Abstract
Increasing knowledge about the organization of proteins into complexes, systems, and pathways has led to a flowering of theoretical approaches for exploiting this knowledge in order to better learn the functions of proteins and their roles underlying phenotypic traits and diseases. Much of this body of theory has been developed and tested in model organisms, relying on their relative simplicity and genetic and biochemical tractability to accelerate the research. In this review, we discuss several of the major approaches for computationally integrating proteomics and genomics observations into integrated protein networks, then applying guilt-by-association in these networks in order to identify genes underlying traits. Recent trends in this field include a rising appreciation of the modular network organization of proteins underlying traits or mutational phenotypes, and how to exploit such protein modularity using computational approaches related to the internet search algorithm PageRank. Many protein network-based predictions have recently been experimentally confirmed in yeast, worms, plants, and mice, and several successful approaches in model organisms have been directly translated to analyze human disease, with notable recent applications to glioma and breast cancer prognosis.
Keywords: Data integration, Function prediction, Humans, Model organisms, Phenotype prediction, Protein interaction networks
1. Introduction: predicting phenotype from proteomics
Model organisms have proven invaluable for better understanding protein function and interactions, both for enabling studies of single proteins via genetic and biochemical tractability, as well as for enabling global surveys of thousands of proteins. Large-scale maps of pair-wise protein interactions [1–3], protein complexes [4–7], genetic interactions [8,9], transcription factor-target interactions [10–13], protein localization [14], and other complementary datasets have accelerated the characterization of protein function on a proteome-wide scale. The bulk of these studies have occurred in yeast and the nematode C. elegans, but increasingly in Arabidopsis, mouse, and fly, leading the way for applications to human cell culture.
These rapidly accumulating large-scale biological data have necessitated a corresponding growth in theoretical methods for interpreting them. One major goal has been to translate such knowledge of proteomic organization into models capable of associating genetic changes with changes to measurable traits, phenotypes, and diseases. In principle, such models will help to better interpret the rapidly growing genotype-based and genome sequence-based data characterizing genetic variation among individuals, whether individual humans or individuals of another species altogether. Models exploiting proteomic organization in order to relate genetic changes to altered traits could, for example, guide the identification of new disease genes, of genes underlying susceptibility to infection, and of genes underlying major crop traits or of many other naturally occurring traits with genetic components. Here, again, model organisms are leading the way: over the past few years, a variety of computational methods, most exploiting large-scale proteomic and genomic datasets, have begun to show strong advances in predicting the phenotypic consequences of mutations and successfully identify genes underlying mutational traits.
One powerful strategy has been to exploit the principle of guilt-by-association (GBA) in protein networks (reviewed in [15]). In this scheme, the function of a protein will more often than not resemble the functions of those proteins with which it interacts, or is co-expressed, or is co-localized, and so on. Thus, knowledge of a few proteins’ functions, in combination with large-scale maps of protein-protein associations, provides substantial traction for characterizing the portions of the proteome that are as yet poorly understood. Interestingly, this strategy, developed initially for inferring protein function [16], has recently proved to be a powerful approach for linking genes to phenotypic traits and diseases.
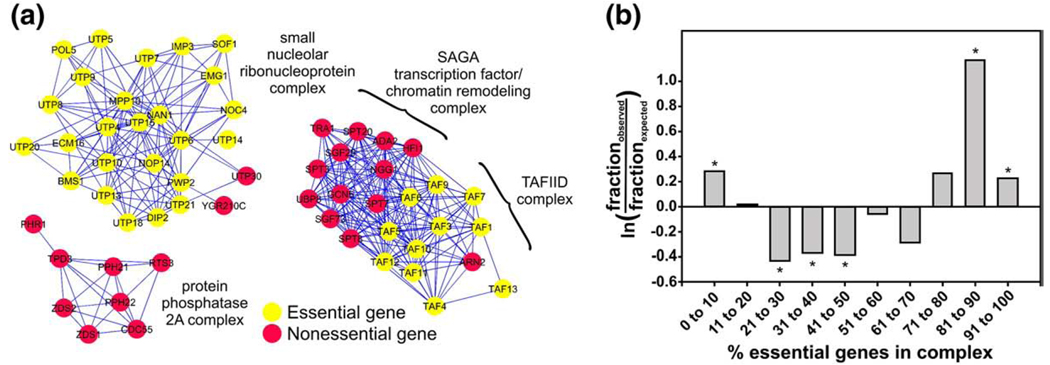
That such an approach might work can be seen intuitively from an examination of arguably one of the simplest “diseases”: lethality of a yeast cell following deletion of an essential gene. Comparisons of the components of yeast protein complexes, measured at large-scale using mass spectrometry [4–7], with genes known from systematic gene deletion experiments to be essential for growth in standard laboratory medium [17], have shown that proteins encoded by essential genes tend to co-occur in the same physical complexes [18,19] (Fig. 1a). There is a general trend for proteins in the same physical complex to be encoded either mostly by essential genes or mostly by non-essential genes; complexes are systematically depleted for intermediate mixtures of essential and non-essential genes [19] (Fig. 1b). Thus, essentiality appears to be a function of the complex — the intact molecular “machine” — rather than the individual gene. Recent observations have further shown that essential complexes tend to be larger than non-essential complexes [20,21], which provides a physical explanation for a long-standing observation about gene essentiality, that proteins encoded by essential genes tend to have more interaction partners than non-essential ones (the “centrality-lethality rule”) [22].
Fig. 1.
The “simplest” disease – lethality – appears tied not to the protein itself, but rather to the molecular complex or module in which that protein participates, as for the examples of yeast protein complexes drawn in (a). Proteins are depicted as colored circles and experimentally detected memberships in the same protein complexes as connecting lines. (b) Across the complete set of yeast protein complexes, a systematic trend is apparent for members of the same complex to either be all essential, or all non-essential, with depletion for intermediate mixtures of essential and non-essential proteins. Figures are adapted from ref. [19].
From a practical perspective, the observation that proteins in the same physical complexes tend to be linked to similar mutational phenotypes suggests that knowing physical complexes and a subset of genes linked to a trait, one could confidently predict additional genes relevant to that trait based upon their interaction partners. Indeed, this strategy works reasonably well [23–25]; even better predictive performance comes from considering broader biological pathways and functional associations, for which this trend also appears to hold, rather than considering physical complexes alone [24,26]. In fact, studies have shown that highly but transiently connected proteins (e.g. kinases) often play key roles in complex disease [27]. Thus, a general consideration of functional associations, whether restricted to the same physical complex or not, appears to be a reasonable strategy for linking genes to traits. This general strategy – exploiting the tendency for genes underlying the same trait to encode functionally associated proteins – has proven generally applicable and has now been tested for a wide variety of traits and phenotypes, and even human diseases (e.g., [23–26,28], among others).
