Abstract
How individual protein subunits assemble into the higher order structure of a protein complex is not well understood. Four proteins dedicated to the assembly of the V0 subcomplex of the V-ATPase in the ER have been identified in yeast, but their precise mode of molecular action remains to be identified. In contrast to the highly conserved subunits of the V-ATPase, orthologs of the yeast assembly factors are not easily identified based on sequence similarity. We show here that two ER-localized Arabidopsis proteins that share only 25% sequence identity with Vma21p can functionally replace this yeast assembly factor. Loss of AtVMA21a function in RNAi seedlings caused impaired cell expansion and changes in Golgi morphology characteristic for plants with reduced V-ATPase activity, and we therefore conclude that AtVMA21a is the first V-ATPase assembly factor identified in a multicellular eukaryote Moreover, VMA21p acts as a dedicated ER escort chaperone, a class of substrate specific accessory proteins so far not identified in higher plants.
Keywords: V-ATPase, assembly factor, ER-export, protein complex, Arabidopsis
Introduction
The V-ATPase is a multisubunit protein complex that acidifies the lumen of intracellular compartments in an ATP dependent way. The acidification of intracellular organelles plays an important role in a number of important cellular processes (1), such as receptor endocytosis and down regulation (2), intracellular membrane trafficking (3) and coupled transport of small molecules (4). The V-ATPase exhibits structural similarity to the F-ATPase of bacteria, mitochondria and chloroplasts (1). Like the F-AT Pase the V-ATPase consists of two subcomplexes; a cytosolic V1 subcomplex, which hydrolyzes ATP, and a membrane bound V0 subcomplex, which translocates protons from the cytosol to the lumen. The V1 subcomplex consists of 8 subunits (A-H) and can be assembled in the cytosol independent of the V0 subcomplex (5, 6). The V0 subcomplex consists of 6 different subunits in fungi (a, c, c’, c”, d & e), but only 5 different subunits (a, c, c”, d & e) in plants and animals (1, 7, 8). The V0 subunits are assembled into the V0 subcomplex in the Endoplasmic Reticulum (ER) membrane.
In yeast four genes have been identified encoding assembly factors that are necessary for assembly of the V0 subcomplex (9, 10). The four V-ATPase assembly factors (Vma12p, Vma21p, Vma22p & Pkr1p) are all localized to the ER membrane, and are not part of the final V0V1 holoenzyme (9, 10). VMA12 (Vma12p) encodes a 25 kDa protein with two predicted transmembrane domains, with its N- and C-termini orientated towards the cytosol (11). VMA22 (Vma22p) codes for a 21 kDa hydrophilic protein (12), that associates with the ER membrane through interactions with Vma12p. Vma12p and Vma22p form a complex and transiently interact with subunit a of the V0 subcomplex during assembly in the ER (13). VMA21 (Vma21p) encodes an 8.5 kDa protein with two predicted transmembrane domains that interacts with the ‘c ring’ of the V0 subcomplex made up of subunits c, c’ and c” (14). After V0 assembly is complete, Vma21p is transported together with the V0 subcomplex to the Golgi complex (14). Vma21p dissociates from the V0 subcomplex in the cis-Golgi and is retrieved back to the ER via a dilysine ER retrieval signal at the C-terminus Vma21p for more rounds of V0 subcomplex assembly.
In plants and other higher eukaryotes the assembly of the V-ATPase is largely unaddressed. Despite the strong conservation in overall composition of the V-ATPase between fungi, plants and animals, there is no evidence implicating the involvement of ER-based assembly factors in any organism except Saccharomyces cerevisiae. We have identified two potential Arabidopsis thaliana Vma21p orthologs. Here we report that one of these putative orthologs is ubiquitously expressed and similar to the yeast V-ATPase assembly factor Vma21p, AtVMA21a localizes to the ER. Remarkably, both AtVMA21a and AtVMA21b can function in place of yeast Vma21p in the assembly of the yeast V-ATPase. FRET analysis revealed that AtVMA21a and AtVHA-c” are intimately associated in plant cells. Finally, reduction of AtVM21a transcript levels led to impaired V-ATPase activity shown by reduced hypocotyl length in Arabidopsis. Our analysis indicates that AtVMA21a is required in the plant cell ER to assemble a functional V-ATPase, and that AtVMA21a is a true ortholog of yeast VMA21.
Results
Identification of two Arabidopsis orthologs of VMA21
The genome of the model plant species Arabidopsis thaliana encodes two paralogous proteins that share 25% protein sequence identity with yeast Vma21p, which we therefore tentatively named AtVMA21a (At2g31710) and AtVMA21b (At1g05780) (Figure 1A). RT-PCR was performed to analyze the expression of both candidate genes in different tissues and developmental stages of Arabidopsis. Whereas the AtVMA21a transcript could be detected in all samples indicating ubiquitous expression, no AtVMA21b transcript was found in etiolated seedlings (Figure 1B). However, low levels of AtVMA21b were found in flowers and siliques.
Figure 1. Arabidopsis has two potential Vma21p orthologs.
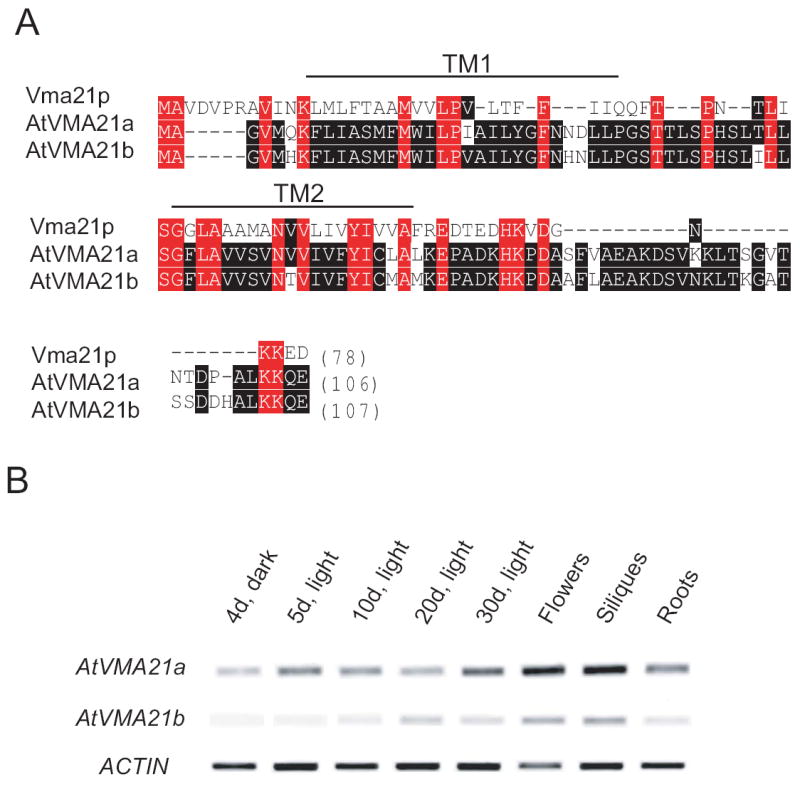
A) Amino acid sequences of yeast Vma21p and the Arabidopsis proteins AtVMA21a (At2g31710) and AtVMA21b (At1g05780) were aligned using the Clustal W algorithm. Vma21p and AtVMA21a share 28% protein identity, whereas Vma21p and AtVMA21b share 27% protein identity. Identity between the two Arabidopsis proteins is highlighted by black boxes, and identity between the yeast protein and the Arabidopsis proteins is highlighted by red boxes. The cleavage site for the predicted N-terminal signal peptide is indicated by an asterisk. B) RT-PCR analysis of AtVMA21a and AtVMA21b expression in Arabidopsis.
Total RNA for RT-PCR analysis was isolated from 4 day old etiolated seedlings, 5 day old light grown seedlings, 10 day and 20 day old plantlets, as well as leaves (leaves are not indicated in Fig 1B, unless “30d, light” are leaves), flowers, siliques and roots of 30d old Arabidopsis thaliana plants. Actin2 was amplified as an internal control.
