Abstract
The main genomic changes in the evolution of host-restricted microbial symbionts are ongoing inactivation and loss of genes combined with rapid sequence evolution and extreme structural stability; these changes reflect high levels of genetic drift due to small population sizes and strict clonality. This genomic erosion includes irreversible loss of genes in many functional categories and can include genes that underlie the nutritional contributions to hosts that are the basis of the symbiotic association. Candidatus Sulcia muelleri is an ancient symbiont of sap-feeding insects and is typically coresident with another bacterial symbiont that varies among host subclades. Previously sequenced Sulcia genomes retain pathways for the same eight essential amino acids, whereas coresident symbionts synthesize the remaining two. Here, we describe a dual symbiotic system consisting of Sulcia and a novel species of Betaproteobacteria, Candidatus Zinderia insecticola, both living in the spittlebug Clastoptera arizonana. This Sulcia has completely lost the pathway for the biosynthesis of tryptophan and, therefore, retains the ability to make only 7 of the 10 essential amino acids. Zinderia has a tiny genome (208 kb) and the most extreme nucleotide base composition (13.5% G + C) reported to date, yet retains the ability to make the remaining three essential amino acids, perfectly complementing capabilities of the coresident Sulcia. Combined with the results from related symbiotic systems with complete genomes, these data demonstrate the critical role that bacterial symbionts play in the host insect’s biology and reveal one outcome following the loss of a critical metabolic activity through genome reduction.
Keywords: genome reduction, minimal genome, genome sequencing, Bacteroidetes, nutritional symbioses
Introduction
Bacteria that have developed obligate symbioses with multicellular hosts often have smaller genomes than those of their free-living relatives (Andersson and Kurland 1998; Andersson and Andersson 1999b; Moran 2002; Moran and Plague 2004; Moran et al. 2008; Moya et al. 2008). Genome reduction is thought to be the result of a combination of factors, including small population sizes, frequent population bottlenecks (due to their strict cytoplasmic inheritance), asexuality that limits the efficacy of selection (Moran 1996; Andersson and Kurland 1998), an unusually stable and metabolically rich growth environment, and the general bacterial mutational bias favoring deletions over insertions (Andersson and Andersson 1999a; Nilsson et al. 2005; Moran et al. 2009). Genes from nearly every cellular process are lost in the genomes of obligate intracellular symbionts, including genes involved in DNA recombination, repair, and uptake (Dale et al. 2003; Silva et al. 2003). The loss of recombinogenic activities results in genomes that are unusually stable, and several examples of symbiont genome pairs diverged by 20–200 My show complete colinearity among shared genes (Tamas et al. 2002; van Ham et al. 2003; Degnan et al. 2005; McCutcheon et al. 2009a), a level of genome stability that is unique in bacteria. It appears that once a bacterial lineage becomes obligately associated with a host cytoplasm and its genome shrinks beyond a certain size threshold (around 1-Mb pairs), the only remaining gross structural change observed is further genomic degradation (Tamas et al. 2002; van Ham et al. 2003; Degnan et al. 2005; McCutcheon et al. 2009a).
Insects that have nutritionally unbalanced food sources often have acquired intracellular symbiotic microorganisms to supplement their diet (Buchner 1965; Douglas 1989). For example, insects that feed exclusively on plant sap—which can have high levels of carbohydrates but extremely low levels of essential amino acids—possess stably associated (coevolving) endocellular bacteria that provision the host insect with essential amino acids and/or vitamin cofactors (Shigenobu et al. 2000; Baumann 2005; Nakabachi et al. 2006; Wu et al. 2006; McCutcheon and Moran 2007; McCutcheon et al. 2009a, 2009b). Insects in the suborder Auchenorrhyncha, which includes sharpshooters (leafhoppers), cicadas, planthoppers, and spittlebugs (froghoppers), have formed particularly elaborate symbioses with diverse groups of microorganisms (Buchner 1965; Moran 2007). These insects typically have multiple—often 2, but sometimes up to 6—diverse nutritional symbionts living in specialized tissues called bacteriomes (Buchner 1965). The most ancient and widely distributed is the Bacteroidetes Candidatus Sulcia muelleri (hereby referred to as Sulcia for simplicity) (Moran et al. 2005). Complete genomes for Sulcia and its coresident symbiont have been sequenced from three insect species: the glassy-winged sharpshooter Homalodisca vitripennis (Sulcia-GWSS) (McCutcheon and Moran 2007), the green sharpshooter Draeculacephala minerva (Sulcia-DMIN) (Woyke et al. 2010) and the cicada Diceroprocta semicincta (Sulcia-DSEM) (McCutcheon et al. 2009b). (The Sulcia-GWSS and Sulcia-DMIN genomes are functionally identical from a nutritional perspective and so only Sulcia-GWSS will be discussed further.) Although Sulcia-GWSS diverged from Sulcia-DSEM at least 200 Ma based on host fossils (Shcherbakov and Popov 2002), the genomes are completely collinear and almost identical in gene content; importantly, they both retain identical and near-complete gene sets for the production of 8 of the 10 amino acids (leucine, isoleucine, valine, threonine, lysine, arginine, phenylalanine, and tryptophan) (McCutcheon et al. 2009a). In both the GWSS and DSEM systems, the remaining 2 essential amino acids (methionine and histidine) are made by Sulcia’s cosymbiont, which in GWSS is the Gammaproteobacteria Candidatus Baumannia cicadellinicola and in DSEM is the Alphaproteobacteria Candidatus Hodgkinia cicadicola (hereby referred to as Hodgknia for simplicity) (Wu et al. 2006; McCutcheon et al. 2009a).
