only one gene. We removed the following TFs as not being transcribed in rich media based on tiling array data (David et al. 2006): DAL80, GAL4, SIP4, and THI2.
For the correlation analysis, we varied the promoter region length from 400 to 1,000 in increments of 200, the posterior probability cutoff from 0.3 to 0.9 in increments of 0.2, and a fold change cutoff from 0 to 0.6 in increments of 0.2. We experimented with other strategies, such as taking the sum or the expectation instead of the max, using two sets of estimated fold changes (Brem and Kruglyak 2005; Wang et al. 2007) and using the estimated activities of the TFs over all segregants, fit using a linear model, similar to previous works (Sun et al. 2007; Ye et al. 2009). None of these strategies resulted in improved correlations though the sum statistic gives similar results to the max statistic (supplementary table S1, Supplementary Material online). We also attempted to divide genes into groups according to which TF regulated them. In theory such a grouping might give an improvement because of the different ways in which changes in PWM score might affect changes in expression. In practice, however, the groups were very small and resulted in high variance in average correlation coefficient, such that of 51 TFs with >2 genes in their group, 21 had negative correlation. For the analysis of the data from Emerson et al. (2010), we used the dependent method of parameter estimation (Emerson et al. 2010) because it has higher accuracy and the correlation between cis and trans effects is not relevant in our application.
Results
A Major Reannotation of the Yeast Transcription Regulatory Network
We started by assembling a catalog of 164 yeast TF PWMs. This catalog was computed using our previously described algorithm (FANTOM Consortium 2009) to combine 141 motifs derived from two recent sets of protein binding microarray experiments (Badis et al. 2008; Zhu et al. 2009) with 79 motifs predicted from genome-wide ChIP-chip data (Harbison et al. 2004; Siddharthan et al. 2005). By comparison, a previous, commonly used data set based only on ChIP-chip data and literature-derived motifs (Macisaac et al. 2006) contained 124 motifs. Our motif set thus contains a large fraction of the ∼200 TFs in the S. cerevisiae genome (Harbison et al. 2004).
Using our updated catalog of motifs, we predicted TFBSs in S. cerevisiae using MotEvo, a Bayesian TFBS prediction algorithm that combines matches to a given PWM with a rigorous analysis of orthologous sequence segments across related species using an explicit statistical model of TFBS evolution (van Nimwegen 2007). That is, while promoter segments that are likely capable of being bound by a given TF are identified based on PWM match, cross-species comparison is used to evaluate the evidence of purifying selection acting to preserve the binding site, and a posterior probability that the site is functional is assigned based on this evidence. Thus, whereas MotEvo is in principle able to detect binding sites that are specific to a single species, higher probability will be assigned to those sites that exhibit evidence of selection acting to preserve them. To facilitate comparison with our previous annotations, we used the same set of parameters and alignments as in our previous analysis (Erb and van Nimwegen 2006). Because it is known that low-affinity (Tanay 2006) and nonconserved (Dermitzakis et al. 2003; Emberly et al. 2003) binding sites can be biologically important, we compared the two annotations over a wide range of posterior probability thresholds (0.05 < Prob < 0.9). Over this range, the new annotations contained roughly twice the number of TF-TFBS relationships and 31–44% more bases in at least one TFBS compared with the old annotations. Moreover, there was substantial overlap between the bases previously annotated to be in TFBSs and those newly annotated to be in TFBSs (table 1). Although ∼22–25% of the bases in the old annotations were reannotated as not in TFBSs in the new annotations (table 1), the large overlap implies that the major change between our annotations was the addition of the new motifs from the protein binding microarray data rather than changes in previously known motifs.
Table 1.