Like VMA21p both Arabidopsis proteins are predicted to have two transmembrane domains. While the AtVMA21a and AtVMA21b regions C-terminal of the last predicted TMD are considerably longer then in yeast Vma21p, both AtVMA21a and AtVMA21b possess a C-terminal dilysine (KK)-motif that has been shown to be responsible for ER retrival of Vma21p (15) (Figure 1A).
To determine the subcellular localization of AtVMA21a, the GFP coding sequence was inserted between the transmembrane domain and the C-terminal KK-motif. A genomic fragment including the endogenous promoter was used to avoid localization artifacts caused by overexpression of AtVMA21a-GFP. Transgenic Arabidopsis lines carrying a single T-DNA insertion site were selected and were found to be indistinguishable from wildtype plants. Confocal laser scanning microscopy of meristematic root tip cells showed the typical ER-pattern of this cell type. (Figure 2A).
Figure 2. Subcellular localization of AtVMA21a.
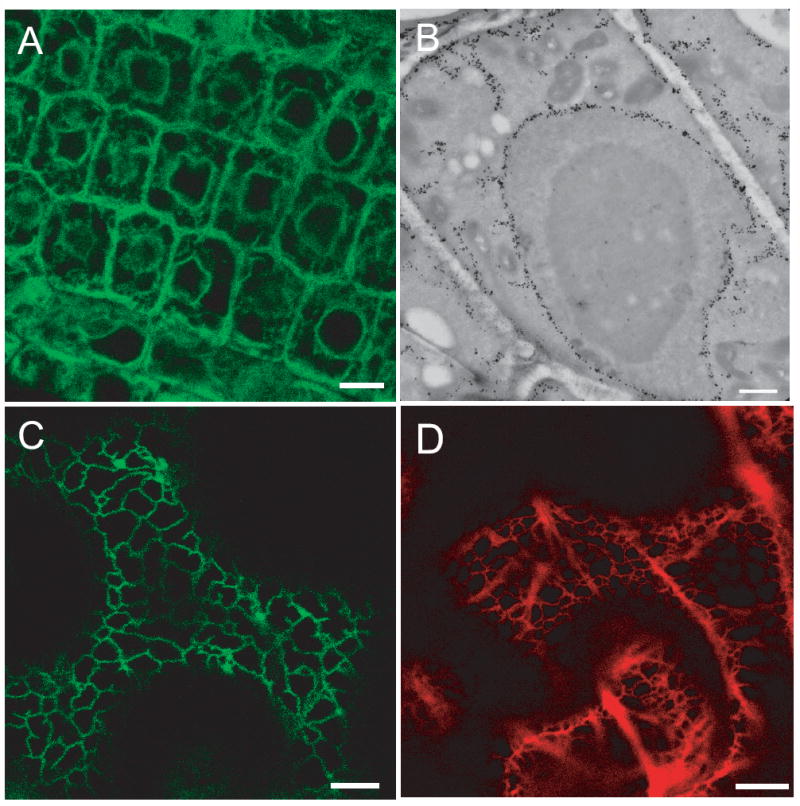
A) AtVMA21a-GFP localizes to the ER in Arabidopsis. CLSM image of root tip cells stably expressing AtVMA21a-GFP under the control of the endogenous promotor, scale bars represent 10μm. B) Overview of an immunogold labeled cryosection of a root tip cortex cell expressing AtVMA21-GFP. Gold markers accumulate in nuclear envelope, ER strands and the cortical ER, scale bar represents 1μm. C) AtVMA21a-GFP localizes to the ER in N. benthamiana. CLSM image of leave epidermal cells transiently expressing AtVMA21a-GFP after Agrobacterium infiltration. D) mRFP-AtVMA21 is also localized to the ER. CLSM image of leave epidermal cells transiently expressing mRFP-AtVMA21a after Agrobacterium infiltration, scale bar represents 10μm.
Immunogold electron microscopy was performed to confirm the ER localization of AtVMA21-GFP. In ultra-thin cryosections of root tissue from AtVMA21-GFP seedlings incubated with a GFP-antibody, gold particles specifically labeled the nuclear envelope, ER strands and a thin layer of cortical ER (Figure 2B).
When the same construct was transiently expressed in Nicotiana benthamiana after Agrobacterium infiltration, GFP fluorescence was associated with the typical reticulate ER found in epidermal cells (Figure 2C). A similar ER pattern was observed when mRFP-AtVMA21a, an N-terminal fusion with monomeric red fluorescence protein, was expressed in N. benthamiana (Figure 2D). We therefore conclude that the ER-localization of AtVMA21a-GFP reflects the endogenous localization of AtVMA21a and is not an artifact of its fusion with GFP.
Arabidopsis AtVMA21a and AtVMA21b localize to the yeast ER
To assess the ability of AtVMA21a and AtVMA21b to function in place of yeast Vma21p, a low-copy, centromere-based plasmid was generated in which the cDNA sequence encoding AtVMA21a or AtVMA21b replaced the coding sequence of Vma21p, placing the AtVMA21 coding sequences under control of the yeast VMA21 promoter and terminator elements. To allow detection of these proteins we inserted either the HA epitope or GFP immediately after the start codon. Immunoblot analysis of protein samples prepared from vma21Δ yeast expressing HA-tagged forms of either AtVMA21a (pLG270) or AtVMA21b (pLG266) was performed to determine if these proteins were expressed and stable in yeast. Enriched yeast membrane protein samples were prepared, proteins were separated by SDS-PAGE, and probed for the presence of HA epitope-tagged proteins. As a control, a sample was also prepared and analyzed from yeast cells expressing the HA epitope-tagged ScVma21p (pLG277). The yeast VMA21 gene encodes a protein of 77 amino acids with a predicted molecular weight of 8.5 kDa (15). The N-terminal 3xHA epitope-tagged ScVma21p is fully functional and migrates as a protein of approximately 15 kDa (Figure 3, top panel). Protein bands also migrating at ~15 kDa were recognized by the anti-HA antibodies in samples of yeast cells expressing either AtVMA21a or AtVMA21b, indicating that these proteins are synthesized and stable in yeast.
Figure 3. Arabidopsis AtVMA21a and AtVMA21b are stably expressed in yeast cells.
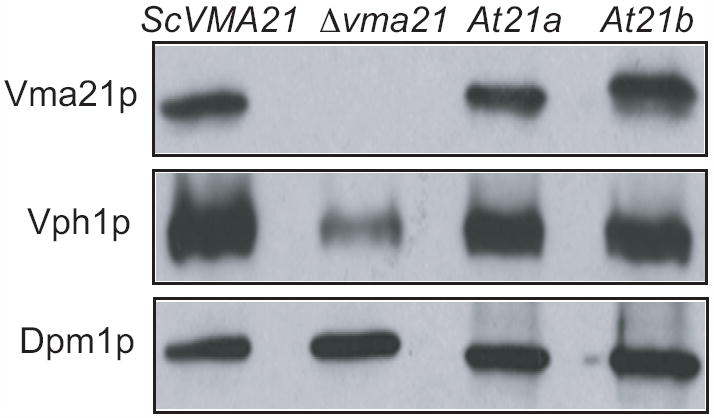
Immunoblot analysis of membrane fractions from vma2Δ yeast cells transformed with N-terminal HA-tagged constructs of Vma21p (pLG277), AtVMA21a (pLG270) or AtVMA21b (pLG266). 20 ug of membrane protein sample was loaded in each lane of a SDS-PAGE gel, resolved by electrophoresis, transferred to nitrocellulose and probed with antibodies recognizing the HA epitope, Vph1p and Dpm1p.
The membrane samples were also probed with antibodies detecting the V-ATPase V0 subunit “a” (Vph1p). Yeast cells lacking Vma21p (or any other V-ATPase assembly factor; (9) fail to assemble the V0 subcomplex, and the V0 subunit Vph1p is degraded rapidly in these cells. As previously reported, vma21Δ cells have significantly lower levels of Vph1p (15). Samples were also probed with antibodies recognizing the ER membrane protein Dpm1 to ensure equal amounts of protein was loaded in each lane (Figure 3, lower panel). The near wild-type levels of Vph1p present in yeast cells expressing AtVMA21a or AtVMA21b suggests the presence of a stable V0 subcomplex (Figure 3, middle panel).