Just as for the initial collection of the underlying large-scale proteomic data, model organisms have served as a productive test-bed for these approaches. Here, we discuss several of the major computational approaches for computationally integrating proteomics and genomics observations into integrated protein networks, then applying these networks in order to identify genes underlying traits. While much work has gone into identifying causal genes for traits, e.g. by association studies (reviewed in [29]) or integrating genotypic and genomic data (e.g., [30–32]), we primarily focus here on GBA methods, reviewing the basic approaches, discussing recent bioinformatics developments (such as a growing recognition of the relationships between these methods and Google’s web search algorithm PageRank), and presenting recent examples of experimental validation of these approaches at linking genes to traits in organisms ranging from yeast to mammals to plants.
2. Protein networks: the basics
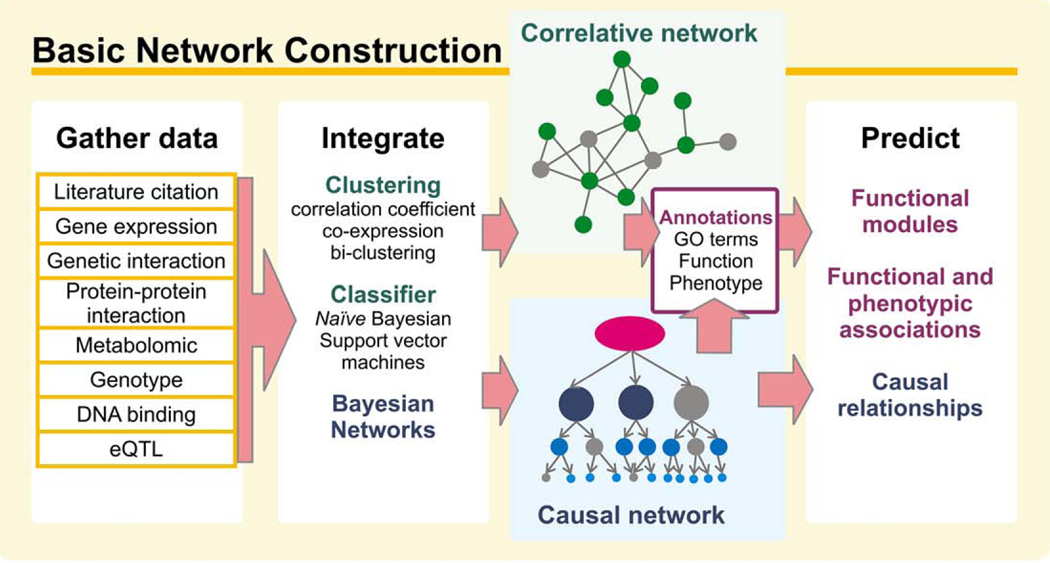
For the purposes of GBA, protein networks can consist solely of direct measurements of protein-protein or genetic interactions, such as might come from a single large-scale yeast two-hybrid or mass spectrometry assay. Typically, though, many such measurements are first computationally integrated into a composite network. Diverse methods exist for constructing integrated protein networks from mixed proteomic and genomic data sets, and rapidly increasing amounts of data from high-throughput experiments have pushed this field to be very active. Networks are increasingly used for modeling a wide variety of biological relationships, often requiring complex construction strategies. Given that we wish to focus here on the value of the resultant networks for discovering protein function and relevance to traits, we provide only a brief high-level overview of the different strategies of network construction (Fig. 2). For more in-depth reviews of this field, see [33–37]; in some cases, step-by-step instructions are available (e.g., [26,38]).
Fig. 2.
The flow from proteomics and genomics datasets to predicting protein function and ultimately associating genes to traits, phenotypes, and diseases involves several major steps, discussed in the main text. One major strategy involves the construction of protein networks from the large-scale datasets, which then serve as theoretical frameworks for generating more specific hypotheses about each protein’s function and the likely downstream impacts of perturbing it.
Building a network begins with the selection of relevant data. Generally, any data which suggest relationships between pairs of proteins can be used. For example, proteins whose corresponding mRNAs exhibit correlated expression levels across different cellular conditions are often likely to be functionally associated [16,39]. Similarly, protein-protein interaction data, such as from mass spectrometry of purified complexes or yeast two-hybrid assays, can often provide strong support for proteins to function together. Such data might be used directly for inferring protein function or organization (e.g., as in Refs. [40,41]), but can also be logically combined with other types of proteomic or genomic data. Naturally, the motivation of the network guides the choice of data used. For example, in order to study host-pathogen interactions between humans and the H. pylori bacteria, Tyagi et al. used predictions of transmembrane protein-protein interactions and virulence factors [42]. In another recent twist, Park et al. applied population-level disease patterns to study disease co-morbidity [43]. For basic protein networks, a common strategy is to integrate a mixture of proteomics and genomics datasets in order to infer protein-protein associations, as the resultant networks tend to be more complete and robust [26,28,38,44–55]. In an early example of a large-scale integrative network, Troyanskaya et al. demonstrate how the combination of various data sources provides for better coverage and accuracy of predicted functional relationships [56].
The integration of multiple types of data may be approached from many avenues, with two general frameworks used most commonly: correlative and causal networks. In the former case, network edges are undirected and represent functional coupling between pairs of associated proteins. For example, such a network might summarize evidence for pairs of proteins to physically interact, but would not indicate which protein in such a pair is upstream in the biological pathway. Methods for constructing correlative networks include combining clustering coefficients of data of varying weights [57] and training support vector machines to identify co-complexed protein pairs [58]. In naïve Bayesian networks, likelihood scores of proteins participating in the same pathway are calculated for each line of evidence and then combined as a weighted sum [59], providing a (weighted) Bayesian estimate for linked proteins to participate in the same cellular processes. Caveats to this approach include the reliance on current annotations (e.g., the Gene Ontology Consortium [60]) for training the networks. As annotations are often incomplete and may serve to propagate errors, there is a danger of introducing circularity. Nonetheless, many approaches may be taken to minimize this, such as using independent annotation test sets, benchmarking, and weighting of data (e.g., [30,44,45,61]). Importantly, networks constructed in this manner have proven strongly predictive for gene functions that lie outside of the annotated gene set, as discussed below (e.g., in the MouseFunc contest [62]).