Comparison between Old and New TFBS Annotations
| Previous Annotation |
New Annotation |
||||
| Threshold | Number of TFBS | Total Bases | Number of TFBS | Total Bases | Bases Overlapping |
| 0.05 | 62,654 | 440,101 | 139,498 | 594,576 | 343,701 |
| 0.5 | 14,003 | 123,863 | 28,147 | 178,160 | 96,668 |
| 0.9 | 5,409 | 55,678 | 9,297 | 72,876 | 41,651 |
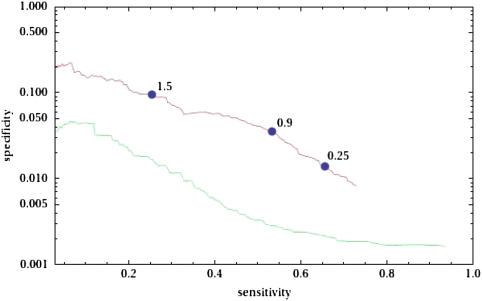
Estimating the absolute sensitivity and specificity of genome-wide TFBS predictions is known to be difficult because there are essentially no comprehensive collections of TFBSs with experimentally demonstrated functionality to use as a reference. In our previous annotation (Erb and van Nimwegen 2006), we used data from the SCPD (Zhu and Zhang 1999) and TRANSFAC (Matys et al. 2003) databases to curate a collection of 452 experimentally determined TFBSs from 184 promoter regions and calculated the sensitivity and specificity of the MotEvo annotations on this small set of known sites. We have repeated this analysis for our new TFBS annotation (fig. 1), and we find that the specificity attained by the new annotation is essentially the same as that of the previous annotation. It should be noted that since the set of known sites represents only a small fraction of all true functional sites, the specificity reported in figure 1 underestimates the true specificity of our predictions by a substantial factor.
FIG. 1.—
Sensitivity and specificity on a reference set of experimentally verified TFBSs of the target promoters predicted by MotEvo (red) and by the ChIP-chip data of Harbison et al. (2004) (green). All putative interactions between TFs and target promoters were sorted by significance (P-value of binding for the ChIP-chip data and predicted number of sites for MotEvo). By varying the cut-off on the significance, we determined how the specificity of the predictions (the fraction of all predictions that correspond to known TF-promoter interactions) depends on their sensitivity (the fraction of all known TF-promoter interactions that are among the predictions. The vertical axis is shown on a logarithmic scale. The blue dots on the red curve show the sensititivies and specificities obtained when the MotEvo predictions are cutoff at 0.25, 0.9 and 1.5 predicted sites (i.e. total posterior probability) in the promoter.
Although it is tempting to use results from genome-wide binding (i.e., ChIP-chip) or microarray experiments to define reference genome-wide target sets and to use these for estimating absolute specificities, we previously demonstrated (Schlecht et al. 2008) that computationally predicted TFBSs show more overlap with target sets obtained by different high-throughput methods than the experimental target sets show with each other. This suggests that computational predictions of the genome-wide targets of TFs may be more accurate than targets inferred from high-throughput experiments. To further analyze this phenomenon, we obtained the genome-wide binding data from the ChIP-chip experiments of Harbison et al. (2004) and used these to predict target promoters, sorted by P value, for each TF analyzed. We then calculated the sensitivity and specificity that the ChIP-chip data attain on the same set of known sites (fig. 1). Strikingly, the specificity attained by the ChIP-chip data is a factor 5–10 lower than that attained by the MotEvo predictions, strongly supporting that MotEvo’s predictions are substantially more accurate than those based directly on ChIP-chip data. In summary, our observations imply that our new annotations represent a significant increase in coverage of the yeast genome TF regulatory network and that these predictions are at least as accurate as predicted targets based on high-throughput experiments.
Selective Constraint on Predicted TFBSs Is Comparable with That on Nonsynonymous Sites
To further validate the accuracy of our TFBS annotations and establish their functional significance, we used a population genomics approach similar to previous works (Fairbrother et al. 2004; Chen and Rajewsky 2006; Chen et al. 2009). The basic idea of this approach is to use the estimated strength of natural selection on predicted cis-regulatory sites as a measure of the functionality of the sites and the accuracy of the predictions. To carry out this analysis, we used data from a recent survey of polymorphism in 39 isolates of S. cerevisiae (Liti et al. 2009). We used two statistics to quantify the level of selective constraint: SNP density and minor allele frequency (MAF). Although the SNP density measure can be biased by heterogeneity in the mutation rate, the allele frequency spectrum is free from such mutation rate biases (Fay et al. 2002) and thus is likely to be a more accurate measure of selective constraint than SNP density. As reported by Liti et al. (2009) and confirmed in our study (data not shown), the DAF spectrum has an anomalously high number of high frequency alleles. Such a pattern is consistent with positive selection acting on those alleles. However, this pattern can also result from misspecification of ancestral alleles, which is likely to occur when the outgroup and ingroup species are separated by a large evolutionary distance, as are S. paradoxus and S. cerevisiae. For this reason, we followed a previous analysis of noncoding SNPs in S. cerevisiae (Fay and Benavides 2005) by using MAF spectra rather than DAF spectra.