The Arabidopsis proteins AtVMA21a and AtVMA21b were GFP tagged and expressed in yeast, and the cellular localization determined by fluorescence microscopy. GFP-tagged ScVma21p, a well-characterized ER membrane protein (15), displays bright green perinuclear staining (with the nucleus defined by Hoechst 33342 staining) in addition to staining ER membranes adjacent to the plasma membrane (Figure 4, top panel). Fluorescence from GFP-tagged AtVMA21a (Fig 3, middle panel) and AtVMA21b-G (Fig 3, bottom panel) also display perinuclear staining, consistent with ER localization of these Arabidopsis proteins expressed in yeast. These results indicate that AtVMA21a and AtVMA21b are stably expressed in yeast, these proteins localize to the ER, and the V0 subunit Vph1p is stabilized by AtVMA21 expression, strongly suggested normal assembly of the V0 subcomplex.
Figure 4. Arabidopsis AtVMA21a and AtVMA21b demonstrate ER localization similar to the yeast V-ATPase assembly factor Vma21p.
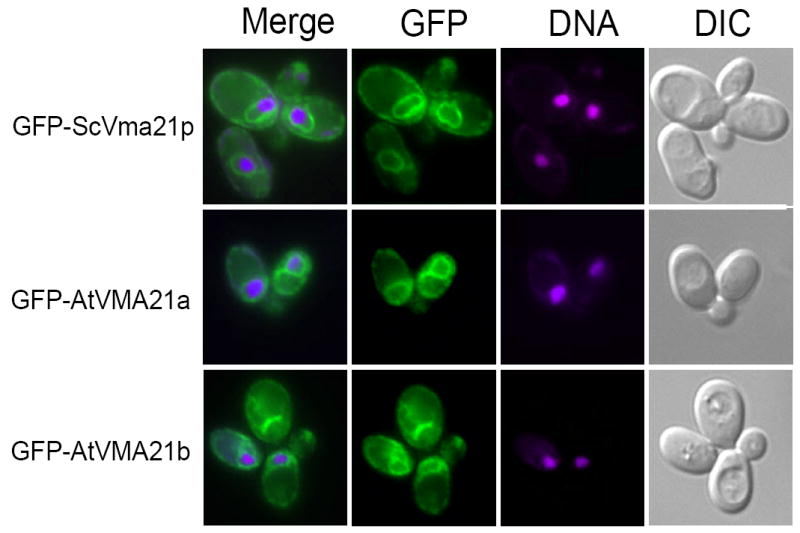
Cell imaging of vma21Δ yeast cells transformed with N-terminal GFP-tagged constructs of ScVma21p (pLG278), AtVMA21a (pLG269) or AtVMA21b (pEB05). Prior to cell imaging samples were fixed (3.7% formaldehyde for 10 minutes) and incubated with 5 mg/ml Hoechst 33342 DNA binding dye. The panels on the right show yeast cell morphology as viewed by differential interference contrast microscopy (DIC). The panels on the far left are a merge of the green GFP fluorescence (second set of panels from the left) with the magenta-stained nucleic acid fluorescence (third set of panels from the left).
AtVMA21a and AtVMA21b function in V-ATPase assembly in yeast
To determine whether AtVMA21a and AtVMA21b are functional orthologs of Vma21p, we tested their ability to functionally replace Vma21p in yeast mutant cells lacking this protein (vma21Δ). Yeast cells lacking a functional V-ATPase, due to disruption of any VMA gene, are unable to grow on media containing elevated levels of calcium, yet are able to grow on media buffered to the acidic pH 5.0 (9). Membrane-bound transport proteins responsible for maintaining ionic homeostasis in the cell cytosol utilize the proton gradient generated by the V-ATPase (4). The growth phenotype of cells lacking the required assembly factor ScVma21p was compared to vma21Δ cells transformed with plasmid expressing ScVma21p or expressing either Arabidopsis AtVMA21a or AtVMA21b. As shown in Figure 5, cells lacking Vma21p (vma21Δ), thus lacking a functional V-ATPase, display compromised growth even on rich media (YEPD) buffered to pH 5.0 and do not grow on rich media in the presence of 60 mM CaCl2. The introduction of the yeast VMA21 gene on a plasmid in vma21Δ cells allowed the cells to grow in the presence of elevated calcium due to the ability of the cells to assemble a functional V-ATPase. The introduction of the plasmid expressing the Arabidopsis AtVMA21a protein in vma21Δ cells also allowed the cells to grow in the presence of 60 mM CaCl2, indicating that AtVMA21a can function in place of ScVma21p to bring about the assembly of a functional V-ATPase (Figure 5). The HA-epitope tagged and GFP forms of AtVMA21a also allowed vma21Δ cells to grow on 60 mM CaCl2, revealing that AtVMA21a function is not compromised as a V-ATPase assembly factor due to the presence of either the HA epitope tag or GFP.
Figure 5. Arabidopsis AtVMA21a and AtVMA21b reverse the compromised growth phenotype in the presence of elevated levels of calcium of yeast cells lacking ScVma21p.
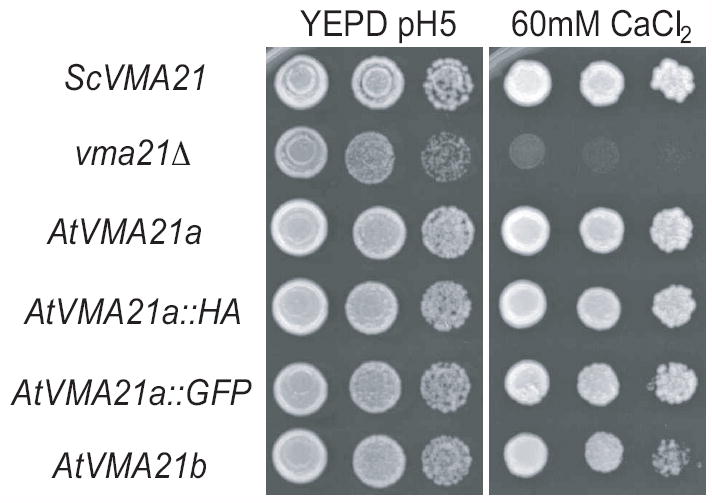
Exponentially growing cultures of vma21Δ yeast cells transformed with various plasmid constructs were serially diluted and spotted onto rich medium buffered to pH 5 (YEPD) or rich medium containing 60 mM CaCl2. ScVMA21, AtVMA21a, AtVMA21a⌧HA, AtVMA21a⌧GFP, AtVMA21b, AtVMA21b⌧HA and AtVMA21b⌧GFP represent vma21Δyeast cells transformed with pLG277, pLG268, pLG270, pLG269, pEB03, pLG266, and pEB05 respectively.
Similarly, the expression of the Arabidopsis AtVMA21b protein in vma21Δ cells rescued growth of these cells on elevated calcium and the growth, and the function of AtVMA21b was also unaffected by the addition of either the HA epitope tag or GFP (Figure 5). These growth data indicate that both AtVMA21a and AtVMA21b can function in yeast in a manner identical to the required V-ATPase assembly factor ScVma21p.