Alternatively, in constructing causal networks describing causal relationships between genes, common strategies include ordering molecular events temporally, or by incorporating prior knowledge as to cause and effect (e.g., DNA mutations may alter a gene’s expression, but the reverse is unlikely). Zhu et al. demonstrated this technique by integrating transcription factor binding and expression QTL data into a probabilistic causal network in yeast [30]. They first estimated joint probability distributions for various models of causality between loci and traits, then identified the most likely model which fit their observed data. Alternatively, Bonneau et al. used time-series DNA microarray measurements of the transcriptional response to different environmental perturbations to construct causal models of interactions between environmental factors and transcription factors [63]. Though often computationally costly in construction, causal networks are of value for simplifying models of complex protein relationships, and in principle can identify early events in regulatory cascades, thereby guiding the selection of useful points of intervention for blocking such cascades (e.g., [64]). To accelerate progress in this field, an annual contest – the DREAM contest – is held dedicated to testing and improving the algorithms for deriving regulatory gene networks [65]. For a more in-depth discussion of causal networks, see [66–68]. Both correlative and causal models capture a wide variety of molecular interactions, ranging from stable physical interactions to transient interactions to genetic, non-physical interactions, all of which may be functionally relevant to the pathway of interest.
3. Propagating information through protein networks
Much of the real value of gene and protein networks lies in their utility for elucidating protein function. Thus, much work has been devoted to forming accurate predictions of protein functions using network information and previously known protein functions, generally by propagating annotations across network edges. Given that all current functional annotations for proteins are incomplete, sometimes woefully so, and that the networks represent an attempt to objectively reconstruct functional relationships among proteins, propagating annotations across a network’s edges is often useful, suggesting new functions for under-annotated proteins. These functional assignments are rarely unambiguous. Instead, functional prediction methods typically produce a score or rank representing how likely each protein is to be involved in the function. In order to provide some intuition for the relative merits of such approaches, we next introduce several methods and compare their abilities to annotate proteins in correlative networks. We focus on a small number of methods which we consider to be distinct and interesting; more comprehensive reviews of methods are available (e.g., [69]).
One of the most straightforward approaches to predict protein function via a protein network is that of neighbor counting (NC) [70]. In this approach, for a particular protein function or pathway, the proteins with the most neighbors associated with that function are themselves deemed the most likely to share that function. In a slightly more sophisticated variation on this method, naïve Bayes label propagation (NB), the sum of the network edge-weights to implicated neighbors is used, rather than the count of interactions [26,38,46]. This latter approach relies on network edge-weights that correspond to log likelihood scores for proteins to participate in the same process; thus, the sum of edge-weights to a set of genes of interest corresponds to a naïve Bayes estimate for a gene to also belong to the gene set of interest. NC and NB are both limited in that they only score direct neighbors of annotated proteins. However, as discussed later, these simple methods have made many experimentally verified predictions.
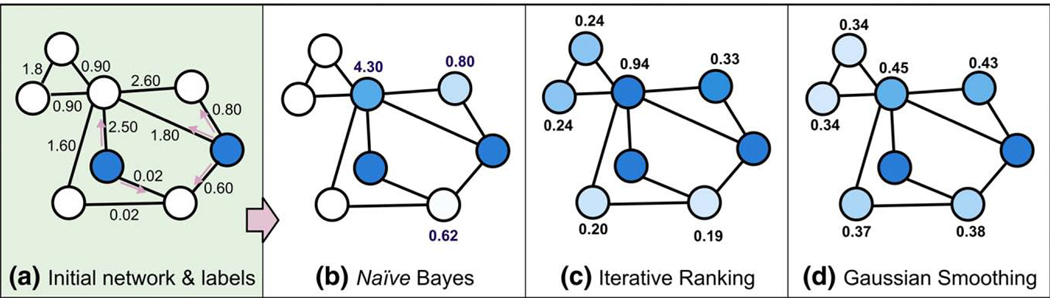
Alternatively, “network diffusion” methods have been developed in order to effectively diffuse information throughout a network, thus overcoming a major limitation of methods such as NC and NB that consider only direct associations. Here we consider two methods which diffuse information from a single node to directly and indirectly connected nodes over the course of several iterations. In the first, which we term iterative ranking (IR), the score for a protein to be linked to a particular function or phenotype consists of an initial score and the normalized scores or weighted “votes” of each neighbor (e.g., [71]). As scores are updated in successive iterations, information about proteins relevant to the function of interest propagates across network edges, “smearing” the initial functional assignments across the network. Another diffusion method we consider is Gaussian field label propagation, which we refer to here simply as Gaussian smoothing (GS). In GS the minimization of two distances is computed: the difference between a protein’s initial and final scores and the weighted score difference between the protein and each neighbor [72]. Box 1 and Fig. 3 present more detailed explanations of network diffusion algorithms, and should provide some intuition regarding their uses.
Box 1 The use of diffusion algorithms in network-based prediction of protein function
Diffusion algorithms are common in the growing body of work applying GBA in functional networks to predict protein function. Generally, network-based prediction algorithms utilize two pieces of information: f0, an initial vector of scores representing each protein's prior known association with a function, and W, the network topology matrix. The initial scores are propagated throughout the network, resulting in a set of final scores indicating each protein's predicted association to a function. The basic format for a diffusion algorithm is
| (1) |
where the vector of scores f is a combination of X, which contains initial scores for nodes, and Y, which contains information on the network topology. This convex combination allows the two components to be weighted differentially, along with some other mathematical conveniences. In contrast, in simpler neighbor counting and naïve Bayes methods, where a protein's score is the count or sum of edge-weights to seeds, the two components are combined (Table 1, Fig. 3b). Diffusion algorithms are advantageous because they assign scores to indirectly connected nodes. Additionally, the final scores are often readily computed. Below we describe two diffusion methods that have been successfully employed in various studies.
Iterative ranking (IR)
This algorithm was first developed in 1941 to model the input–output flows of economic industries by Nobel Prize winner Wassiliy Leontief [95]. It was more recently popularized by Larry Page and Sergey Brin as a method (PageRank) to rank internet search query results using the link topology of the web [96]. With minor adjustments, IR has been applied to numerous biological problems, including prioritizing functionally associated proteins [97–100], identifying protein clusters [101,102], identifying genes responsible for adverse drug reactions [103], and improving protein identification in high-throughput methods [71,104]. In the context of predicting protein function, the IR score of a protein is the combination of the initial seeds and the weighted average of IR scores of the protein's neighbors. Since each protein's score depends on that of its neighbors, the computation is iterative:
| (2) |
where ft are the scores at time t and U is the matrix of normalized network edges. The final scores are obtained when f stabilizes to within some threshold, or as the solution to the linear equation (Table 1, Fig. 3c). The type of network edge normalization performed is dependent on the application. In ranking internet search results, where each edge is equal weight, a page which points to a multitude of other sites, such as a home page, is likely unspecific in topic. Thus, each edge is normalized by the total number of outgoing edges from the node. In predicting protein function, where network edges are usually weighted, we wish to normalize each neighbor's contribution to a node's score. Thus, edges here are normalized by the total weight of incoming edges to the node. A more in-depth description of normalization differences is available [105].