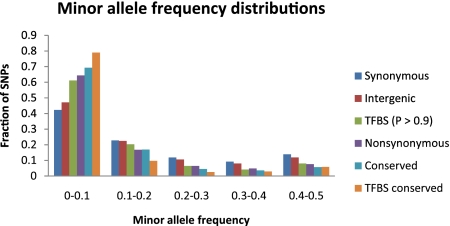
We observed that selective constraint as measured by SNP density was greater on predicted TFBSs than on synonymous sites or on sites in intergenic regions (which include TFBSs) (table 2). This is likely to be a conservative test for purifying selection because many synonymous sites are under selective constraint in S. cerevisiae (Zhou et al. 2010), and intergenic regions are likely to contain constrained sequences other than TFBSs (e.g., nucleosome positioning elements, noncoding RNAs etc.). When comparing the new and old sets of TFBS predictions, we found that the selective constraint was virtually identical across the full range of posterior probabilities (table 2). This result further supports that the new TFBS predictions achieved essentially the same specificity as the old TFBS predictions while significantly improving the overall coverage of the yeast transcriptional network. The MAF spectra exhibited similar patterns to those observed in the SNP density analysis (fig. 2). That is, the frequency spectrum was more skewed toward rare alleles for predicted TFBSs than for synonymous sites and intergenic regions.
Table 2.
SNP Density in Different Functional Classes of Sites across the Genome
| Class of sites | SNP density | |
| Nonsynonymous | 0.012 | |
| Synonymous | 0.048 | |
| Intergenic | 0.030 | |
| Conserved intergenic | 0.016 | |
| TFBS | New annotations | Old annotations |
| TFBS (conserved pos) | 0.0150 | 0.0145 |
| TFBS (P > 0.9) | 0.0192 | 0.0196 |
| TFBS (P > 0.5) | 0.0205 | 0.0203 |
| TFBS (P > 0.05) | 0.0228 | 0.0227 |
Note.—TFBS (conserved pos) refers to positions exactly conserved in five species in TFBS with posterior probability > 0.9 (see Materials and Methods).
FIG. 2.—
MAF distributions in different classes of sites across the genome. The distribution is shown as the fraction of SNPs in each MAF bin for each class of sites, as indicated.
However, when we compared TFBSs with nonsynonymous sites and with 7-mers in intergenic regions that were completely conserved across the five species (hereafter “conserved elements”; see Materials and Methods), we found that the selective constraint on TFBSs was lower than on either of these two classes of sites (table 2, fig. 2). One reason for this result could be that many positions in TF motifs are degenerate and therefore expected to evolve under relatively low selective constraint. The results presented in the next section provide support for this interpretation. We restricted our attention to positions in TFBS conserved across the five species and found that these positions were indeed highly constrained as measured by SNP density, similar to positions in conserved elements. According to the MAF distribution analysis, conserved positions in TFBSs were even more strongly constrained than conserved elements. Overall, these data suggest that selective constraint on conserved positions in TFBS is at least as high as that on nonsynonymous sites or conserved elements.
We confirmed these patterns by examining the SNP density and MAF spectra in 35 isolates of the closely related species S. paradoxus (Liti et al. 2009) (data not shown). Although these data were very similar to the S. cerevisiae results overall, the MAF distribution for nonsynonymous sites was more strongly skewed toward low-frequency alleles in S. cerevisiae than in S. paradoxus. This result is likely due to the draft nature of the S. paradoxus gene annotations, which were simply lifted over from S. cerevisiae based on the genome alignment (http://www.sanger.ac.uk/research/projects/genomeinformatics/sgrp_manual.pdf). For example, over 1,000 annotated genes in S. paradoxus have a coding-sequence length that is not a multiple of three. Thus, a significant fraction of putative nonsynonymous sites are likely to be actually synonymous or intergenic sites.
