Abstract
The Genomics Education Partnership offers an inclusive model for undergraduate research experiences incorporated into the academic year science curriculum, with students pooling their work to contribute to international data bases.
Teaching students to think like scientists has traditionally been accomplished by immersing undergraduates in a full-time summer research program, an experience that enhances professional development and provides career clarification (1, 2). We believe that conducting research is the best way to learn how knowledge is defined and created in a field. Furthermore, students acquire a deeper understanding of fundamental concepts as they apply what they have learned to accomplish defined research goals. Undergraduate research experiences can sustain student interest in a science career, providing an opportunity to work collaboratively with colleagues while making novel contributions to the community (1).
Most undergraduates begin their research with mentoring by a faculty member, graduate student, or postdoc during a summer spent in the lab. This approach, while usually successful, excludes many students who do not have the summer free or funded. Other barriers include limited funding and facilites as well as a lack of experienced mentors (1, 3). Thus we must incorporate student research experiences into our regular academic-year curriculum to make such experiences more broadly available. Our experiences as founding members of the Genomics Education Partnership encourage us to believe that a curriculum in genomics can train students to think like scientists (4).
Genomics is an attractive area for student-scientist partnerships. First, genomics represents an exciting advance in the life sciences. With genomics, we can analyze not only one gene but many genes, and can address questions ranging from patterns of gene expression to evolution of a species. Second, the teaching and research materials can be obtained for little or no cost: DNA sequences and microarray results are freely available on the web (5). Third, genomics research projects can range from simple to complex, allowing engagements to match a variety of academic calendars.
Mindful of these advantages, a number of faculty groups and institutions have launched projects to bring genomics into the undergraduates curriculum. These range from primarily utilizing computer-based resources for annotation and analysis to having a major wet-lab focus (6). Two major projects engage students in analyzing genomes from phage and from bacteria or archaea (7). (See SOM for further descriptions.) The Genomics Education Partnership (GEP) was formed in 2006 as a collaboration between faculty from various institutions and the Biology Dept. and Genome Sequencing Center at Washington Univ. in Saint Louis (15). The group currently consists of over 40 faculty members who teach undergraduate courses in biology, statistics, mathematics, and computer science at a diverse set of institutions, including primarily undergraduate, research, state regional, and private schools (4). The goal of the GEP is to provide opportunities for undergraduate students to participate in original research experiences within the framework of course curricula.. The GEP pools student efforts across institutions to improve DNA sequence quality and to generate hand-curated gene models.. Developing a coordinated research effort allows us to tackle large projects together, making work in eukaryotes accessible. The first project undertaken by GEP members was sequence improvement and annotation of the dot chromosomes of several Drosophila species. The distal 1.2 Mb of this unusual small chromosome are of interest because the domain appears to be heterochromatic, a packaging state normally associated with gene silencing, but has the same gene density of more transcriptionally active euchromatic regions (8). Because of the high density of repetitious elements, improving the sequence to attain NHGRI mouse standard (1 error in 1000 bases) provides greater credibility to the analysis. Annotation then follows, with students generating the most plausible gene models based on available evidence, and then defending their models (4). (See SOM for a more detailed description.)
Economies in mentoring can be realized because the students are all using similar tools and strategies as they work independently on a specific region. The amount of sequence assigned to a student can be adjusted according to the available time. During a typical semester lab course one student can improve and annotate 40 kb of Drosophila DNA. By organizing the raw data in 40 kb packages that can be “claimed” from a web site, the GEP can undertake fairly large projects requiring sequence improvement and annotation.
Participating course instructors integrate this research into their courses using a variety of formats. Results from the individual students are then checked and pooled at Washington University to create the final dot chromosome assembly that is used to generate a variety of statistics documenting the nature of the chromosome and the genes present. Comparing dot chromosomes of several Drosophila species allows us to address issues of chromosome evolution.
Assessment and sustainability
Students know that their work will be used for a scientific publication and deposited in a public database for other scientists to build from. Sequence data are submitted to Genbank, and annotation results to FlyBase. One scientific paper has been published based on the student work (9); contributing students are co-authors. We anticipate an accumulation of publications and database contributions with time. The significance of this is illustrated by some of the comments from students and teaching assistants (SOM).