In all studied cases for bacteriome-associated symbionts in sap-feeding insects, the relationship is mutually obligate: the host and its symbionts are completely dependent on each other to survive (Douglas 1989; Nakabachi and Ishikawa 1999; Moran et al. 2008; Moya et al. 2008). This presents a seemingly tenuous situation, where the host is dependent on its symbiotic bacteria to survive, but these same bacteria have irrevocably lost the genomic dynamism typical of bacteria and are incapable of restoring genes that are lost through genome reduction. Here, we show one possible outcome of the complete loss of an essential amino acid pathway in dual symbiotic system containing Sulcia and a coresident symbiont: the lost pathway in Sulcia is completely retained in the cosymbiont, precisely conserving the collective production of all ten essential amino acids by the bacterial partners.
Materials and Methods
DNA Preparation
Spittlebugs were collected from Rosmarinus officinalis (rosemary) bushes on the University of Arizona campus. Red (containing Sulcia) and yellow (containing Zinderia) portions of the bacteriomes were dissected from 4 to 5 insects in phosphate-buffered saline. The volume was reduced to 10 μl, and the bacteriome tissue was lysed in 10 μl of cell lysis buffer (400 mM KOH, 10 mM ethylenediaminetetraacetic acid, 100 mM dithiothreitol) on ice for 10 min, followed by the addition of 10 μl of neutralization buffer (400 mM HCl, 600 mM Tris–HCl, pH 7.5). Five separate whole-genome amplification reactions were performed using 3 μl of the neutralized sample as template following the instructions in the Amersham GenomiPhi V2 kit. This procedure was repeated several times over the course of 2 years to generate DNA for Roche 454 FLX, Roche 454 FLX Titanium, and Illumina sequencing.
Genome Sequencing
The Sulcia genome was assembled into 10 contigs (274,670 nts) from a Roche 454 FLX run of 457,666 reads totaling 113,860,631 bases in Newbler version 1.1.02.15 and closed by polymerase chain reaction (PCR) and Sanger sequencing. The Zinderia genome was highly fragmented (50+ contigs) and of poor quality at this stage, but it was clear that the guanine + cytosine (GC) content was extremely low.
Due to the low GC content of the Zinderia genome, an amplification-free Illumina protocol was used, as this was reported to prevent biasing the sequencing library toward GC-rich sequences (Kozarewa et al. 2009). A paired-end run on an Illumina Genome Analyzer IIx generated 4,653,772 fifty-nine nt reads totaling 274,572,548 nts. The Illumina data were used to 1) correct homopolymer errors in the Sulcia genome generated by 454 FLX (by mapping the Illumina reads to the 454 genome with BlastN; parameters: -G 2 -E 1 -F F -e 1e-15 -W 7 -b 1 -v 1) and 2) attempt a de novo assembly of the Zinderia genome. After removing the 355,976 Illumina reads that mapped to the Sulcia genome, the remaining 4,297,796 reads were assembled using velvet (Zerbino and Birney 2008) (velveth parameters: k = 41 –shortPaired; velvetg parameters: –ins_length 200 –cov_cutoff 20 –exp_cov 255). The Illumina/velvet Zinderia assembly had 8 contigs totaling 212,107 nts with an average GC content of 13.4%, and these contigs formed the core of the Zinderia genome.