SNPs in Predicted TFBSs Are Biased to Conserve Binding Affinity to the Cognate TF
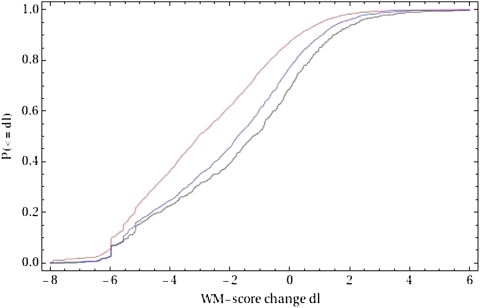
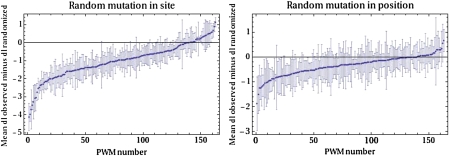
So far we have considered only the SNP density and MAF spectrum within predicted TFBSs. These analyses demonstrated that positions within TFBS are under negative selection but they do not address the specific function of these nucleotides since conceivably they could have a function other than acting as a TFBS. To test for evidence that positions in TFBSs are specifically under selection for binding to the corresponding TF, we compared the distribution of PWM score changes of the observed SNPs with those resulting from randomly mutating a randomly chosen position in the same TFBS. We also performed a more stringent test in which we compared the observed PWM score changes with those that result from randomly mutating the same position in the TFBS. As shown in figure 3, the PWM score changes observed in the SNPs are very significantly biased to maintain the affinity of the TFBSs to their cognate TF (P values of 4 × 10−131 and 5 × 10−18, respectively, Kolmogorov–Smirnov test). These results strongly suggest that the predicted TFBSs are indeed under selection for maintaining their affinity to the cognate TF. Supplementary Figure S1 (Supplementary Material online) shows the analogous results for predicted TFBSs of three individual TFs. To summarize the results of this analysis for individual TFs, figure 4 shows the average and standard error of the difference between PWM score changes for the observed SNPs and PWM score changes in the randomized data sets for each TF separately. Although in many cases, the number of SNPs in predicted TFBSs is too small for a meaningful statistical analysis, for the large majority of individual TFs, the observed PWM score changes are biased toward maintaining the affinity of the TFBS (i.e., the vertical coordinate in fig. 4 is negative), demonstrating that selection for maintaining TFBS affinity applies across most of the TFs for which we provide predictions.
FIG. 3.—
Reverse-cumulative distribution of the changes in PWM scores induced by SNPs in predicted TFBSs. For all predicted TFBS in the BY strain with a single SNP in the RM strain, the difference in log-likelihood (dl) of the sequences for the corresponding PWM was determined (black line). For comparison, the red line shows the reverse-cumulative distribution of log-likelihood differences (dl) that would be obtained by randomly mutating a single position in the same TFBS. The blue line shows the analogous distribution for random mutations in the same position in the TFBS as the observed SNPs.
FIG. 4.—
Effects of random mutations on PWM scores. For each TF, the difference between the average PWM score change (dl) induced by the observed SNPs and the average PWM score change (dl) induced by random mutations is shown, both for random mutations at any position in the TFBS (left panel) and random mutations at the same position as the observed SNP (right panel). The error bars show the standard errors for these differences in mean PWM score change. In each panel, the TFs are ordered from left to right by the difference of means.
Correlation of Sequence and Expression Variation for a Restricted Set of Genes
Having examined the selective constraint on our predicted TFBSs, we next tested if changes in the TFBSs correlated with changes in gene expression between S. cerevisiae strains. We thus analyzed genome-wide gene expression and genotype data from 112 haploid segregants from a cross of two parental S. cerevisiae strains (Brem and Kruglyak 2005), a wild strain, RM11-1a, and a standard laboratory strain, BY4716 (hereafter RM and BY, respectively). Treating gene expression level as a quantitative trait, roughly a quarter of all gene expression levels were significantly associated with a marker close to the gene itself. We will refer to these genes as cis-expression QTLs (eQTLs) or “CE genes,” with the understanding that in some cases the variation may in fact be due to a nearby trans factor. CE genes are also referred to as genes with local eQTLs (Rockman and Kruglyak 2006).
For the remainder of the analysis, we restricted our attention to genetic variation relevant to these eQTL data (i.e., we considered only SNPs between the BY and RM strains). Likewise, we only considered TFs expressed by cells growing in rich media (David et al. 2006), the experimental condition used for the microarray measurements of gene expression. First, we confirmed that the upstream cis-regulatory regions (hereafter referred to as promoter regions) of CE genes were significantly enriched for SNPs between RM and BY in TFBS when compared with the promoter regions of all genes (Chi-square test, P value 0.0147), consistent with a previous result that used a different set of TFBS predictions (Ronald et al. 2005). For this analysis, we restricted our attention to the most confident set of TFBS predictions (posterior probability > 0.9) because we previously observed that the degree of selective constraint correlated well with the posterior probability cutoff on the TFBSs.