Gaussian smoothing (GS)
This Gaussian field label propagation algorithm [72] minimizes the Euclidean distance between (1) the initial and final scores of a protein and (2) a protein's score and that of each of its neighbors:
| (3) |
where wij is the edge weight between protein i and its neighbor j. This can be derived from the assumptions that the error between initial and final scores f – f0 is normally distributed, f follows a multivariate normal distribution, and the covariance matrix Σ is equivalent to the inverse graph Laplacian matrix:
| (4) |
Network edge normalization may also be useful when implementing this algorithm. The authors of GS normalize each edge by the square root of the sum of incoming edges and the square root of the sum of outgoing edges for each node [72]. Similar to IR, the GS score of a protein depends on an initial score and the scores of neighboring proteins. The solution to this minimization problem also reduces to a linear equation (Table 1, Fig. 3d).
Table 1.
Components of diffusion algorithms.
| Algorithm | X term | Y term | U | ffinal | |
|---|---|---|---|---|---|
| naïve Bayes | Uf0 | - | W | Uf0 | |
| Iterative ranking |
f0 | Uf | α[I − (1 − α)U]−1f0 | ||
| Gaussian smoothing |
(f − f0)T(f − f0) | fTUf |
D–W where Dii = Σj wij and Dij = 0 if i≠j |
Fig. 3.
A comparison of diffusion algorithm-based methods for predicting genes associated with a function. In (a) two proteins are known to have a particular function, indicated by their color. The strength of association between proteins is indicated along each edge. (b) Naïve Bayes assigns scores to neighboring nodes. The ranking of scores is indicated by the shade of color: higher ranked proteins are more darkly colored. Note that several proteins have no score because they are not directly linked. In (c) and (d), all proteins are assigned to a score, but the overall rankings differ.
A wide variety of approaches can be imagined for propagating information across a network, and many such approaches, often initially developed in fields outside of biology, are proving useful for linking proteins to functions or traits. For example, Markov clustering (MCL) groups nodes based on simulation of stochastic flow in the network [73]. This method was originally applied to predict protein families based on sequence similarity. While this method is useful for identifying clusters of functionally related proteins, it does not directly identify proteins of a particular function. Instead, it identifies clusters containing proteins of interest, but does not rank candidate genes within each cluster. However, proteins can then be prioritized by a variety of other approaches, such as by considering the sum of a protein’s edge-weights within a cluster relative to all of its edge-weights, with larger sums indicating more relevance to the functions captured by that cluster.
Finally, another interesting method analyzes the flow through a network using concepts from electric circuit analysis [74]. In this circuit-based method (CB), the protein network is represented by an electrical circuit, where edge-weights are analogous to conductance (1/resistance) and implicated proteins are assigned as ground nodes. A current is simultaneously applied to each protein, and the nodes emerging with the highest current flowing through are predicted to be most likely to be associated with the ground nodes.
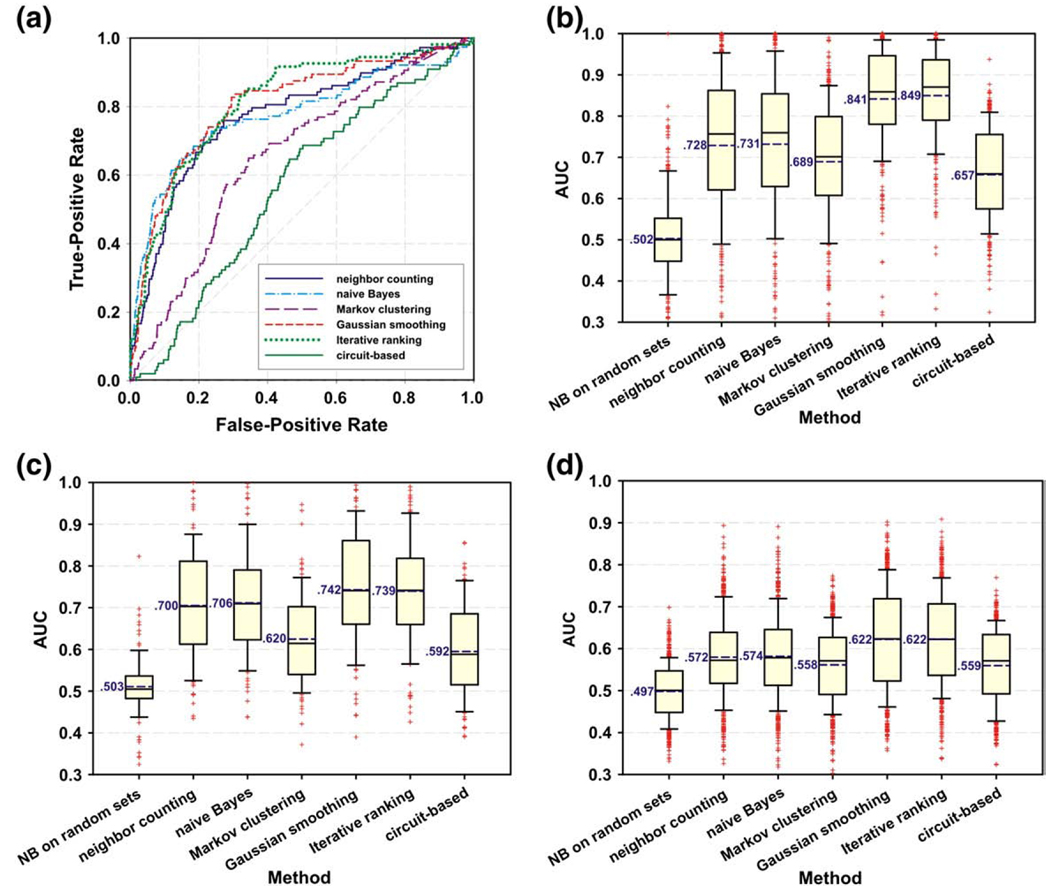
Because strongly connected nodes in a functional protein network are likely to work together in the same biological processes, they are also likely to share similar loss-of-function phenotypes. This can be demonstrated using correlative functional networks available for C. elegans [26] and S. cerevisiae [46] along with 318 RNAi phenotype gene sets available from WormBase [75], 100 loss-of-function phenotype sets from McGary et al. [24], and statistics of 282 morphological parameters for 4718 yeast gene deletion mutants from the Saccharomyces Cerevisiae Morphological Database (SCMD) [76]. In order to analyze the quantitative data from SCMD, we assigned the genes corresponding to the 40 largest and smallest values for each morphological feature as phenotype sets, resulting in 564 total sets. Fig. 4 illustrates the relative performance of the various algorithms discussed above at identifying genes underlying traits, focusing on RNAi knockdowns in C. elegans (Fig. 4b) and loss-of-function mutational phenotypes (Fig. 4c) and morphological phenotypes (Fig. 4d) in yeast. A standard strategy for evaluating such algorithms is to perform 10-fold cross-validation, separating known examples into distinct training and test sets of proteins. Using this approach, for each phenotype, we calculated the true-positive rates (TPR) and false-positive rates (FPR) as a function of a method’s score or rank and plotted the corresponding ROC curve. Fig. 4a provides an example ROC curve illustrating the predictive ability of each method for correctly identifying genes responsible for abnormal locomotion of C. elegans following RNAi knockdown. The area under a ROC curve (AUC) provides a convenient summary of a method’s predictive ability on that phenotype; a curve along the diagonal line and an AUC near 0.5 has no predictive ability, while one pushed to the top left of the plot with an AUC closer to 1 has strong predictive ability.