The majority of the V-ATPase assembled in yeast is localized to the vacuolar membrane, where it functions in ATP-driven translocation of protons into the lumen of the vacuole. This translocation of protons results in acidification of the vacuole lumen. Vacuolar acidification can be visualized in live yeast cells using quinacrine, a fluorescent dye that accumulates in acidic cellular compartments. As shown in Figure 6, vma21Δ cells expressing ScVma21p exhibit highly fluorescent vacuoles, seen as bright green disks when visualized under the microscope. Cell morphology was viewed by differential interference contrast microscopy (DIC) where the vacuole can be visualized as a large indentation within the yeast cell. The outline of the yeast cells, depicted in red, is visualized due to concanavalin A-tetramethylrhodamine bound to the yeast cell wall (Figure 6). Yeast cells lacking Vma21p (vma21Δ) do not assemble a functional V-ATPase and thus do not contain acidic vacuoles. As expected, vma21Δ cells failed to accumulate the quinacrine fluorescent dye as seen by a lack of any green fluorescence (Figure 6). Cells lacking endogenous Vma21p (vma21Δ) but expressing either AtVMA21a or AtVMA21b exhibit bright green vacuolar fluorescence, indicating highly acidified vacuoles due to a functional V-ATPase complex present on the vacuolar membranes (Figure 6A). Additionally, vacuolar membranes were isolated from cells lacking ScVma21p but expressing AtVMA21a and the ATPase activity was determined to be 67% of the wild-type activity (Figure 6B). These data confirm that either AtVMA21a or AtVMA21b can function in the yeast ER to replace ScVma21p in the assembly of the V0 subcomplex and a fully functional V-ATPase complex.
Figure 6. Yeast cells expressing Arabidopsis AtVMA21a or AtVMA21b exhibit wild-type levels of V-ATPase dependent vacuolar acidification.
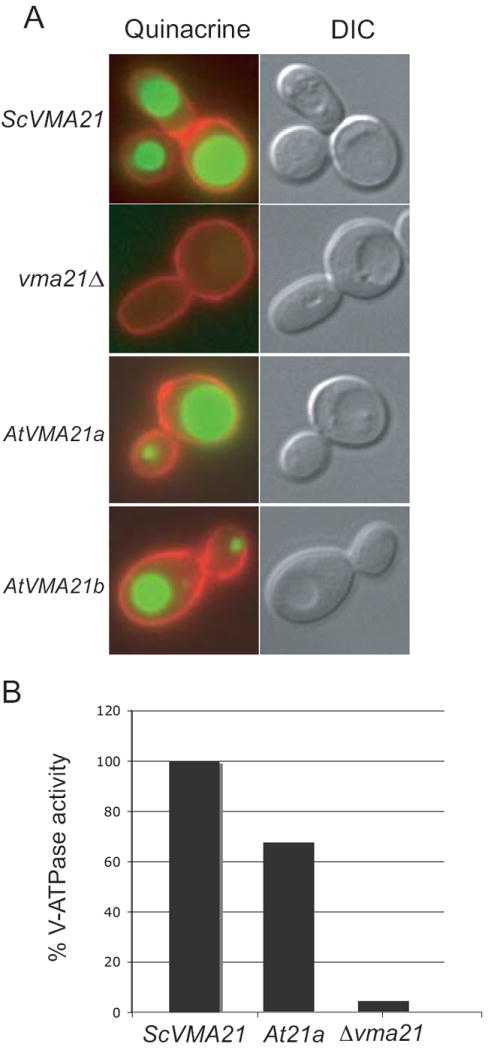
A) Cells lacking ScVma21p (vma21Δ) and transformed with plasmid expressing either AtVMA21a (pLG268; At21a) or AtVMA21b (pEB03; At21b) were stained with quinacrine and concanavalin A tetramethylrhodamine as described in the Materials and Methods. The images on the right show cells morphology as observed by differential interference contrast microscopy (DIC). The panels on the left depict fluorescence of the quinacrine in green and of the concanavalin A tetramethylrhodamine in red. B) The ATPase activity measured from vacuolar membranes isolated from vma21Δ cells expressing ScVma21p was 0.405 +/-0.006 μmol of ATP/min/mg and from vacuolar membranes isolated from vma21Δ cells expressing AtVMA21a was 0.270 +/- 0.007 μmol of ATP/min/mg.
AtVMA21a interaction with VHA-c” as detected by FRET analysis
Having shown that AtVMA21a and AtVMA21b can replace Vma21p in V-ATPase assembly in yeast, it remained to be shown, if V-ATPase assembly also is their in planta function. We therefore used FRET analysis to determine if a direct physical interaction of AtVMA21 and subunits of the Arabidopsis V0 complex could be detected. To this end the proteolipid subunit VHA-c” was fused to CFP and AtVMA21a to YFP. Radiationless energy transfer was measured by quantitative imaging of Arabidopsis protoplasts transiently co-expressing both VHA-c”-CFP and AtVMA21a-YFP (Figure 7A). The calculated FRET efficiency was 27.7 ± 1.5 (mean ± SE, n=246), the distribution of FRET efficiencies revealed two maxima of occurrence: i) low FRET efficiency below 10% and ii) medium FRET efficiency in the range of 20-30% (Figure 7C). The co-existence of two maxima was not caused by fluctuations in fluorophor stoichiometry. The stoichiometry index was approximately 0.66, thus displaying a slight excess of CFP and a correlation between stochiometry and FRET efficiency was not detectable (R2=0.01; Figure 7B). Regardless of fluorophor orientation, a FRET-efficiency below 10% corresponds to a distance of more than 7.2 nm, which exceeds the dimensions of the V0-sector. Recently, VHA-c” and VHA-e2 were identified at the ER and VHA-e1 in endosomal compartments (16). A corresponding isoform- and compartment-specific association of VHA-c” and VHA-e isoforms was observed by FRET measurements. The ER-specific subunits VHA-e2 and VHA-c” were shown to associate (20% FRET-efficiency) but not the putative endosomal VHA-e1 and the ER-specific VHA-c” (3.3% FRET-efficiency) (16). Within the present study, the first maximum resembles the results obtained with VHA-e1 and VHA-c”. Hence, it is concluded, that the first maximum is caused by the absence of interaction between AtVma21a and VHA-c”, whereas the second maximum reveals the association of V-ATPase and AtVma21a. These results indicate that AtVMA21a and the V0 subunit VHA-c” are partially in very close proximity in plant cells, consistent with the proposal that AtVMA21a functions as an ER-localized V-ATPase assembly factor that interacts with V0 subunits only during assembly.
Figure 7. FRET between VHA-ch”-CFP and AtVMA21a-YFP indicates that these proteins interact closely.
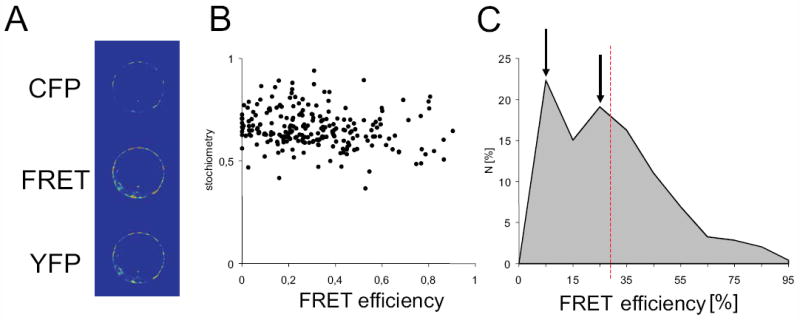
A) representative FRET-images. CFP and YFP emission was detected by CLSM of transiently transfected mesophyll protoplasts. Emission was recorded in three channels comprising CFP-, FRET- and YFP-channel. CFP and YFP channels were essential for estimation of CFP-crosstalk and YFP-direct excitation. The average intensity was 2779 ± 953 in the YFP-channel, 1843 ± 982 in the CFP-channel and 3448 ± 650 in the FRET channel (mean ± SD, n=246). B) Relationship of fluorophor stochiometry index and FRET-efficiency. The average stoichiometry index of CFP and YFP was 0.66. C) Distribution of calculated FRET efficiency. Maxima are indicated by arrows, average FRET efficiency is shown by dashed red line.