Next, we turned to the more difficult problem of correlating the magnitude of the changes in PWM scores with the expression fold change. Because the annotation of TFs as activators or repressors is not currently complete, and the role of a TF as activator or repressor can be dependent on the binding of cofactors or the cellular condition, we examined the correlation of the absolute value of the changes in PWM scores with the absolute log fold change of mRNA expression. Manually annotating TFs as activators or repressors based on their SGD annotations (http://www.yeastgenome.org/) and considering the signs of the changes in PWM scores and mRNA expression did not result in any improvement (data not shown). We made the further approximation of taking the maximum PWM score change over all promoter region SNPs for promoters with multiple SNPs. However, the results were very similar when taking the sum instead of the maximum (supplementary table S1, Supplementary Material online). Because the observed correlations depended on the parameters (e.g., promoter length, posterior probability of a TFBS etc.), we explored the parameter space thoroughly (see Materials and Methods). We then compared the distribution of Pearson correlation coefficients over the parameter space for different sets of genes and TFBS predictions.
For the set of CE promoter regions, the average correlation over all parameter settings was moderately strong (average Pearson’s R = 0.301). In comparison, when using the previous TFBS predictions, we found a lower average correlation (average Pearson’s R = 0.203). This difference in correlation corresponds to approximately a doubling of the variance in expression-level change explained by TFBS changes and thus strongly supports the conclusion that our new TFBS annotations are a significant improvement. To estimate the statistical significance of the correlation results, we computed the same statistic over 1,000 random permutations of the fold change to gene assignments. The highest correlation observed among the permutations was 0.0947 for the old TFBSs and 0.116 for the new TFBSs, implying an empirical P value of less than 0.001.
Many of the CE genes had only a small difference in expression between the two parental strains, even though this difference was significantly linked to a genomic locus. We thus further constrained the CE set to only those genes with a statistically significant expression difference based on a larger set of microarray experiments for the two parental strains (Wang et al. 2007). We further removed all genes with trans-eQTLs to minimize non-cis sources of variation (Yvert et al. 2003). The remaining set of 305 genes, which we call our “restricted cis-eQTL” or RCE genes (supplementary table S2, Supplementary Material online), is small but contains the most confident genes for the purposes of identifying causal cis-regulatory SNPs. Using GO term enrichment analysis (Berriz et al. 2009), we found that the genes in the RCE set are significantly enriched for genes functioning in processes related to the cell wall (corrected P = 0.001) and plasma membrane (corrected P = 0.005). Furthermore, the expression of one gene associated with the plasma membrane, FLO11, was previously shown to correlate with the brightness difference of the cell wall in the two relevant strains, BY and RM (Nogami et al. 2007). Cell wall organization and biogenesis were also observed as an overrepresented functional category for genes with experimentally measured variation in binding of the TF Ste12 in segregants of a cross of a BY-related laboratory strain with a different divergent strain, HS959 (Zheng et al. 2010). Thus, the RCE set of genes contains functionally coherent subsets of genes that may contribute to phenotypic differences between the BY and RM strains.
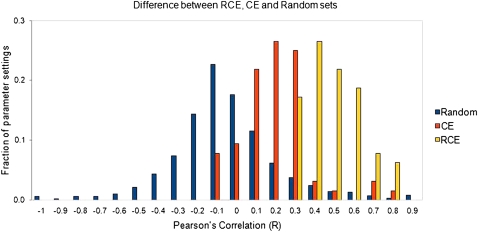
When we repeated the correlation analysis on the set of RCE genes, we found significantly stronger correlations than for the CE genes (fig. 5, average Pearson’s R = 0.314 for the old predictions and 0.514 for the new predictions). Our analysis thus demonstrates the feasibility of correlating sequence divergence at individual nucleotides with expression divergence, at least for a restricted set of genes. It also demonstrates that our new TFBS predictions improve on our previous TFBS predictions because they correlate more strongly with gene expression changes.
FIG. 5.—
Correlation of change in gene expression with change in PWM score. The Pearson correlation of the absolute log fold change of mRNA expression with the absolute value of the change in PWM score was computed for a range of values of the promoter region length, posterior probability cutoff, and fold change cutoff. The fraction of these parameter settings with a given correlation coefficient is shown as a histogram for randomized (blue), CE (orange), or RCE genes (yellow).