A 454 FLX Titanium run was done in parallel to the Illumina sequencing, which produced an additional 52,484 reads with an average length of 378 nts. These reads were assembled with Newbler version 2.0.00.22, resulting in 34 Zinderia genome contigs with an average GC content of 13.7%. The eight Illumina/Velvet contigs were assembled with the thirty-four 454 FLX Titanium contigs in phrap (Felsenstein 1989), and the results were hand checked for consistency. A few minor Velvet misassemblies were corrected, and some contigs from each separate assembly were joined, resulting in four supercontigs. These four supercontigs were closed by PCR and Sanger sequencing. Both genomes were annotated as described previously (McCutcheon and Moran 2007), except that Infernal (Nawrocki et al. 2009) was used to predict the boundaries of the ribosomal RNA genes.
Transmission Electron Microscopy
Yellow portions of the bacteriome, which exclusively contains Zinderia, were dissected from spittlebugs and fixed in room temperature 2.5% glutaraldehyde in 0.1 M 1,4-piperazinediethane-sulfonic acid (PIPES) buffer pH 7.4 for 1 h. The bacteriomes were washed in 0.1 M PIPES, postfixed in 1% osmium tetroxide, and washed for 10 min in deionized water. The sample was dehydrated by 5–10 min washes in 50, 70, 90, and 100% ethanol. Microwave resin infiltration was done using a 1:1 mix of Spurr’s resin to ethanol at 250 W at 20 °C for 3 min in a vacuum; two additional infiltrations were done in pure Spurr’s resin, 25 W at 20 °C for 3 min in a vacuum. The samples were then left in Spurr’s resin for 30 min at room temperature and finally embedded with an overnight incubation at 60 °C. Sixty nm sections were cut onto uncoated copper mesh grids, stained with 2% uranyl acetate for 20 min followed by 2% lead acetate for 2 min. Sections were viewed in an FEI CM12 transmission electron microscope operated at 80 kV.
Phylogenetics
Individual protein alignments (FusA, RplB, TufA, MnmG, and RpoB) for the protein-based tree were produced using the linsi module of MAFFT (Katoh et al. 2005), hand-edited to remove poorly aligned regions, and concatenated. The resulting data set had 73 species and 3,255 columns. Maximum likelihood trees were generated using RAxML (relevant parameters: -d -m PROTGAMMAJTT -x 12345 -# 200 -f a). A list of species (and GenBank accession numbers) used in the generation of the protein tree can be found in the supplemental materials (Supplementary Material online).
The 16S rRNA-based tree was generated by aligning the Zinderia 16S rDNA sequence to the Ribosomal RNA Database (RDP) bacterial model using the RDP-based Infernal aligner (Cole et al. 2009; Nawrocki et al. 2009), collecting 49 other high-quality Betaproteobacterial 16S rDNA sequences in the RDP database, and using this alignment to generate a maximum likelihood tree in RAxML (relevant parameters: -f a -x 12345 -# 100 -m GTRCAT).
Results
Spittlebugs Have Two Long-term Symbionts
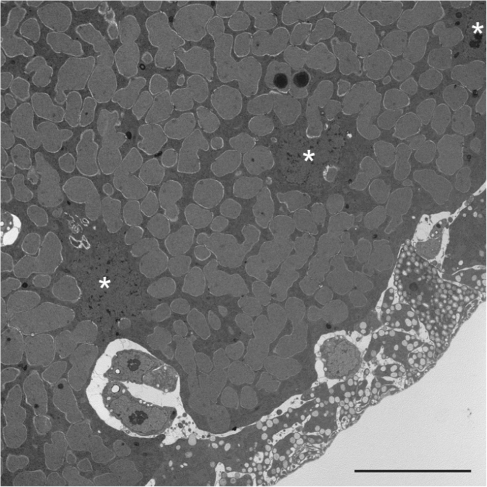
Previous work showed that the spittlebug Clastoptera arizonana contains Sulcia as a symbiont (hereby referred to as Sulcia-CARI) (Moran et al. 2005). We identified a second symbiont by universal 16S rDNA PCR (data not shown) and transmission electron microscopy (fig. 1). Together, these experiments revealed that the second symbiont was a novel member of the Betaproteobacteria with large amorphous cells. This cell shape often indicates a bacterium that has undergone substantial genome reduction (Moran et al. 2005; Nakabachi et al. 2006; McCutcheon et al. 2009b). Therefore, in an ongoing effort to understand genome reduction in multiple bacterial lineages and to further document complex symbioses containing diverse bacterial partners, we sequenced the genomes from Sulcia-CARI and the novel Betaproteobacteria for which we propose the name Candidatus Zinderia insecticola (and will refer to as Zinderia for simplicity).
FIG. 1.—
Transmission electron microscopy of a Zinderia-containing bacteriocyte cell from Clastoptera arizonana. Three insect nuclei are indicated with white asterisks. The scale bar is 10 μm.