AtVMA21a-RNAi lines exhibit a V-ATPase deficient phenotype
Total loss of V-ATPase activity as caused by null alleles of Arabidopsis VHA-genes results in gametophyte or embryo lethality (17), whereas cell expansion is compromised in Arabidopsis plants with reduced V-ATPase activity in the Golgi/TGN (18, 19). We therefore chose an inducible RNAi approach to interfere with AtVMA21a expression and determine the effect on cell expansion. The AtVMA21 coding sequence was cloned as an inverted repeat and brought under the control of the ethanol-inducible AlcA promotor (20). When etiolated seedlings from homozygous AtVMA21a-RNAi lines were grown in the presence of 0.2% ethanol (EtOH) hypocotyl length was reduced to 30% of the control without EtOH but wild type seedlings did not show any significant change in hypocotyl length (Figure 8). This reduction in hypocotyl growth reflects a reduction in V-ATPase function (17, 18).
Figure 8. AtVMA21a-RNAi lines exhibit a V-ATPase deficient phenotype.
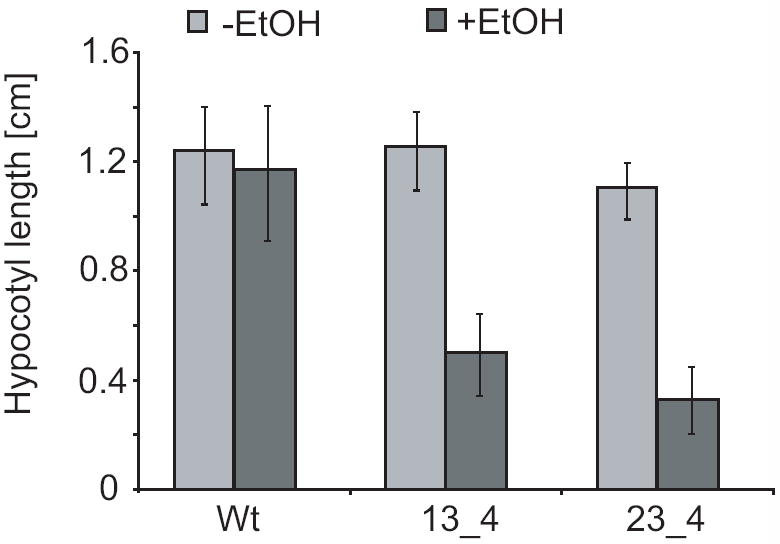
Hypocotyl length of 4 day old etiolated seedlings grown in the absence or presence of ethanol. Shown are the results for the wildtype background Col-0 and two independent transgenic lines expressing the RNAi-construct. At least 30 seedlings of each line were measured, error bars represent standard error.
On the subcellular level, in agreement with the presence of a Golgi/TGN-specific isoform of VHA-a (22), V-ATPase inhibition in Arabidopsis causes characteristic changes in the morphology of Golgi stacks and their associated trans-Golgi networks (19, 21, 22). We therefore used electron microscopy of high-pressure frozen and freeze-substituted seedlings to examine the ultrastructural effects of AtVMA21a inhibition. In contrast to the normal Golgi morphology of control seedlings (Figure 9A), Golgi stacks of seedlings grown in the presence of ethanol displayed bending and swelling of cisternae (Figure 9B), typical for reduced V-ATPase function (20). These results indicate that AtVMA21 is required for V-ATPase function in Arabidopsis.
Figure 9. Altered Golgi morphology in AtVMA21a-RNAi seedlings.
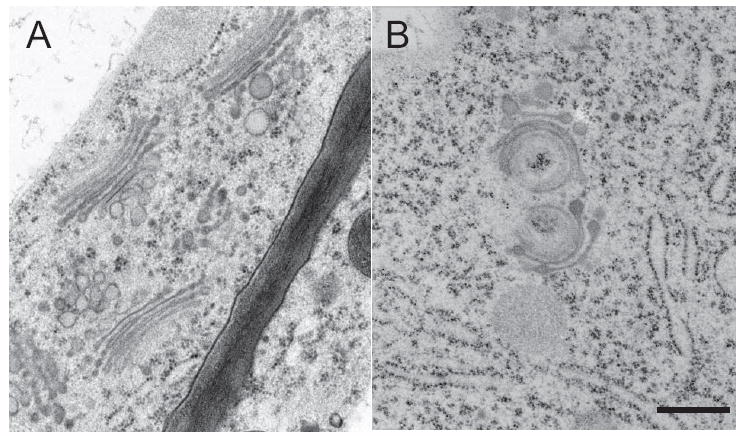
Electron micrographs of Golgi stacks and their associated TGNs in high-pressure frozen and freeze substituted root tip cortex cells of uninduced (A) and induced AtVMA21-RNAi seedlings (B), scale bar represents 0.25μm.
Discussion
Protein complexes are likely to be the most important units of cellular function and yet in most cases it remains elusive how assembly of the individual protein subunits into a higher order structure is achieved. The yeast V-ATPase is exceptional as four proteins dedicated to the assembly of the Vo subcomplex have been identified but the exact molecular mechanisms underlying assembly of the Vo complex in the ER membrane remain to be determined. Moreover, it is still unclear, how coordination between assembly and vesicular transport out of the ER is achieved. The identification of functional orthologs of V-ATPase assembly factors would not only demonstrate that their presence is not limited to yeast, but could also provide important insight into structural and functional determinants which in turn might provide insight into the molecular mode of action of Vo assembly.
Given the ubiquitous presence of the V-ATPase it may seem likely that assembly factors are also found in higher eukaryotes. However, in contrast to genes encoding V-ATPase subunits, which are easily identified in any eukaryotic genome, potential orthologs of the yeast assembly factors are substantially less well conserved.
We have shown here that two Arabidopsis proteins that are functional orthologs of yeast VMA21p. Like in most higher eukaryotes, many subunits of the Arabidopsis V-ATPase are encoded by multiple genes (7). Assuming that at least some of the isoforms are co-expressed, a multitude of V-ATPase complexes with different subunit composition could be formed, adding an additional layer of complexity to the question of assembly.
AtVMA21a and AtVMA21b function in V-ATPase assembly in yeast
There are four dedicated V-ATPase assembly factors in yeast, and all of these proteins are localized to the ER membrane. Yeast cells lacking the assembly factor Vma21p exhibit phenotypes that are indistinguishable from the phenotypes resulting from deletion of V-ATPase subunit genes (9). In addition, Vma21p exits the ER together with the V0 subcomplex in COPII vesicles, and blocking entry of Vma21p into COPII vesicles blocks sorting of the V0 subcomplex into these vesicles (14). These studies point to the central role played by Vma21p in the assembly of the V0 subcomplex in the ER, and in the transport of the V0 subcomplex out of the ER.
We expressed the Arabidopsis proteins AtVMA21a and AtVMA21b in yeast cells lacking the endogenous Vma21p (vma21Δ cells) to test whether either of these plant proteins could functionally replace Vma21p in V-ATPase assembly and transport. We found that HA-tagged and GFP-tagged forms of AtVMA21a and AtVMA21b were stable in vma21Δ yeast and localized to the ER membrane. Phenotypic analysis of these cells revealed that either AtVMA21a or AtVMA21b could functionally replace yeast Vma21p; these yeast cells exhibited normal ability to grow on media containing elevated Ca2+ (requires V-ATPase function), their vacuoles were acidified normally as revealed by quinacrine accumulation. These studies reveal that AtVMA21a and AtVMA21b are both functional orthologs of the fungal Vma21 protein.
Arabidopsis VMA21a functions in V-ATPase assembly
The V-ATPase of Arabidopsis thaliana has been well characterized and found to reside throughout the dynamic endomembrane system of different tissues of the plant, including both the endocytic and secretory membrane network (22). V-ATPase function has been implicated in both endocytic and exocytic functions in plants, in addition to the anticipated functions in the tonoplast/vacuole. Despite the wealth of information on higher eukaryotic V-ATPase composition, localization, and function, there is a complete lack of information about the assembly of this multisubunit complex (1).
V-ATPase assembly has been well characterized in yeast, where it has been shown that the ER membrane protein Vma21p interacts with newly synthesized V0 subunits and functions in V0 subcomplex assembly. We show here that Arabidopsis has two genes (AtVMA21a and AtVMA21b) predicted to encode proteins with ~25% amino acid identity with yeast Vma21p. Whereas AtVMA21a is expressed ubiquitously in different tissues and developmental stages, the expression of AtVMA21b was more limited. Both AtVMA21a and AtVMA21b are predicted to carry the C-terminal dilysine motif, which functions in ER retention/retrieval in plants and fungi. Expression of a GFP-tagged AtVMA21a revealed that AtVMA21a is indeed an ER membrane protein.