Fig. 4.
Results of a comparative test of the performance of various protein network-based algorithms for linking genes to phenotypic traits. (a) Presents a ROC curve illustrating the relative predictive abilities of six algorithms for identifying the genes associated with abnormal locomotion in worm following RNAi knockdown. A method assigns each gene in the genome a score or rank, and this is used to calculate the true-positive rate (TP/(TP + FN)) and false-positive rate (FP/ (FP + TN)), using 10-fold cross-validation to assess the performance. Performing this test systematically across many distinct phenotypes allows the relative merits of the different algorithms to be measured. (b), (c), and (d) present the distributions of the areas under ROC curves (AUCs) obtained from each method for identifying the causal genes underlying 318 C. elegans RNAi phenotypes, predicted using a worm gene network [26], and 100 yeast deletion mutant phenotypes and 564 morphological phenotypes, predicted using a yeast gene network [46], respectively. 10, 25, 50, 75, and 90 percentiles are plotted. Dashed blue lines and corresponding blue text denote mean AUCs.
The overall performance of each algorithm at predicting loss-of-function phenotypes in worm and yeast is shown as distributions of AUC values in Fig. 4b–d. NC and GBA methods perform quite similarly, presumably because a minimum edge weight threshold applied in network construction [26,46] causes the NC method to return similar rankings to the NB method. The MCL and CB methods, originally developed for different purposes, did not adapt well to task of phenotype prediction. However, MCL performance would most likely improve with further refinements to the ranking of proteins within clusters and to the optimization of clustering parameters. Overall, the two diffusion methods outperform the others by a notable margin. Some guides for effective use come from these analyses: In some cases, the diffusion methods perform poorly for small FPR and extremely well for higher FPR. Therefore, when choosing a predictive algorithm, the false-positive cost for the particular experiment should be considered; NC or NB methods are appropriate when false-positives are costly and diffusion methods suit cases where a more exhaustive set of predictions is desired. Notably, the relative performance of each method was robust to choice of organism and test set. The tests in C. elegans yielded higher average AUCs and a stronger performance boost from diffusion methods. These methods can also be further adapted to predict quantitative gene-pathway based traits (e.g., predicting quantitative yeast phenotypes with a modified NB method [77]). Finally a general caveat is merited: we have observed that the various network label propagation methods tend to perform differently for different applications, and it is often advisable to test several to see which performs best for a particular test of interest.
4. Validated applications to model organisms
While much work on the computational analysis of gene networks has relied on computational tests, such as the cross-validation employed above, the last few years have seen increasing direct experimental validation of network-based predictions of protein functions and involvement in phenotypes. Here again, model organisms have proven invaluable for enabling rapid in vivo tests of the validity of these methods. In some studies, simple loss-of-function experiments have confirmed very striking phenotypes. For example, protein networks have successfully predicted genes whose RNAi knockdown suppresses the loss of the retinoblastoma tumor suppressor, validated in C. elegans (Fig. 5a) [26]. In A. thaliana, novel regulators of drought sensitivity and lateral root development were discovered using NB predictions (Fig. 5b) [38]. Similarly, in E. coli, many proteins were predicted to play roles in cell envelope biogenesis. Cells in which these proteins were deleted exhibited differential sensitivity to peptidoglycan assembly inhibitors [78]. In another study, Qi et al. construct a yeast synthetic lethal genetic interaction network in order to predict pathway memberships and genetic interactions. Using a diffusion method similar to IR (described above), they identified and confirmed 18 novel genetic interactions for the transcriptional cofactor Ada2 and 20 for Esa1, a subunit of the histone acetyltransferase complex [79].
Fig. 5.