Sulcia-CARI Does Not Encode the Tryptophan Biosynthetic Pathway
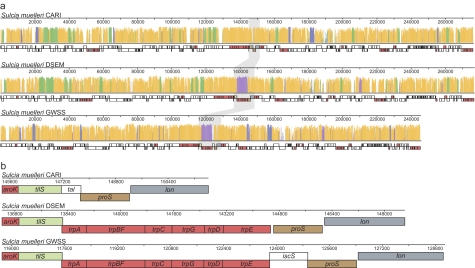
The Sulcia-CARI genome is 276,511 nts in length, has a 21.1% GC content, and codes for 246 protein-coding genes, 29 tRNAs, and one copy each of tmRNA, RNase P RNA, 5S, 16S, and 23S rDNA. A three-way alignment of the Sulcia genomes from GWSS, DSEM, and CARI shows that the only gross genomic changes between the lineages are differential losses of various genes; there is complete colinearity among retained genes (fig. 2a). This striking pattern of conservation was also observed in previous whole-genome alignments between Sulcia-GWSS and Sulcia-DSEM (McCutcheon et al. 2009a), as well as in previous comparisons of other highly reduced bacterial symbiont genomes (Tamas et al. 2002; Degnan et al. 2005), and this result provides further confirmation of genome stability as the prevailing pattern in reduced-genome symbionts. More surprising was the complete loss of all six genes in the tryptophan biosynthetic operon in Sulcia-CARI (fig. 2b), as this is one of the eight essential amino acids produced by Sulcia-GWSS and Sulcia-DSEM. All other genes involved in the production of the remaining seven essential amino acids (leucine, isoleucine, valine, threonine, lysine, arginine, and phenylalanine) are conserved among all three Sulcia genomes (fig. 2a).
FIG. 2.—
Whole-genome alignment for three Sulcia species. Each complete Sulcia genome is shown in a linear representation where each gene is represented by a box. Boxes for genes involved in the synthesis of essential amino acids are colored red; all others are represented by white boxes except for selected genes flanking the tryptophan biosynthetic pathway region. (a) The histogram above the linear genome schematic indicates the level of conservation, where a higher bar represents greater sequence identity. Regions that are shared between all three genomes are colored orange; those that are shared between Sulcia-CARI and Sulcia-DSEM are green; those shared between Sulcia-CARI and Sulcia-GWSS are blue; and those shared between Sulcia-DSEM and Sulcia-GWSS are purple. The region flanking the tryptophan biosynthetic pathway is shaded in gray. (b) Zooming in on the tryptophan biosynthesis region shows the precise nature of genome reduction, as no fragment of any gene in the tryptophan pathway remains.
Zinderia: A Betaproteobacterial Symbiont with Extreme Genomic Features
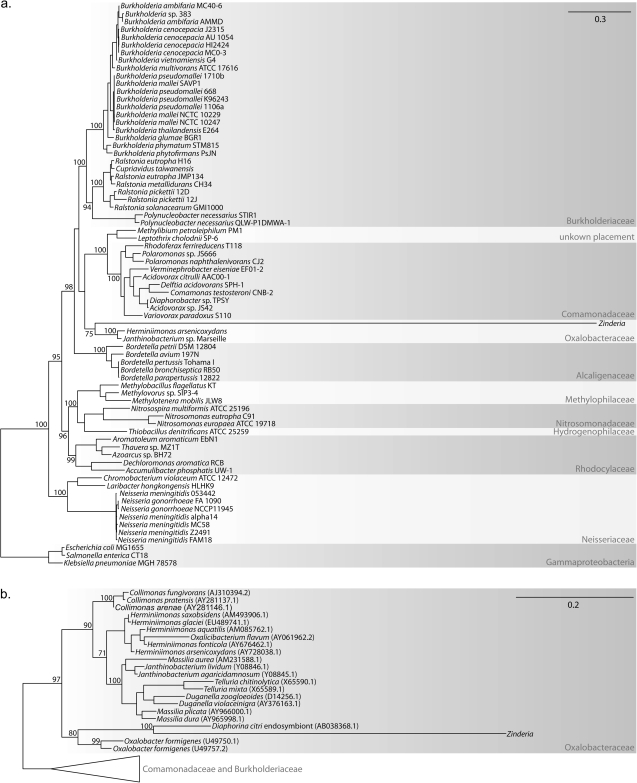
The Zinderia genome is 208,564 nts and codes for 202 protein-coding genes, 25 tRNAs, and one copy each of tmRNA, 5S, 16S, and 23S rDNA. Similar to other insect symbionts with highly reduced genomes, Zinderia does not code for genes involved in making a cell membrane, peptidoglycan, nucleotides, or vitamin cofactors. Zinderia is not capable of doing most reactions involved in carbohydrate metabolism—including glycolysis and the citric acid cycle—and seems incapable of making adenosine triphosphate (ATP), as no F1F0 ATPase is present and no obvious pathway for substrate-level phosphorylation exists. The Zinderia genome does however encode homologs for all 14 genes of the minimal NADH:ubiquinone oxidoreductase I proton translocation machinery (nuoABCDEFGHIJKLMN), all four genes in the cytochrome bo terminal oxidase complex (cyoABCD), and various electron transfer proteins, but the functions of these genes are unclear in the context of such a limited gene repertoire. Similar to other highly reduced symbiont genomes (McCutcheon 2010), the Zinderia genome contains a minimal set of gene orthologs involved in genome replication, transcription, and translation (supplementary fig. S1, Supplementary Material online). Phylogenetic analyses with concatenated protein (fig. 3a) and 16S rDNA (fig. 3b) sequences clearly define Zinderia as member of the Betaproteobacteria and suggest that genera in the Oxalobacteraceae (e.g., Oxalobacter, Herminiimonas, and Janthinobacterium) are its closest free-living relatives, although this should be interpreted with some caution as the branch lengths on the Zinderia lineage are extremely long (fig. 3a and b).