The yeast Vma21 protein has been shown to be in close contact with the so-called proteolipid ring composed of the V0 subunits c, c’ and c” but subunit c’ has been implicated as the primary binding surface that allows VMA21p to interact with other Vo subunits (14). Interestingly, orthologs of subunit c’ are not found in higher eukaryotes indicating that although the AtVMA21 proteins are able to functionally replace VMA21p, their function in Arabidopsis requires interaction with another Vo subunit. We therefore labeled Arabidopsis AtVMA21a with YFP and the Arabidopsis proteolipid subunit VHA-c” with CFP and expressed these proteins in plant protoplasts, and measured fluorescence energy transfer between these hybrid proteins. The strong FRET efficiency indicated that the ER-localized putative V-ATPase assembly factor AtVMA21a and the V0 subunit VHA-c” are very closely associated in plant cells. Since AtVMA21a is restricted in localization to the ER membrane, we conclude that AtVMA21a and VHA-c” come into close proximity during V0 subcomplex assembly in the ER.
Previous analysis of V-ATPase function in plants has revealed that null alleles of Arabidopsis VHA genes results in embryo and gametophyte lethality (17, 21). We used an inducible RNAi approach to interfere with AtVMA21a expression, and found that interference with AtVMA21a expression resulted in significant changes in hypocotyl length, as expected for loss of V-ATPase function in etiolated seedlings. At the cellular level AtVMA21a inhibition caused bending and swelling of Golgi cisternae typically found in plant cells with reduced V-ATPase function. These studies indicate that reducing AtVMA21a function in Arabidopsis interferes with V-ATPase activity, revealing that AtVMA21a is required for normal V-ATPase function in Arabidopsis just as Vma21p is required for function of the V-ATPase in yeast.
Conclusion
We have shown here that functional orthologs of the ER-localized V-ATPase assembly factor Vma21p are present in higher eukaryotes. We therefore propose that the mechanism of ER-localized assembly of the V-ATPase complex is conserved across all eukaryotic cells and that comparative analysis of assembly factors form diverse eukaryotes will thus be important to determine the underlying molecular mechanism. Moreover, the presence of two isoforms of AtVMA21 indicates that the potential of higher eukaryotes to form multiple V-ATPase complexes might be reflected at the level of complex assembly.
Materials and Methods
Plant Materials and Growth Conditions
All plants used were Arabidopsis thaliana ecotype Col-0. Seedlings used for microscopy were grown on Murashige and Skoog (MS) medium + 1% sucrose at 22°C, with cycles of 16 h of light and 8h of dark for 3 to 5 d. Etiolated plants were grown on MS medium + 1% sucrose and + 0.2% Ethanol at 22°C in darkness for 3 to 5 d. A 4 h light stimulus was given after plating the seeds on MS medium to ensure even germination.
Plasmid Constructs and Plant Transformation
For AtVMA21a (At2g31710), a 1,312-bp genomic fragment was amplified using primers AtVMA21.FOR (5’-CAG CTC TTC TAG CTC TAC TGC-3’) and AtVMA21.REV (5’-TGT CAA GAG AAT GCG GAA GAG-3’) and cloned into pCR2.1-TOPO (Invitrogen). A NheI site was introduced into Exon 2 without changing the amino acid sequence. GFP5-S65T was inserted into the NheI site. The resulting fragment was cloned into the binary vector pTkan, a derivative of pPZP221 (23). The resulting binary plasmids were introduced into Agrobacterium tumefaciens strain GV3101:pMP90 and selected on 5 μg/mL rifampicin, 10 μg/mL gentamycin, and 100 μg/mL spectinomycin. Col-0 plants were transformed using standard procedures, and transgenic plants were selected on MS medium + 1% sucrose plates containing 50 μg/mL kanamycin. To amplify the cDNA of AtVMA21-GFP, RNA was isolated from transgenic seedlings expressing the genomic GFP-fusion described above using the RNeasy plant mini kit (Qiagen, Hilden, Germany). cDNA was generated from total RNA using MMLV-RTase (Fermentas, St. Leon-Rot, Germany) and oligo dT primer. AtVMA21∷GFP cDNA was amplified using the primers AtVMA21cds.FOR (5’- ATG GCT GGG GTT ATG CAC AAG-3’) and AtVMA21cds.REV (5’- CTC TTG TTT CTT CAG CGC AG-3’). To create AtVMA21a-RNAi lines, the coding sequence of AtVMA21 was amplified from wildtype cDNA using AtVMA21cds.FOR and AtVMA21cds.REV and cloned as an inverted repeat into a derivative of pHANNIBAL (24) in which the 35S-Promotor has been replaced by the ethanol inducible promoter pAlcA (20). The pAlcA:VMA21-RNAi cassette was then cloned into the NotI site of the plant transformation vector pBart_AlcR, that contains the coding sequence of the alcR transcription factor required to activate the pAlcA promoter in the presence of ethanol. Transgenic plants were selected based on the phosphinothricin (BASTA) resistance conferred by the bar gene contained in pBART_AlcR. Homozygous lines were established and hypocotyl length of etiolated seedlings grown on MS plates containing 0.2% ethanol was compared to seedlings grown in the absence of ethanol. For transient expression in protoplasts, the cDNA of AtVMA21a was amplified using primers 5’-atggctggggttatgcag-3’ and 5’-tcactcttgtttcttcag-3’. Flanking BamHI and AgeI restriction sites were added by PCR using primers 5’-aaaaggatccaatggctggggttatgcag-3 ’ and 5 ’-ttttaccggtctctcttgtttcttcag-3’. The digested fragment was introduced into 35S-CFP-NosT plasmid (25).
RT-PCR
Total RNA was extracted from 4-d-old etiolated, 5-d-old, 10-d-old and 20-d-old light grown seedlings using the RNeasy plant mini kit (Qiagen) and used as a template for cDNA synthesis with MMLV-RTase (Fermentas, St. Leon-Rot, Germany) according to the manufacturer’s instructions. Actin 2 (At3g18780) was used as a control.
Confocal laser scanning microscopy
Confocal laser scanning microscopy was executed using a Leica TCS SP2 confocal laser-scanning microscope (Leica, Germany). All CLSM images were obtained using the Leica Confocal software and a 63x water-immersion objective. The excitation wavelength was 488 nm; emission was detected for GFP between 500 and 530 nm, for RFP between 565 and 600 nm. Images were processed using Adobe Photoshop.
FRET-analysis
Protoplasts were isolated and transiently transfected as described before (25). For FRET-analysis a Leica TCS SP2 confocal system with 63-fold magnification (oil immersion objective, NA=1.4) and the double dichroic filterset DD458/514 was used. The scan speed was 400 Hz, the image resolution 1024*1024 pixels and background noise was eliminated by line averaging and a PMT offset value of -20. CFP was detected in the range of 470-510 nm (CFP-intensity ICFP) and FRET (FRET raw intensity IFRET) in the range of 530-600 nm after 458 nm excitation. The YFP-reference was obtained in the range of 530-600 nm after 514 nm excitation (YFP-intensity IYFP). The signal amplification was identical for CFP and YFP-detection. The detection of YFP direct excitation by the 458 nm laserline depended on the intensity of the 514 nm laserline. The correction factors kdir for YFP direct excitation were 1.027 (5% laser intensity) and 0.452 (7% laser intensity). The FRET images were quantified using the Quantify modul of the Leica confocal software and processed using Zeiss Image Browser. Maxima were read out and FRET efficiencies were calculated. Direct excitation intensities of YFP and CFP-crosstalk were subtracted from the signal recorded in the FRET-channel. The remaining intensity of the FRET channel was finally normalized to the sum of the emission intensities in the donor channel and the FRET channel: Equation 1: E = (IFRET − 0.61 * ICFP − kdir * IYFP) / (IFRET + ICFP). The fluorophor stochiometry index S was calculated as suggested for ALEX (alternating laser excitation) measurements (26): Equation 2: S = (ICFP + IFRET) / (ICFP + IFRET + IYFP) Arithmetic mean as well as standard error were calculated.