Because the maximum or sum of PWM changes is positively correlated with the number of TFBSs and SNPs in the promoter, it is plausible that either of these is the underlying signal in our experiments. To exclude this possibility, we examined the relationship between expression change and either number of TFBSs or SNPs over the same parameter space (see Materials and Methods). However, we found only weak correlations in both cases (supplementary table S1, Supplementary Material online), suggesting that it is necessary to combine both TFBS and SNP information, and consider PWM score changes, to achieve reasonable correlations with gene expression. Another plausible explanation for our results is that the new predictions are enriched for binding sites of a small number of TFs that, for an unknown reason, show exceptionally good correlations between PWM score and expression change. However, we do not observe a limited set of TFs regulating the genes in the RCE set: there are 41 TFs that regulate at least one gene in the RCE set and of these TFs, 21 are associated with TFBS SNPs after taking the maximum over each promoter individually. Thus, the improved correlation combines contributions from a large number of TFs.
In a recent study of cis and trans effects between the BY and RM strains using next-generation transcriptome sequencing (Emerson et al. 2010), the authors identified a “cis only” set (61 genes) for which only cis effects were detected and a “cis major” set (an additional 371 genes) for which both cis and trans effects were detected but the cis effects were significantly larger. We found an average correlation of R = 0.212 for the cis major set, similar to what we obtained for CE genes (R = 0.301). For the cis only set, we found an average correlation of R = 0.450, similar to our result for the RCE set (R = 0.514) (see Materials and Methods). We conclude that our gene sets and those of Emerson et al. (2010) are both enriched for cis effects but we used our sets of genes because they are larger and produced slightly higher correlations.
In addition to showing a statistical association between gene-expression changes and TFBS changes, another important goal is to identify specific nucleotide changes that underlie functional divergence. Examination of the TFBS SNPs in the RCE promoter regions (supplementary table S3, Supplementary Material online) reveals several cases where our predictions align with additional biological information to generate hypotheses that can be tested experimentally. In particular, there are several RCE promoter regions that contain TFBS SNPs for two or more TFs with related functions. Some such cases are trivial because two TFs (e.g., INO4/INO2, MSN2/MSN4, PBF1/PBF2) have identical or nearly identical PWMs and so share the same TFBS. However, there are several nontrivial cases of multiple TFBS SNPs. One example involves SKN7 and MCM1, both of which have TFBS SNPs in the promoter region of AMN1, which encodes a protein involved in exit from mitosis (Wang et al. 2003). These two TFs function together in osmoregulation (Li et al. 1998). For both the SKN7 TFBS and the MCM1 TFBS, the PWM score is higher in BY relative to RM, perhaps indicating a larger connection between mitotic exit and osmotic stress in BY than in RM. Another example involves the glucose-dependent repressors MIG1, MIG2, and MIG3 (Westholm et al. 2008), all three of which have TFBS SNPs in the promoter region of YKL187C, which encodes a protein of unknown function. In this case, the PWM score for the MIG1 TFBS changes in the opposite direction from the other two. This might therefore be an example of compensatory evolution leading to the same total amount of repression. We previously predicted such compensatory coevolution of inputs to the same gene on theoretical grounds (Siegal et al. 2007). In the promoter region of APT2, on the other hand, the PWM scores of TFBS for MIG1, MIG2, and MIG3 are all higher in BY, implying greater glucose-mediated repression of APT2 in BY than in RM.
Discussion
Identifying the nucleotides responsible for gene expression variation is an important problem in genetics and evolution. Although advances have been made using genome-wide association studies to map complex phenotypes (Hindorff et al. 2009), the mapping resolution of these studies is typically too low to pinpoint causal genes let alone individual nucleotides (Altshuler et al. 2008). One promising idea is to use eQTL analysis to predict causal genes, under the assumption that candidate genes in a disease-related locus that also have cis-eQTLs are more likely to be causal genes for the phenotype (Cookson et al. 2009). This approach has been used to map a number of causal genes for obesity in mice (Schadt et al. 2005; Yang et al. 2009).
Here, we have taken the eQTL mapping paradigm to the single nucleotide level by predicting specific nucleotide differences that cause gene expression divergence using computational predictions of TFBSs. Several groups previously explored the relationship between sequence divergence and gene expression divergence across yeast species (Doniger and Fay 2007; Tirosh et al. 2008). Our analysis differs from those analyses in several ways. First, we produced a set of TFBS predictions that is significantly larger than those used in previous studies yet maintains specificity. Beyond the analyses presented here, we believe that our comprehensive TFBS annotations will be of independent interest to the community.