FIG. 3.—
Phylogenetic analysis indicates that Zinderia is a Betaproteobacteria in the family Oxalobacteraceae. Select bootstrap values greater than 70 are shown on each maximum likelihood tree. The tree in (a) was calculated from a concatenated alignment of several protein sequences and that in (b) was calculated from the 16S rDNA sequence alone.
The genome sequence strongly suggests that Zinderia uses an alternative genetic code, in which UGA codes for tryptophan instead of stop. This code change has been reported in certain lineages of Mollicutes, such as Mycoplasma (Yamao et al. 1985), the Alphaproteobacteria Candidatus Hodgkinia cicadicola (McCutcheon et al. 2009b), some ciliate nuclear genomes (Lozupone et al. 2001), and in several mitochondrial lineages (Knight et al. 2001). Evidence for the mapping of UGA to tryptophan in Zinderia comes from multiple sequence alignments of proteins, which show tryptophan occurring in several highly conserved positions in other Proteobacteria that are coded for by UGA in Zinderia (fig. 4). If UGA is assumed to be reassigned, 360 of 374 (96%) of the putative tryptophans encoded in Zinderia open-reading frames use the UGA codon, whereas only 14 use the standard tryptophan codon UGG. This usage pattern is consistent with that of other degenerate codon families in Zinderia, where the more AT-rich codon is always used preferentially over the more GC-rich codon (supplementary table S1, Supplementary Material online).
FIG. 4.—
Alignment of conserved regions of proteins suggests that UGA codes for tryptophan in the Zinderia genome. The numbers indicate the position in the Zinderia protein. Zinderia, Janthinobacterium (GenBank accession: NC_009659), Herminiimonas (NC_009138), Burkholderia (NC_008784, NC_008785), and Neisseria (NC_003112) are all Betaproteobacteria; Sinorhizobium (NC_003047) is an Alphaproteobacteria and Escherichia (NC_000913) is a Gammaproteobacteria.
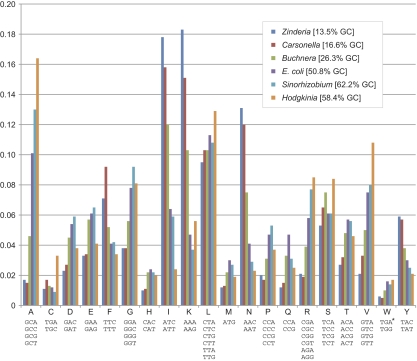
The genomic GC content of Zinderia is 13.5%, the lowest yet observed in any cellular genome. This is also lower than any reported viral genome (at 17.8%, the Entomopoxvirus Amsacta moorei [Bawden et al. 2000] has the lowest GC content of all 3,573 viral genomes listed in the GenBank Viral Genomes Resource, http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/viruses.html) and lower than the vast majority of organellar genomes (only 4 out of the 3,472 genomes listed in the GOBASE Organelle Genome Database [O'Brien et al. 2009] have a lower GC content; the lowest is 10.9% GC in the mitochondrion of the yeast Kluyveromyces bacillisporus [Bouchier et al. 2009]). Although the GC content of the protein-coding regions is 13.2% overall, the GC content of first, second, and third codon positions are 17.5, 18.8, and 3.3%, respectively. This pattern suggests a strong mutational bias toward AT in the genome, with purifying selection acting on the first and second codon position to maintain amino acid residues, similar to what has been observed in other reduced genomes with strong compositional bias (Moran and Wernegreen 2000). As expected from previous work (Moran 1996; Clark et al. 1999), this extreme GC bias has a profound effect on the amino acid profile of the Zinderia proteome; 36.1% of all amino acid residues are either isoleucine (58.9% ATA, 40.6% ATT, 0.5% ATC) or lysine (97.9% AAA, 2.1% AAG), and amino acids with GC-rich codons are greatly underrepresented compared with organisms with more balanced genomic GC contents (fig. 5 and supplementary table S1, Supplementary Material online).