Transmission Electron Microscopy
For structural analysis, root tips were either chemically fixed with 2.5% glutaraldehyde, 1% aqueous uranyl acetate, and 1% osmium tetroxide, dehydrated, and embedded in Epon or high-pressure frozen (Bal-Tec HPM 010; Balzers) in hexadecane (Merck Sharp and Dohme), freeze-substituted (72 h, − 90°C; 8 h, −60°C; 8 h, −35°C; 4 h, 0°C) in acetone containing 2% osmium tetroxide and 0.5% uranyl acetate, washed at 0°C, and embedded in Epon. Immunogold labeling was performed on ultrathin thawed Tokuyasu cryosections of formaldehyde-fixed (8%, 3h) and sucrose-infiltrated (2.1 M) root tips using rabbit anti-GFP serum (1:25; Abcam Ltd, Cambridge, UK) and silver-enhanced (HQ Silver, 6 min; Nanoprobes, New York, USA) goat (Fab′) anti-rabbit IgG coupled to Nanogold (#2004, Nanoprobes, New York, USA)
Yeast Strains, Plasmids and Culture Conditions
Strains and plasmids used in the study are listed in Table I. Yeast were cultured in YEPD, YEPD buffered to pH 5.0 using 50 mM succinate/phosphate, or yeast nitrogen base synthetic complete minimal medium supplemented with dextrose (SD) and amino acids as needed using standard techniques. To test the growth phenotype of various yeast strains, exponentially growing yeast cells cultured in SD medium plus appropriate amino acids were diluted to cell density measuring 1.0 O.D.600 and then further serially diluted 10 fold and spotted on agar medium. 5 ml of the starting culture and each dilution were spotted onto a YEPD pH 5.0 agar or YEPD + 60 mM CaCl2 agar and incubated 48 hours at 30°C.
Table 1.
Yeast Strains and plasmids used in this study
| Description | Origin | |
|---|---|---|
| Yeast Strains | ||
| SF838-1Dα | MATα ade6, leu2-3,112, ura3-52, pep4-3, his4-519, gal2 | (30) |
| TASY006 | SF838-1Dα̣ vma21Δ∷Kanr | (31) |
| Plasmids | ||
| pJET1-AtVMA21a | cDNA insert encoding AtVMA21a (At2g31710) | This work |
| pENTR221-AtVMA21b | cDNA insert encoding AtVMA21b (At1g05780) | This work |
| pRS316 | CEN ARS URA3 AMP | (32) |
| pEB03 | pRS316 ScVMA21pro-AtVMA21b-ScVMA21 3’UTR | This work |
| pEB05 | pRS316 ScVMA21pro-GFP-AtVMA21b-ScVMA21 3’UTR | This work |
| pLG263 | pRS316 ScVMA21 Nt Bgl II site codons 3 & 4 | This work |
| pLG266 | pRS316 ScVMA21pro-3xHA-AtVMA21b-ScVMA21 3’UTR | This work |
| pLG277 | pRS316 3XHA-ScVMA21 | This work |
| pLG278 | pRS316 GFP-ScVMA21 | This work |
| pLG268 | pRS316 ScVMA21pro-AtVMA21a-ScVMA21 3’UTR | This work |
| pLG269 | pRS316 ScVMA21pro-GFP-AtVMA21a-ScVMA21 3’UTR | This work |
| pLG270 | pRS316 ScVMA21pro-3xHA-AtVMA21a-ScVMA21 3’UTR | This work |
| pSMY92 | 123bp 3XHA Bgl II fragment | (33) |
| CY3714 | GFP(S65T)-HIS3MX6 | (34) |
Cloning and GFP or HA tagging
A PCR fragment was generated containing the AtVMA21a open reading frame (ORF), including start and stop codons, using pJET1-AtVMA21a as a template along with primers that also possessed sequence homologous to 40 bp 5’ and 3’ of the ScVMA21 ORF. Also present in the PCR 5’ primer was a Mlu1 restriction site immediately following the start codon and introducing two additional amino acids to the protein sequence (Thr, Arg). Similarly, a PCR fragment was also generated containing the AtVMA21b ORF and sequence homologous to 5’ and 3’ of the ScVMA21 ORF. Plasmid constructs were generated by homologous recombination mediated in vivo ligation using yeast where AtVMA21a and AtVMA21b completely replaced the ScVMA21 ORF carried on a plasmid. These plasmid constructs also contain approximately 600 bp of 5’ UTR and 250 bp 3’ UTR yeast sequence flanking the ScVMA21 ORF. Briefly, vma21D yeast cells were co-transformed with pLG263 digested with BglII and MscI and a PCR fragment containing AtVMA21a or AtVMA21b plus 40 bp ScVMA21 sequence. Yeast cells that repaired the plasmid by homologous recombination where able to grow in SD medium minus uracil. Plasmids were recovered from the yeast culture, transformed into E. coli, and prepared plasmid DNA was screened for the presence of the unique MluI restriction site generating plasmids pLG268 and pEB03. PCR fragments encoding either 3XHA epitope using pSMY92 as a template or GFP using plasmid CY3714 as template were generated using primers that also introduce 5’ and 3’ MluI flanking restriction sites. MluI digested 3XHA or GFP fragments were ligated with MluI digested pLG268 or pEB03 to generate epitope tagged versions of AtVMA21a (pLG269 and pLG270) or AtVMA21b (pEB05 and pLG266). Correct orientation of the epitope tag inserts was first confirmed using PCR, then by DNA sequencing.
In order to epitope tag ScVMA21 a PCR fragment was generated using either pSMY92 (3xHA) or CY3714 (GFP) with primers that also possessed sequence homologous to 40 bps 5’ noncoding region of the ScVMA21 ORF including start codon and the first 40 bps of the ScVMA21 coding sequence. Co-transformation in yeast of the PCR fragments with Bgl II digested pLG263 followed by homologous recombination and selection for intact plasmid resulted in the introduction of the epitope tags immediately following the start codon generating plasmids pLG277 and pLG278.
Immunoblot analysis
Yeast membranes were prepared in a similar manner as described previously (26). Briefly, yeast strains were grown to mid-log phase in YEPD at 30°C. Cells were treated with 10 mM DTT plus 50 mM Tris pH 9.5, resuspended in spheroplast buffer (1.2 M sorbitol, 50 mM KPO4 pH 7.4, 1 mM MgCl2) plus 250 μg/ml Zymolyase 100T, and lysed in PBS (Phosphate Buffered Saline) plus protease inhibitors (0.5 mM PMSF, 1 μg/mL leupeptin, and 1 μg/mL pepstatin) with vigorous pipetting. Unbroken cells were separated from the cell extracts by centrifugation at 1000 × g. Samples were centrifuged at 18,000 × g for 20 min to pellet membrane fractions, supernatants were removed, and membrane samples were resuspended in Thorner buffer minus b-mercaptoethanol (8M urea, 10% SDS, 40 mM Tris pH 6.8) and solubilized by mixing aggressively. The protein concentration of the membrane samples was determined by using the method of Markwell (27) Prior to SDS-PAGE analysis, b-mercaptoethanol and Bromophenol blue dye were added to final concentrations of 5% and 0.001% respectively. SDS-PAGE analysis was performed on the various protein preparations. Proteins were transferred to 0.2 mm-nitrocellulose membrane and probed with antibodies recognizing HA epitope (HA.11: Covance Research Products), Vph1p (Molecular Probes, 10D7) and Dpm1p (Molecular Probes, 5C5). Bands were visualized by incubation with anti-mouse Horse Radish Peroxidase conjugated secondary antibodies (HRP; Jackson ImmunoResearch Laboratories, Inc.), chemiluminescence reagents and imaged using X-ray film.