Second, we used eQTL and microarray data to focus our attention on a restricted set of 305 genes whose differences in expression are likely caused by changes in cis-regulatory elements. Thus, although it remains difficult to correlate sequence divergence with gene expression change for all genes in the genome, we have shown that such an analysis is possible at least for a restricted set of genes. Third, we focused on closely related yeast strains as opposed to different yeast species to minimize the occurrence of complex changes in promoter organization. A recent study showed that variation in TF binding between S. cerevisiae strains can often be associated with specific mutations in TF binding sites (Zheng et al. 2010), but they studied only one TF, whereas we analyzed sites for 164 TFs. Together, these improvements allowed us to make progress on the problem of identifying sequence determinants of gene expression variation (Yuan et al. 2007).
There have also been several attempts to correlate sequence and expression divergence in metazoans. Castillo-Davis et al. (2004) examined the correlation of cis-regulatory region divergence and gene expression in Caenorhabditis elegans and C. briggsae. Because there does not exist a set of TFBS annotations for C. elegans of comparable accuracy with those for yeast, the authors estimated promoter region divergence using alignment programs. Andersen et al. (2008) took a similar approach to ours using TFBS predictions in humans. Several other authors have explored the relationship of sequence change and gene expression change. Segal et al. (2007) showed computationally that a single nucleotide change in a TFBS can change the mechanism of TF binding and thereby significantly change gene expression. Lapidot et al. (2008) examined specific nucleotide changes in TFBSs and found that changes involving adenine were more likely to maintain the expression pattern, whereas changes involving guanine were more likely to change it. Swamy et al. (2009) studied the impact of one or two nucleotide changes in TFBSs on gene expression in S. cerevisiae cells grown in different conditions, unlike our study which focused on changes between strains of S. cerevisiae under one growth condition. They found that 1/3 of variable positions in TFBS motifs and 20% of dependent position pairs in TFBS motifs are correlated with gene expression. Many of these TFBS positions were also evolutionarily conserved and condition dependent.
The complexity of transcriptional regulation in metazoan genomes poses significantly greater difficulties than in the relatively simple yeast genome. For example, promoter and enhancer regions are much larger and not characterized as well. It is also more difficult to control the environmental conditions and to assay the cell-type specificity of gene expression change in a metazoan compared with unicellular organisms. Nonetheless, we expect that the availability of more functional genomics data sets for humans, such as protein binding microarray data (Badis et al. 2009) and tissue-specific eQTL data, will likely make the problem of mapping DNA sequence change to gene expression change in humans more tractable in the near future.
Supplementary Material
Supplementary figure S1 and tables S1–S3 are available at Genome Biology and Evolution online ((http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Rachel Brem for sharing the eQTL data with us. We also thank Ian Ehrenreich, Sasha Levy, and Matt Rockman for comments on the manuscript as well as Nicholas Socci and the Siegal laboratory for helpful discussions. This work was partially supported by the National Institutes of Health (grant numbers F32HG 004590-01, K99HG004515-01, R00HG004515-02 to K.C.), National Science Foundation (grant number IOS-0642999 to M.L.S.), and Swiss National Science Foundation (grant number SNF: 3100A0-118318 to E.v.N.).
References
- Altshuler D, Daly M, Lander E. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M, et al. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput Biol. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz G, Beaver J, Cenik C, Tasan M, Roth F. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Pachter L. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci U S A. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis C, Hartl D, Achaz G. cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004;14:1530–1536. doi: 10.1101/gr.2662504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Maaskola J, Siegal M, Rajewsky N. Reexamining microRNA site accessibility in Drosophila: a population genomics study. PLoS One. 2009;4:e5681. doi: 10.1371/journal.pone.0005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Cliften P, et al. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis E, Bergman C, Clark A. Tracing the evolutionary history of Drosophila regulatory regions with models that identify transcription factor binding sites. Mol Biol Evol. 2003;20:703–714. doi: 10.1093/molbev/msg077. [DOI] [PubMed] [Google Scholar]
- Doniger S, Fay J. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol. 2007;3:e99. doi: 10.1371/journal.pcbi.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberly E, Rajewsky N, Siggia E. Conservation of regulatory elements between two species of Drosophila. BMC Bioinformatics. 2003;4:57. doi: 10.1186/1471-2105-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J, et al. Natural selection on cis and trans regulation in yeasts. Genome Res. 2010;20:826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb I, van Nimwegen E. Statistical features of yeast's transcriptional regulatory code. In: Shanghai, editor. IEEE Proceedings First International Conference on Computational Systems Biology. 2006. pp. 111–118. [Google Scholar]