FIG. 5.—
Amino acid frequency distributions for six bacterial genomes reveal extreme biases in GC-poor symbiont genomes. The asterisk on TGA indicates that this codon has been reassigned to tryptophan in the Hodgkinia and Zinderia genomes. The most GC-poor codons (e.g., phenylalanine [F], isoleucine [I], lysine [K], aparagine [N], and tyrosine [Y]) are all overrepresented in reduced symbiont genomes that are GC-poor such as Zinderia, Carsonella, and Buchnera. The opposite pattern of GC-rich codons being overrepresented in tiny GC-rich symbiont genomes such as Hodgkinia is apparent in some (e.g., alanine [A]) but not all (e.g., proline [P] and glutamine [Q]) codon families.
Zinderia Is Able to Produce Three Essential Amino Acids
It is thought that all animals have lost the ability to make ten amino acids (the essential amino acids) and that these compounds are not present in high levels in xylem sap (Redak et al. 2004). Therefore, because the tryptophan biosynthetic pathway was lost in Sulcia-CARI and the previously studied cosymbionts of Sulcia in GWSS and DSEM produced methionine and histidine (Wu et al. 2006; McCutcheon et al. 2009a), it was of interest to see if Zinderia had gene homologs for the production of tryptophan, methionine, and histidine. Genome analysis indicates that this is the case—Zinderia makes exactly this set of amino acids (fig. 6). In the production of methionine, Zinderia uses the direct sulfhydrylation pathway (metXY), which is unique in insect nutritional symbionts; all previous examples use the transsulfuration pathway (metABC) (Hacham et al. 2003). In the production of both histidine and tryptophan, Zinderia has gene homologs for all the standard reactions, although shikimate is needed as a precursor in the production of tryptophan, as homologs of genes for the conversion of phosphoenolpyruvate (PEP) to shikimate (aroABDE) are missing in Zinderia. Sulcia-CARI has retained all genes for the conversion of PEP to phenylalanine, including aroABDE, and may be the source of shikimate for tryptophan production in Zinderia.
FIG. 6.—
The essential amino acid metabolisms of three Sulcia-containing dual symbiont systems. Complete pathways for the production of essential amino acids (red font) or related cofactor compounds (blue font; Hodgkinia uses the B12-dependent version of methionine synthase in the last step of methioine production [McCutcheon et al. 2009a]) are shown. Missing genes are represented in a light gray font. Note the three different pathways taken in the production of methionine in Sulcia’s cosymbionts.
Discussion
The Tryptophan Operon Is Precisely Excised in Sulcia-CARI
Bacterial mutation exhibits an inherent deletional bias (Andersson and Andersson 1999a, 2001; Mira et al. 2001), and this bias has two important consequences. The first is that intergenic regions are typically small, resulting in a coding density that is stable (about 1 gene every 1,000 bases) across the entire two orders of magnitude range of bacterial genome size (Bentley and Parkhill 2004; Ochman and Davalos 2006). The second is that a gene that no longer provides a selective advantage in a certain environment does not persist over long periods of time; when selection is no longer strong enough to maintain a gene it is pseudogenized and eventually the DNA is removed from the genome (Andersson et al. 1998; Mira et al. 2001; Ochman and Davalos 2006). Because obligate endosymbionts have small effective population sizes (which limits the efficacy of selection) and live exclusively in a stable and metabolically rich environment, they lose many genes that are retained in free-living bacterial genomes, even genes that are slightly beneficial (Andersson and Andersson 1999a, 2001; Moran 2003). The process of genome reduction in obligate endosymbionts seems to have two phases, where early in the symbiosis large blocks of sequence corresponding to many unrelated genes can be lost (Moran and Mira 2001; Nilsson et al. 2005), followed by a later phase of DNA loss in smaller blocks of sequence (Moran et al. 2009). The pattern of loss seen in the region of the Sulcia-CARI genome that codes for the tryptophan pathway in the other Sulcia genomes demonstrates the precision of the later phase of genome reduction (fig. 2b). No remnant of any gene in the tryptophan pathway remains in the Sulcia-CARI genome, and intergenic regions between the flanking genes that are conserved in all three Sulcia genomes (tilS and proS) are nonexistent, as the coding region of remaining intervening gene in Sulcia-CARI (tal) overlaps the coding region of both flanking genes.