Fluorescence Microscopy
Localization of GFP tagged proteins and stained DNA were determined by fluorescence microscopy. 1 ml of exponentially growing cells at 0.25 OD600 were fixed by treatment with 3.7% formaldehyde for 10 minutes. Cells were harvested by centrifugation for 30 seconds at 4000 × g, washed once with 100 mM MES pH 7.7 plus 2% glucose, and resuspended in buffered glucose plus 5 mg/ml Hoechst 33342 DNA dye. Cells were incubated with the DNA dye for 10 minutes at room temperature, collected by centrifugation, and washed twice with buffered glucose prior. The acidification of vacuoles in various yeast strains was visualized using the lysosomotropic fluorescent dye quinacrine. Quinacrine staining of live yeast cells was conducted as previously described (8). Quinacrine was used at a final concentration 200 μM prepared in 100 mM MES pH 7.7 to stain acidified vacuoles, while concanavalin A-tetramethylrhodamine (Molecular Probes) was added at a final concentration of 50 μg/mL to allow for fluorescent visualization of the cell surface. Images were acquired on a Zeiss Axioplan 2 fluorescence microscope using 100X objective and manipulated using AxioVision software (Zeiss).
Enzyme Assay
Enriched yeast vacuolar membranes were isolated by osmotic lysis and gradient centrifugation as previously described (28). The protein concentration of the isolated vacuole samples was determined using the modified Lowry protocol (27). ATPase activity rates were measured using the coupled enzyme assay of Lotscher et al (29) in the presence and absence of 1.0 uM concanamycin A.
Acknowledgments
The authors are grateful to Zhao-Xin Wang and Dagmar Ripper for excellent technical assistance. Support for this work was provided by a grant in the DAAD-NSF PPP to K.S., a grant from the National Institutes of Health (GM38006) to T.H.S and a National Institutes of Health Ruth Kirschstein NRSA award (1F32GM083572-01) to E.M.C.
Footnotes
Accession Numbers Arabidopsis Genome Initiative numbers for the genes discussedin this article are as follows: AtVMA21a, At2g31710; AtVMA21b, At1g05780.
References
- 1.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8(11):917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Nishi T, Forgac M. The vacuolar (H+)-ATPases--nature’s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3(2):94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 3.Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 4.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70(1):177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty RD, Kane PM. Partial assembly of the yeast vacuolar H(+)-ATPase in mutants lacking one subunit of the enzyme. J Biol Chem. 1993;268(22):16845–16851. [PubMed] [Google Scholar]
- 6.Tomashek JJ, Sonnenburg JL, Artimovich JM, Klionsky DJ. Resolution of subunit interactions and cytoplasmic subcomplexes of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1996;271(17):10397–10404. doi: 10.1074/jbc.271.17.10397. [DOI] [PubMed] [Google Scholar]
- 7.Sze H, Schumacher K, Muller ML, Padmanaban S, Taiz L. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H(+)-ATPase. Trends Plant Sci. 2002;7(4):157–161. doi: 10.1016/s1360-1385(02)02240-9. [DOI] [PubMed] [Google Scholar]
- 8.Chavez C, Bowman EJ, Reidling JC, Haw KH, Bowman BJ. Analysis of strains with mutations in six genes encoding subunits of the V-ATPase: eukaryotes differ in the composition of the V0 sector of the enzyme. J Biol Chem. 2006;281(37):27052–27062. doi: 10.1074/jbc.M603883200. [DOI] [PubMed] [Google Scholar]
- 9.Graham LA, Flannery AR, Stevens TH. Structure and assembly of the yeast V-ATPase. J Bioenerg Biomembr. 2003;35(4):301–312. doi: 10.1023/a:1025772730586. [DOI] [PubMed] [Google Scholar]
- 10.Davis-Kaplan SR, Compton MA, Flannery AR, Ward DM, Kaplan J, Stevens TH, Graham LA. PKR1 encodes an assembly factor for the yeast V-type ATPase. J Biol Chem. 2006;281(42):32025–32035. doi: 10.1074/jbc.M606451200. [DOI] [PubMed] [Google Scholar]
- 11.Hirata R, Umemoto N, Ho MN, Ohya Y, Stevens TH, Anraku Y. VMA12 is essential for assembly of the vacuolar H(+)-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J Biol Chem. 1993;268(2):961–967. [PubMed] [Google Scholar]
- 12.Hill KJ, Stevens TH. Vma22p is a novel endoplasmic reticulum-associated protein required for assembly of the yeast vacuolar H(+)-ATPase complex. J Biol Chem. 1995;270(38):22329–22336. doi: 10.1074/jbc.270.38.22329. [DOI] [PubMed] [Google Scholar]
- 13.Graham LA, Hill KJ, Stevens TH. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol. 1998;142(1):39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malkus P, Graham LA, Stevens TH, Schekman R. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol Biol Cell. 2004;15(11):5075–5091. doi: 10.1091/mbc.E04-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill KJ, Stevens TH. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol Biol Cell. 1994;5(9):1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidel T, Schnitzer D, Golldack D, Sauer M, Dietz KJ. Organelle-specific isoenzymes of plant V-ATPase as revealed by in vivo-FRET analysis. BMC Cell Biol. 2008;9:28. doi: 10.1186/1471-2121-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strompen G, Dettmer J, Stierhof YD, Schumacher K, Jurgens G, Mayer U. Arabidopsis vacuolar H-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. Plant J. 2005;41(1):125–132. doi: 10.1111/j.1365-313X.2004.02283.x. [DOI] [PubMed] [Google Scholar]
- 18.Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 1999;13(24):3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brux A, Liu TY, Krebs M, Stierhof YD, Lohmann JU, Miersch O, Wasternack C, Schumacher K. Reduced V-ATPase Activity in the trans-Golgi Network Causes Oxylipin-Dependent Hypocotyl Growth Inhibition in Arabidopsis. Plant Cell. 2008 doi: 10.1105/tpc.108.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roslan HA, Salter MG, Wood CD, White MR, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, Caddick MX. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 2001;28(2):225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- 21.Dettmer J, Schubert D, Calvo-Weimar O, Stierhof YD, Schmidt R, Schumacher K. Essential role of the V-ATPase in male gametophyte development. Plant J. 2005;41(1):117–124. doi: 10.1111/j.1365-313X.2004.02282.x. [DOI] [PubMed] [Google Scholar]
- 22.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18(3):715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25(6):989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 24.Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27(6):581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 25.Seidel T, Golldack D, Dietz KJ. Mapping of C-termini of V-ATPase subunits by in vivo-FRET measurements. FEBS Lett. 2005;579(20):4374–4382. doi: 10.1016/j.febslet.2005.06.077. [DOI] [PubMed] [Google Scholar]
- 26.Ross J, Buschkamp P, Fetting D, Donnermeyer A, Roth CM, Tinnefeld P. Multicolor single-molecule spectroscopy with alternating laser excitation for the investigation of interactions and dynamics. J Phys Chem B. 2007;111(2):321–326. doi: 10.1021/jp066082g. [DOI] [PubMed] [Google Scholar]
- 27.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 28.Conibear E, Stevens TH. Studying yeast vacuoles. Methods Enzymol. 2002;351:408–432. doi: 10.1016/s0076-6879(02)51861-9. [DOI] [PubMed] [Google Scholar]
- 29.Lotscher HR, deJong C, Capaldi RA. Interconversion of high and low adenosinetriphosphatase activity forms of Escherichia coli F1 by the detergent lauryldimethylamine oxide. Biochemistry. 1984;23(18):4140–4143. doi: 10.1021/bi00313a020. [DOI] [PubMed] [Google Scholar]
- 30.Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47(6):1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- 31.Compton MA, Graham LA, Stevens TH. Vma9p (Subunit e) is an integral membrane V0 subunit of the yeast V-ATPase. J Biol Chem. 2006 doi: 10.1074/jbc.M600890200. [DOI] [PubMed] [Google Scholar]
- 32.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J Biol Chem. 1994;269(19):14064–14074. [PubMed] [Google Scholar]
- 34.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13(11):1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]