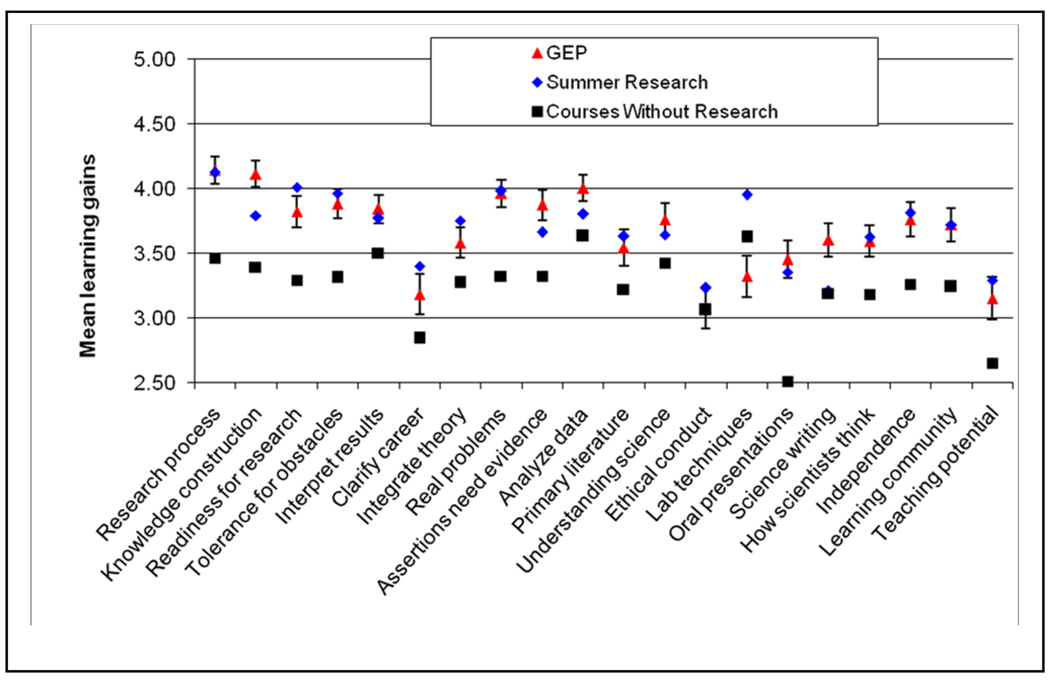
Students completed a post-course survey of their perceptions. This survey, a variation of the Classroom Undergraduate Research Experiences survey (CURE, 4), parallels one evaluating summer undergraduate research experiences (SURE, 1, 10). GEP students report learning and professional gains similar to those reported by students who have spent a summer in the lab (Figure 1). High ratings from GEP students on “knowledge construction”, “assertions need evidence”, “analyze data”, and “science writing” are similar to other research-like experiences (11, 12). From the perspective of students, the GEP course experience is more like summer research than like a standard science course without research. As the GEP matures, more assessment protocols are being added to our efforts. Currently the faculty are devising content tests on annotation and finishing. The GCAT has reported that students in that program make significant gains in comprehension of topics in functional genomics and show increased interest in research (13).
Figure 1.
Student ratings of learning gains. Mean learning gains for common survey items on the SURE, CURE, and GEP surveys. The SURE survey data represent 1973 undergraduate summer research experiences (“Summer Research” data points). The CURE survey data represent 560 student surveys from science courses that did not include a research-like experience (Courses Without Research” data points). The GEP survey data represent 308 students in GEP courses. Error bars represent 2 SEM. See SOM for data on student demographics. Genomics Education Partnership 14
Research opportunities in genomics are likely to increase. There are now 11 sequenced Drosophila species in addition to D. melanogaster, the commonly used lab species (14). By choosing regions of biological interest, such that the results contribute to research papers as well as to the databases, much can be accomplished by a student-scientist partnership that otherwise might be difficult to attain.