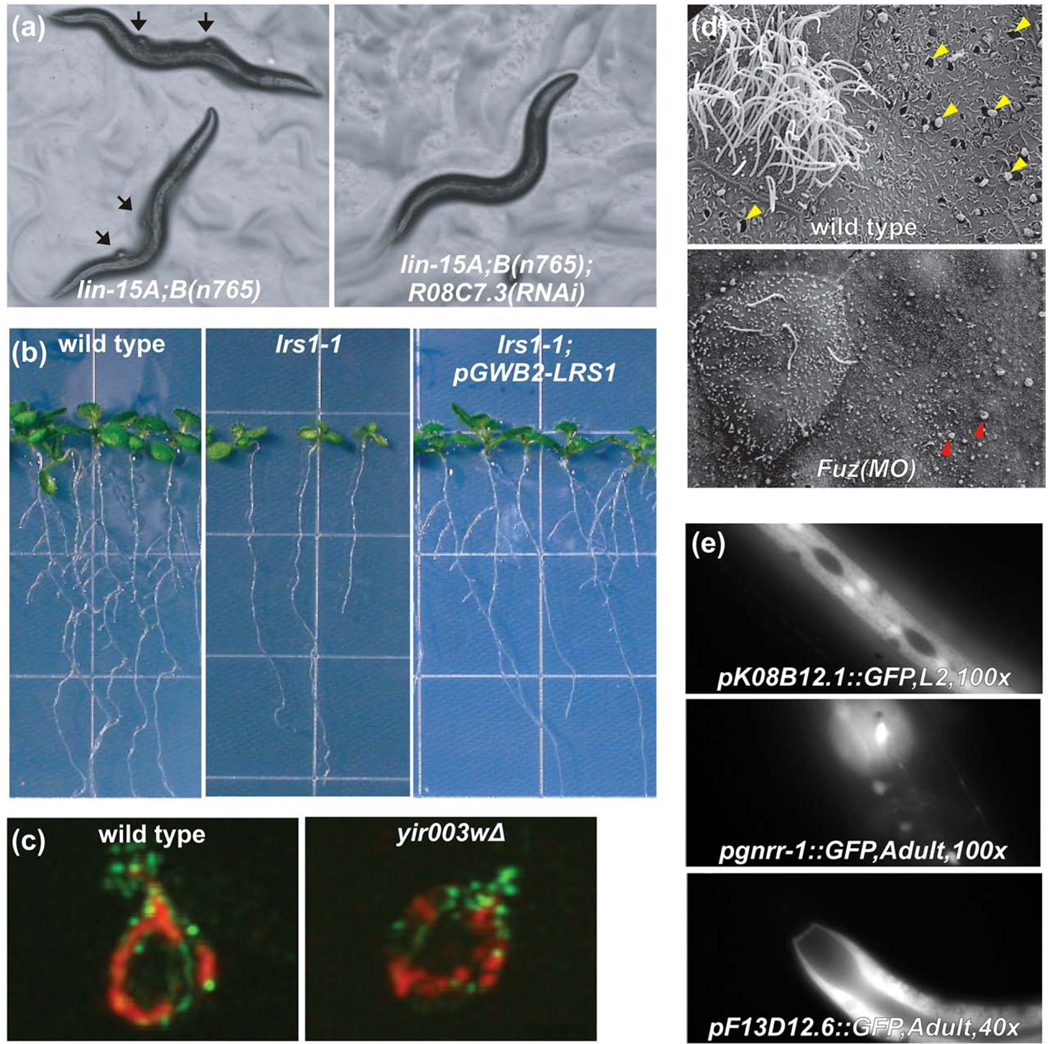
Examples of experimentally validated network-guided predictions of genes underlying specific traits in yeast, plants, and animals. (a) The lin-15A;B(n765) strain of C. elegans has inactivated synMuv A and synMuv B retinoblastoma tumor suppressor pathways, causing a synthetic multivulval phenotype, in which altered cell fate specification causes normally epithelial cells to adopt a vulval cell fate, creating ectopic vulvae that are a C. elegans model for tumor formation [105]. RNAi against genes that antagonize the pathways, such as R08C7.3, suppress the phenotype, as predicted by network guilt-by-association. Figure adapted with permission from Macmillan Publishers Ltd: [26], 2008. (b) When the Arabidopsis gene lrs1-1 is disrupted by a T-DNA insertion, the number of lateral roots is strongly reduced (middle panel) compared to wild-type plants (left panel). Reintroduction of the functional gene as a transgene restores wild-type phenotype (right panel), confirming a role in root development predicted from an Arabidopsis gene network. Figure adapted with permission from Macmillan Publishers Ltd: [38], 2010. (c) A confirmed roles for candidate yeast protein YIR003W in mitochondrial biogenesis, predicted using computational methods from proteomics and genomics datasets, can be seen reflected in mitochondrial motility defects in a YIR003W deletion strain, measured by dual channel immunofluorescence of mitochondria (red) and actin (green) (adapted from ref. [81]). (d) A mouse protein network [48] and results of large-scale computational predictions of mouse protein function [62] suggested a role for the birth-defect relevant gene Fuz in vesicle trafficking and biogenesis of cilia that was confirmed by knockdown in developing Xenopus embryos [89]. The top panel shows a wild-type multiciliated cell flanked by secretory cells in X. laevis. Note the exocytotic pits indicated by the yellow arrowheads. Fuz morphants show ciliogenesis defects as well as failure of exocytosis in secretory cells (bottom); note the apical membrane blebs indicated by green arrowheads. Figure adapted with permission from Macmillan Publishers Ltd: [89], 2009. (e) Using GFP-reporter constructs, C. elegans gene network-predicted tissue-specific gene expression was verified in the hypodermis, intestine, hypodermis, neurons, and muscle of worms by Chikina et al. (adapted from Ref. [82]).
Beyond the simple scheme of mapping single genes to single phenotypes is the goal of understanding how complex phenotypes arise. For example, a time-dependent yeast protein interaction network revealed the role of Cdk1 in protein complex formation throughout the cell cycle [80]. In a separate study employing an integrative yeast network, Hess et al. confirmed 140 of 235 predicted mitochondrial biogenesis genes (one such example is reprinted in Fig. 5c) [81]. In another study, a tissue-specific functional interaction network is constructed in order to study tissue-specific regulation patterns in worm. Several genes predicted to express in the hypodermis, muscle, or neurons were confirmed using promoter-GFP constructs (Fig. 5e) [82]. Similarly, using a co-expression gene network, Ghazalpour et al. predicted factors which influence the body weight of mice [83]. The addition of non-proteomics datasets, including genome sequence or genotype data, has proven to often extend the scope of studies considerably. Applying genome-wide association data to a yeast functional network facilitated the identification of thousands of genetic interactions between protein complexes [84], and a model of protein dosage sensitivity [85]. Using an extensive collection of chemical genetics datasets, Venancio et al. built a chemical-protein complex network and identified potential interactions between drugs and protein complexes [86].
Efforts to exploit proteomics and genomics data in order to better annotate model organism genes recently culminated in an international contest, MouseFunc, to annotate mouse genes with Gene Ontology (GO) annotations. Nine teams from around the world independently developed computational methods to predict gene function from a large collection of M. musculus data [62]. The data included protein sequence pattern annotations, experimentally determined protein-protein interactions, mRNA expression across multiple tissues, gene-phenotype associations, disease associations of human orthologs, and phylogenetic distributions of mouse genes. Each team predicted blinded GO annotations for mouse genes, and were assessed on withheld annotations and annotations newly identified since the start of the contest. The strengths and weaknesses of each algorithm were assessed, and several methods emerged as strong performers. For example, the Gaussian smoothing algorithm GeneMANIA (Box 1, [72]) performed well. Performance using a computational approach known as support vector machines, combined with GO annotations over a Bayesian framework, was robust to the number of genes in the test set [87]. Finally, Funckenstein, composed of two methods combined by logistic regression, produced high precision predictions for a wide range of GO annotations [88]. This method uses guilt-by-association in gene networks in addition to guilt-by-profiling: exploiting the correlation between gene function and other gene characteristics. The ultimate result of this contest was a unified set of predictions over all teams’ approaches that averaged 41% precision over all GO annotations. Moreover, 26% of GO terms achieved a precision > 90%. Many new predictions emerged for 5000 previously uncharacterized genes. Predictions for one of these, the gene Fuz, implicated in vertebrate birth defects, were recently confirmed experimentally in transgenic mice and knockdown experiments in frogs (Fig. 5d) [89].