- Fairbrother W, Holste D, Burge C, Sharp P. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J, Benavides J. Hypervariable noncoding sequences in Saccharomyces cerevisiae. Genetics. 2005;170:1575–1587. doi: 10.1534/genetics.105.042283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J, Wyckoff G, Wu C. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc Natl Acad Sci U S A. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2004;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Mizrahi-Man O, Pilpel Y. Functional characterization of variations on regulatory motifs. PLoS Genet. 2008;4(3) doi: 10.1371/journal.pgen.1000018. e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach L, Zhang Z, Lu C, Kearsey M, Luo Z. The role of cis-regulatory motifs and genetical control of expression in the divergence of yeast duplicate genes. Mol Biol Evol. 2007;24:2556–2565. doi: 10.1093/molbev/msm188. [DOI] [PubMed] [Google Scholar]
- Li S, et al. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macisaac K, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S, Ohya Y, Yvert G. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet. 2007;3:e31. doi: 10.1371/journal.pgen.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Ronald J, Akey J. The evolution of gene expression QTL in Saccharomyces cerevisiae. PLoS One. 2007;2:e678. doi: 10.1371/journal.pone.0000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J, Brem R, Whittle J, Kruglyak L. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 2005;1:e25. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner J. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht U, et al. Genome-wide expression profiling, in vivo DNA binding analysis, and probabilistic motif prediction reveal novel Abf1 target genes during fermentation, respiration, and sporulation in yeast. Mol Biol Cell. 2008;19:2193–2207. doi: 10.1091/mbc.E07-12-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal L, et al. Nucleotide variation of regulatory motifs may lead to distinct expression patterns. Bioinformatics. 2007;23:i440–i449. doi: 10.1093/bioinformatics/btm183. [DOI] [PubMed] [Google Scholar]
- Siddharthan R, Siggia E, van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput Biol. 2005;1:e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal ML, Promislow DE, Bergman A. Functional and evolutionary inference in gene networks: does topology matter? Genetica. 2007;129(1):83–103. doi: 10.1007/s10709-006-0035-0. [DOI] [PubMed] [Google Scholar]
- Sun W, Yu T, Li K. Detection of eQTL modules mediated by activity levels of transcription factors. Bioinformatics. 2007;23:2290–2297. doi: 10.1093/bioinformatics/btm327. [DOI] [PubMed] [Google Scholar]
- Swamy KB, Cho CY, Chiang S, Tsai ZT, Tsai HK. Impact of DNA-binding position variants on yeast gene expression. Nucleic Acids Res. 2009;37:6991–7001. doi: 10.1093/nar/gkp743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N. On the relation between promoter divergence and gene expression evolution. Mol Syst Biol. 2008;4:159. doi: 10.1038/msb4100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E. Finding regulatory elements and regulatory motifs: a general probabilistic framework. BMC Bioinformatics. 2007;8:S4. doi: 10.1186/1471-2105-8-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, et al. Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res. 2007;17:1161–1169. doi: 10.1101/gr.6328907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shirogane T, Liu D, Harper JW, Elledge SJ. Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell. 2003;112:697–709. doi: 10.1016/s0092-8674(03)00121-1. [DOI] [PubMed] [Google Scholar]
- Westholm J, et al. Combinatorial control of gene expression by the three yeast repressors Mig1, Mig2 and Mig3. BMC Genomics. 2008;9:601. doi: 10.1186/1471-2164-9-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Galbraith S, Liao J, Eskin E. Using network component analysis to dissect regulatory networks mediated by transcription factors in yeast. PLoS Comput Biol. 2009;5:e1000311. doi: 10.1371/journal.pcbi.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Guo L, Shen L, Liu J. Predicting gene expression from sequence: a reexamination. PLoS Comput Biol. 2007;3:e243. doi: 10.1371/journal.pcbi.0030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Dietrich F. Mapping of transcription start sites in Saccharomyces cerevisiae using 5' SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gu J, Gu X. How much expression divergence after yeast gene duplication could be explained by regulatory motif evolution? Trends Genet. 2004;20:403–407. doi: 10.1016/j.tig.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Zheng W, Zhao H, Mancera E, Steinmetz L, Snyder M. Genetic analysis of variation in transcription factor binding in yeast. Nature. 2010;464:1187–1191. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Gu W, Wilke C. Detecting positive and purifying selection at synonymous sites in yeast and worm. Mol Biol Evol. 2010;27:1912–1922. doi: 10.1093/molbev/msq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]