We invite scientists to consider what might be accomplished by a distributed community of undergraduate scientists collaborating on a particular set of genes or genomes. At present the GEP has over 40 members, and membership is growing (4). GEP faculty and their (usually undergraduate) Teaching Assistants (TAs) come to Washington Univ. for a one-week workshop to learn the relevant software and discuss pedagogical issues. As the program develops at the institution, the previous year’s students can be recruited as the next year’s TAs. This strategy fosters sustainability and allows class size to grow. Efficient implementation favors one experienced person (the TA or faculty member) for every 6–7 novices to provide support for students learning to use software and evaluate data. Faculty report that students, committed to a group effort, engage in spontaneous peer teaching, which allows a higher novice-to-expert ratio than is possible in many lab situations.
While most institutions already maintain computer labs, thus minimizing start-up costs, training for faculty and TAs is essential. Faculty and project leaders must agree on a common rubric for sequence improvement and annotation goals and on a common reporting mechanism. Good on site Information Technology support is important, but the basic strategy is very portable. The opportunity to teach about genes, genomes and the flow of biological information using this hands-on approach provides entry into a rich domain of data. We have found that involving undergraduates in a genomics research project is a rewarding way for faculty to teach and for undergraduates to learn.
Supplementary Material
References and Notes
- 1.Lopatto D. CBE Life Sci Educ. 2007;6:297. doi: 10.1187/cbe.07-06-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seymour E, et al. Sci. Educ. 2004;88:493. [Google Scholar]
- 3.Russell SH. In: Creating Effective Undergraduate Research Programs in Science. Taraban R, Blandon RL, editors. New York: Teachers College Press; 2008. pp. 53–80. [Google Scholar]
- 4.Genomics Education Partnership (http://gep.wustl.edu). This site includes the list of member institutions and a facsimile of the GEP assessment instrument.
- 5.http://www.ornl.gov/sci/techresources/Human_Genome/research/bermuda.shtml#1; http://www.genome.gov/Pages/Research/WellcomeReport0303.pdf
- 6.For examples see “Teaching Big Science at Small Colleges” http://serc.carleton.edu/genomics/index.html); “Genome Consortium for Active Teaching” (GCAT) introduces students to microarray experiments (http://www.bio.davidson.edu/projects/gcat/gcat.html.
- 7.The Science Education Alliance of HHMI works on phage (http://www.hhmi.org/grants/sea/) while the DOE Joint Genome Institute sponsors work on bacteria (http://www.jgi.doe.gov/education/genomeannotation.html).
- 8.Riddle NC, Elgin SCR. Chromosome Res. 2006;14:405. doi: 10.1007/s10577-006-1061-6. [DOI] [PubMed] [Google Scholar]
- 9.Slawson EE, et al. Genome Biology. 2006;7:R15. doi: 10.1186/gb-2006-7-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopatto D. In: Creating Effective Undergraduate Research Programs in Science. Taraban R, Blandon RL, editors. New York: Teachers College Press; 2008. pp. 112–132. [Google Scholar]
- 11.Denofrio LA, et al. Science. 2007;318:1872. doi: 10.1126/science.1150788. Genomics Education Partnership 12. [DOI] [PubMed] [Google Scholar]
- 12.Trosset C, et al. In: Creating Effective Undergraduate Research Programs in Science. Taraban R, Blandon RL, editors. New York: Teachers College Press; 2008. pp. 33–49. [Google Scholar]
- 13.Campbell MA, et al. CBE-Life Sci.Edu. 2007;6:109. doi: 10.1187/cbe.06-10-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosophila 12 Genomes Consortium. Clark A, et al. Nature. 2007;450:219. [Google Scholar]
- 15.We thank Frances Thuet (Washington Univ.) for management of the GEP CURE assessment web site; Lynn Crosby and William Gelbart of FlyBase (Harvard Univ.) and the staff of the Washington Univ. Genome Sequencing Center for assistance; and the Howard Hughes Medical Institute for financial support (grant #52005790 to SCRE, HHMI Professor). Genomics Education Partnership 13
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.