5. Prospects for humans
Overall, the work in integration of large-scale data sets and functional prediction in model organisms builds toward the ultimate goal of understanding complex human traits and phenotypes. Linding et al. approached this goal on a signaling level and developed an in vivo phosphorylation network, modeling kinase and phosphoprotein relationships [90]. They identified substrates of kinases previously overlooked by motif-based methods alone, including those of ATM (a primary regulator of DNA damage response) and CDK1 (a driver of cell cycle progression). On a different level, genes or proteins associated with human diseases can be predicted through the network propagation methods discussed earlier, and such methods are increasingly being applied to human protein networks. For example, Ostlund et al. recently discovered genes with abnormally high connectivity to cancer genes and defined a novel method for ranking these new cancer gene candidates [91]. Other studies have focused on specific diseases and understanding certain properties of interest. One commonly used approach is to identify network characteristics which map to such properties. For example, Huttenhower et al. first build small networks of biological relationships between genes, then extract disease level information from these models [92]. Sun et al. identified modules of genes which they predicted to be responsible for metastasis of oral cavity tumors [47].
In two striking recent examples, the construction of gene interaction networks has led directly to predictions in patient prognosis. First, Carro et al. built a transcriptional regulatory network which models glioma cancer cell transition into an aberrant mesenchymal phenotype [64]. Using gene expression profiles and array comparative genomic hybridization of 76 high-grade gliomas, they were able to infer C/EBPβ and STAT3 to be transcription factors responsible for initiating and regulating mesenchymal transformation. As a validation of their model, they found that patients with tumors double-positive for C/EBPβ and STAT3 were associated with worse clinical outcome than patients with either single- or double-negative tumors (Fig. 6a). Second, Taylor et al. studied breast cancer patient outcome by analyzing the hub proteins in a human protein interaction network in the context of genome-wide expression data in 79 human tissues [93]. They identified two classes of protein network hubs: intermodular hubs, which display low correlation of co-expression with neighbors, and intramodular hubs, which display high correlation of co-expression with neighbors. Mutations of intermodular hubs were more strongly associated with cancer phenotypes. Using a cohort of breast cancer patients, they defined correlation of co-expression signatures corresponding to good and poor prognosis patients. The model strongly predicts patient outcome, as demonstrated in the Kaplan-Meier survival curves (Fig. 6b). On a broader level, patient records have recently been integrated into models in order to build disease networks [43,94]. These networks elucidate disease-disease relationships and offer insight into disease progression and co-morbidity, and it is reasonable to expect that such models can be usefully integrated with protein association networks to better characterize the genetic basis for human diseases.
Fig. 6.
Examples of experimentally validated network-guided predictions of genes underlying outcomes of human disease. (a) shows a network of transcription factors which regulate mesenchymal transformation, as predicted from a glioma-specific regulatory network inferred from DNA microarray datasets [64]. Activation of transcription factors C/EBPβ and STAT3 correlates with aggressive gliomas and poor clinical outcome, as shown in the Kaplan-Meier plot at the right (Figure adapted with permission from Macmillan Publishers Ltd: [64], 2010). (b) Modularity of the human protein interactome can be used as an indicator of breast cancer prognosis. Taylor and colleagues calculated a subnetwork of hubs with either low or high correlation of co-expression with their interaction partners, termed intermodular (red) and intramodular (blue) hubs, respectively [93]. Patients with hubs that had highly altered correlation of co-expression had a high probability of poor prognosis, as plotted at right (Figure adapted with permission from Macmillan Publishers Ltd: [93], 2009).
The preceding network models and network methods reveal just small portions of the black box of interactions underlying complex phenotypes, but models that integrate multiple types of experimental datasets (e.g., combinations of proteomics and gene expression data) clearly perform best. Importantly, the wide availability of proteomics and genomics data is significantly boosting the ability to link genes to traits. These methods, proven initially in model organisms, are beginning to show utility for human diseases, in spite of a still significant lack of human proteomics data. As a result, most models are verified using high-throughput experiments on model organisms or cell lines, various annotation databases, and occasionally cohort data. Given reasonable expectations for technology developments in proteomics and genome and transcript sequencing to improve data quality and reduce the cost barriers to producing data, it seems safe to expect that the proteomics efforts proven in model organisms will increasingly translate into human studies, dramatically improving our ability to link genes to human diseases and traits.
Acknowledgments
We thank Martin Blom and Smriti Ramakrishnan for helpful discussions. This work was supported by grants from the Texas Advanced Research Program, the N.S.F., N.I.H., the Welch Foundation (F-1515), and a Packard Fellowship. The SCMD database has been provided freely by the University of Tokyo for use in this publication/correspondence only.