Evolutionary Origins of the Observed Genomic Patterns
Given the long time periods involved and the lack of known intermediate steps, it is hard to delineate a model that describes the evolutionary processes required to arrive at the genome structures observed in the three Sulcia-containing insect systems sequenced to date. A few facts are known, however, that fix some points along the paths taken to arrive at these structures: 1) based on the insect fossil record and phylogenetic reconstructions, the initial Sulcia infection was acquired by the auchenorrhynchan ancestor at least 260 Ma (Moran et al. 2005), 2) again based on the insect fossil record, the split between cicadas, spittlebugs, and sharpshooters was at least 200 Ma (Shcherbakov and Popov 2002), and 3) the extreme size and features of the Zinderia genome suggest that it is an ancient association not a recent symbiont acquisition. Therefore, by 200 Ma, the genomic structure of Sulcia was likely similar to what is observed in Sulcia-GWSS and Sulcia-SDEM, with Sulcia producing 8 of the 10 essential amino acids. This is consistent with extensive gene loss in the Sulcia lineage early in the symbiosis and suggests that Sulcia had lost the ability to produce methionine and histidine early in its association with insects, prior to the common ancestor of cicadas, sharpshooters, and spittlebugs, at least 200 Ma. Furthermore, this means that Sulcia has likely had a cosymbiont for at least 200 Ma because all extant examples of Sulcia-containing symbioses collectively make all ten essential amino acids, which implies that all ten are required.
Ten Amino Acids by Any Means Necessary: Sulcia and Its Cosymbionts in Three Insect Systems
Whatever the timing and mechanism of genome reduction that lead to the current metabolic contributions schematized in figure 6, the myriad mechanisms that Sulcia’s diverse cosymbionts have evolved to complement the respective Sulcia genome are remarkable. The three distinct paths taken by Zinderia, Baumannia, and Hodgkinia in the production of methionine exemplify this point. Zinderia uses the direct sulfhydrylation pathway (metXY) in the production of methionine, whereas Baumannia and Hodgkinia use the transsulfuration pathway (metABC). Despite using the same pathway in the production of homocysteine, Baumannia and Hodgkinia use different enzymes for its conversion to methionine; Baumannia uses the cobalamin (vitamin B12)-independent version of methionine synthase (MetE), whereas Hodgkinia uses the cobalamin-dependent version (MetH). Hodgkinia is therefore obliged to retain a large number of genes responsible for the production of the complex vitamin cofactor cobalamin (cobAJMLHNSTOQDP), a gene complement that corresponds to about 7% of its proteome (McCutcheon et al. 2009a). It is sometimes unclear whether these differences reflect the phylogenetic origin of the symbiont or the random nature of genome reduction (McCutcheon et al. 2009a), but in the case of Zinderia’s use of the direct sulfhydrylation pathway in the production of methionine, it seems largely a phylogenetic signal, as Herminiimonas arsenicoxydans and Janthinobacterium sp. Marseille, Zinderia’s closest free-living relatives, both encode copies of metX and metY but not metA or metB (both however do encode metC, which can also be used in cysteine degradation) (Audic et al. 2007; Muller et al. 2007).
When examined at the level of essential amino acid production, the three systems shown in figure 6 present a striking case of convergent evolution in the context of a common selection pressure to retain the capacity for a full complement of amino acids. In all three pairs of symbionts, no genes involved exclusively in essential amino acid production overlap in a genome pair, with the exception of aroKAC in Sulcia-CARI and Zinderia (a few genes that function in two or more pathways, such as carAB, which have roles in amino acid and nucleotide production, are present in both Sulcia-GWSS and Baumannia). Our new findings for Zinderia reveal the first reported case of extreme genome reduction for a member of the Betaproteobacteria. (As members of the Alpha-, Beta-, and Gammaproteobacteria, Hodgkinia, Zinderia, and Baumannia are estimated to have diverged from each other at least 2 billion years ago [Battistuzzi et al. 2004]). Each cosymbiont developed a separate symbiosis with the insect host and Sulcia, underwent massive amounts of genome reduction, and converged on a gene set that perfectly complements their Sulcia partner; this finding highlights the critical role that these symbionts play in the biology of their insect hosts.
These results are similar to what has been observed in the pea aphid Cinara cedri, where part of the tryptophan pathway is retained on a plasmid of the endocellular symbiont Buchnera, and the remaining genes are present in the coresiding symbiont Candidatus Serratia symbiotica (Gosalbes et al. 2008). In this case, however, the level of complementarity is not precisely known because the Serratia genome is not yet complete.
Candidatus Zinderia Insecticola, a Novel Symbiont of Spittlebugs
We propose the name Candidatus Zinderia insecticola for the Betaproteobacterial symbiont of spittlebugs described here. The genus refers to the geneticist Norton D. Zinder (born 1928), and the species name refers to Zinderia’s exclusive presence in insect hosts. Distinctive features include large amorphous cells, an existence restricted to the host cell cytoplasm, a low GC content, a recoding of UGA from stop to tryptophan, and the unique 16S rDNA sequence CTAGTTATTAAATTAAAAATAAAATTTAGTAACG (positions 826–858, Escherichia coli numbering).
Supplementary Material
Supplementary figure S1 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank E. Nawrocki for help in using Infernal to predict the boundaries of the ribosomal RNA genes and T. Day for performing the electron microscopy experiments. This work was supported by the National Science Foundation (Microbial Genome Sequencing award 0626716 to N.A.M.) and the University of Arizona’s Center for Insect Science through the National Institutes of Health (Training Grant 1K 12 GM00708 to J.P.M.].
References
- Andersson JO, Andersson SG. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999a;16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Andersson SG. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev. 1999b;9:664–671. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Andersson SG. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol. 2001;18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Andersson SG, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Audic S, et al. Genome analysis of Minibacterium massiliensishighlights the convergent evolution of water-living bacteria. PLoS Genet. 2007;3:e138. doi: 10.1371/journal.pgen.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Bawden AL, et al. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology. 2000;274:120–139. doi: 10.1006/viro.2000.0449. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- Bouchier C, et al. Complete mitochondrial genome sequences of three Nakaseomyces species reveal invasion by palindromic GC clusters and considerable size expansion. FEMS Yeast Res. 2009;9:1283–1292. doi: 10.1111/j.1567-1364.2009.00551.x. [DOI] [PubMed] [Google Scholar]
- Buchner P. Endosymbiosis of animals with plant microorganisms. New York: Interscience; 1965. [Google Scholar]
- Clark MA, Moran NA, Baumann P. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Wang B, Moran N, Ochman H. Loss of DNA recombinational repair enzymes in the initial stages of genome degeneration. Mol Biol Evol. 2003;20:1188–1194. doi: 10.1093/molbev/msg138. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Lazarus AB, Wernegreen JJ. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989;64:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Gosalbes MJ, Lamelas A, Moya A, Latorre A. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol. 2008;190:6026–6029. doi: 10.1128/JB.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Gophna U, Amir R. In vivo analysis of various substrates utilized by cystathionine gamma-synthase and O-acetylhomoserine sulfhydrylase in methionine biosynthesis. Mol Biol Evol. 2003;20:1513–1520. doi: 10.1093/molbev/msg169. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight RD, Landweber LF. The molecular basis of nuclear genetic code change in ciliates. Curr Biol. 2001;11:65–74. doi: 10.1016/s0960-9822(01)00028-8. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP. The bacterial essence of tiny symbiont genomes. Curr Opin Microbiol. 2010;13:73–78. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009a;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009b;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol. 2003;6:512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:1–2. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Muller D, et al. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 2007;3:e53. doi: 10.1371/journal.pgen.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 1999;45:1–6. doi: 10.1016/s0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AI, et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci U S A. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien EA, et al. GOBASE: an organelle genome database. Nucleic Acids Res. 2009;37:D946–D950. doi: 10.1093/nar/gkn819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- Redak RA, et al. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu Rev Entomol. 2004;49:243–270. doi: 10.1146/annurev.ento.49.061802.123403. [DOI] [PubMed] [Google Scholar]
- Shcherbakov DE, Popov YA. Superorder Cimicidea Laicharting, 1781; Order Hemiptera Linne, 1758. The bugs, cicadas, plantlice, scale insects, etc. In: Rasnitsyn AP, Quicke DLJ, editors. History of insects. Dordrecht (The Netherlands): Kluwer; 2002. pp. 143–157. [Google Scholar]
- Shigenobu S, et al. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Silva FJ, Latorre A, Moya A. Why are the genomes of endosymbiotic bacteria so stable? Trends Genet. 2003;19:176–180. doi: 10.1016/S0168-9525(03)00041-6. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- van Ham RC, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, et al. One bacterial cell, one complete genome. PLoS One. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F, et al. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]