REFERENCES
- 1.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simonis N, Rual J, Carvunis A, Tasan M, Lemmens I, Hirozane-Kishikawa T, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 5.Gavin A, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 6.Gavin A, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 7.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 8.Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 9.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 10.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 11.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 12.Zhu C, Byers KJRP, McCord RP, Shi Z, Berger MF, Newburger DE, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kainth P, Sassi HE, Peña-Castillo L, Chua G, Hughes TR, Andrews B. Comprehensive genetic analysis of transcription factor pathways using a dual reporter gene system in budding yeast. Methods. 2009;48:258–264. doi: 10.1016/j.ymeth.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Huh W, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 15.Ideker T, Sharan R. Protein networks in disease. Genome Research. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 17.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 18.Dezso Z, Oltvai ZN, Barabási A. Bioinformatics analysis of experimentally determined protein complexes in the yeast Saccharomyces cerevisiae. Genome Res. 2003;13:2450–2454. doi: 10.1101/gr.1073603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart GT, Lee I, Marcotte EM. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinform. 2007;8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Kakaradov B, Collins SR, Karotki L, Fiedler D, Shales M, et al. A Complex-based Reconstruction of the Saccharomyces cerevisiae interactome. Mol Cell Proteomics. 2009;8:1361–1381. doi: 10.1074/mcp.M800490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zotenko E, Mestre J, O’Leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong H, Mason SP, Barabasi A, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 23.Fraser HB, Plotkin JB. Using protein complexes to predict phenotypic effects of gene mutation. Genome Biol. 2007;8:R252. doi: 10.1186/gb-2007-8-11-r252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGary KL, Lee I, Marcotte EM. Broad network-based predictability of Saccharomyces cerevisiae gene loss-of-function phenotypes. Genome Biol. 2007;8:R258. doi: 10.1186/gb-2007-8-12-r258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lage K, Karlberg EO, Størling ZM, Olason PI, Pedersen AG, Rigina O, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 26.Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 27.Amit I, Wides R, Yarden Y. Evolvable signaling networks of receptor tyrosine kinases: relevance of robustness to malignancy and to cancer therapy. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linghu B, Snitkin E, Hu Z, Xia Y, DeLisi C. Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol. 2009;10:R91. doi: 10.1186/gb-2009-10-9-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonneau R. Learning biological networks: from modules to dynamics. Nat Chem Biol. 2008;4:658–664. doi: 10.1038/nchembio.122. [DOI] [PubMed] [Google Scholar]
- 34.Christensen C, Thakar J, Albert R. Systems-level insights into cellular regulation: inferring, analysing, and modelling intracellular networks. IET Syst Biol. 2007;1:61–77. doi: 10.1049/iet-syb:20060071. [DOI] [PubMed] [Google Scholar]
- 35.Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson BØ. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franzosa E, Linghu B, Xia Y. Computational reconstruction of protein-protein interaction networks: algorithms and issues. Methods Mol Biol. 2009;541:89–100. doi: 10.1007/978-1-59745-243-4_5. [DOI] [PubMed] [Google Scholar]
- 37.Lee I, Narayanaswamy R, Marcotte EM. Bioinformatic prediction of yeast gene function. Yeast Gene Analysis. 2nd ed. Academic Press; 2007. [Google Scholar]
- 38.Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol. 2010;28:149–156. doi: 10.1038/nbt.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinform. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Zimmerman LJ, Park B, Tabb DL, Liebler DC, Zhang B. Network-assisted protein identification and data interpretation in shotgun proteomics. Mol Syst Biol. 2009;5:303. doi: 10.1038/msb.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyagi N, Krishnadev O, Srinivasan N. Prediction of protein-protein interactions between Helicobacter pylori and a human host. Mol Biosyst. 2009;5:1630–1635. doi: 10.1039/b906543c. [DOI] [PubMed] [Google Scholar]
- 43.Park J, Lee D, Christakis NA, Barabási A. The impact of cellular networks on disease comorbidity. Mol Syst Biol. 2009;5:262. doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan Y, Myers CL, Lu R, Lemischka IR, Bult CJ, Troyanskaya OG. A genomewide functional network for the laboratory mouse. PLoS Comput Biol. 2008;4:e1000165. doi: 10.1371/journal.pcbi.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee I, Li Z, Marcotte EM. An improved, bias-reduced probabilistic functional gene network of baker’s yeast, Saccharomyces cerevisiae. PLoS ONE. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z, Luo J, Zhou Y, Luo J, Liu K, Li W. Exploring phenotype-associated modules in an oral cavity tumor using an integrated framework. Bioinformatics. 2009;25:795–800. doi: 10.1093/bioinformatics/btp057. [DOI] [PubMed] [Google Scholar]
- 48.Kim WK, Krumpelman C, Marcotte EM. Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol. 2008;9 Suppl 1:S5. doi: 10.1186/gb-2008-9-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costello JC, Dalkilic MM, Beason SM, Gehlhausen JR, Patwardhan R, Middha S, et al. Gene networks in Drosophila melanogaster: integrating experimental data to predict gene function. Genome Biol. 2009;10:R97. doi: 10.1186/gb-2009-10-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hibbs MA, Myers CL, Huttenhower C, Hess DC, Li K, Caudy AA, et al. Directing experimental biology: a case study in mitochondrial biogenesis. PLoS Comput Biol. 2009;5:e1000322. doi: 10.1371/journal.pcbi.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suthram S, Dudley JT, Chiang AP, Chen R, Hastie TJ, Butte AJ. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol. 2010;6:e1000662. doi: 10.1371/journal.pcbi.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taşan M, Tian W, Hill DP, Gibbons FD, Blake JA, Roth FP. An en masse phenotype and function prediction system for Mus musculus. Genome Biol. 2008;9 Suppl 1:S8. doi: 10.1186/gb-2008-9-s1-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexeyenko A, Sonnhammer ELL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–1116. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl Acad Sci USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 58.Qiu J, Hue M, Ben-Hur A, Vert J, Noble WS. A structural alignment kernel for protein structures. Bioinformatics. 2007;23:1090–1098. doi: 10.1093/bioinformatics/btl642. [DOI] [PubMed] [Google Scholar]
- 59.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 60.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peña-Castillo L, Tasan M, Myers CL, Lee H, Joshi T, Zhang C, et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9 Suppl 1:S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 64.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stolovitzky G, Monroe D, Califano A. Dialogue on reverse-engineering assessment and methods: the DREAM of high-throughput pathway inference. Ann NY Acad Sci. 2007;1115:1–22. doi: 10.1196/annals.1407.021. [DOI] [PubMed] [Google Scholar]
- 66.Ross J. From the determination of complex reaction mechanisms to systems biology. Annu Rev Biochem. 2008;77:479–494. doi: 10.1146/annurev.biochem.77.102507.115132. [DOI] [PubMed] [Google Scholar]
- 67.Margolin AA, Califano A. Theory and limitations of genetic network inference from microarray data. Ann NY Acad Sci. 2007;1115:51–72. doi: 10.1196/annals.1407.019. [DOI] [PubMed] [Google Scholar]
- 68.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 71.Ramakrishnan SR, Vogel C, Kwon T, Penalva LO, Marcotte EM, Miranker DP. Mining gene functional networks to improve mass-spectrometry-based protein identification. Bioinformatics. 2009;25:2955–2961. doi: 10.1093/bioinformatics/btp461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9 Suppl 1:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. eQED: an efficient method for interpreting eQTL associations using protein networks. Mol Syst Biol. 2008;4:162. doi: 10.1038/msb.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.WormBase web site, release WS204. 2009. [Google Scholar]
- 76.Saito TL, Ohtani M, Sawai H, Sano F, Saka A, Watanabe D, et al. SCMD: Saccharomyces cerevisiae morphological database. Nucl Acids Res. 2004;32:D319–D322. doi: 10.1093/nar/gkh113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fenaroli A. Propagating quantitatve traits in gene networks, Eidgenössische Technische Hochschule Zürich. 2009 [Google Scholar]
- 78.Hu P, Janga SC, Babu M, Díaz-Mejía JJ, Butland G, Yang W, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi Y, Suhail Y, Lin Y, Boeke JD, Bader JS. Finding friends and enemies in an enemies-only network: a graph diffusion kernel for predicting novel genetic interactions and co-complex membership from yeast genetic interactions. Genome Res. 2008;18:1991–2004. doi: 10.1101/gr.077693.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Lichtenberg U. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- 81.Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Comput Biol. 2009;5:e1000417. doi: 10.1371/journal.pcbi.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hannum G, Srivas R, Guénolé A, van Attikum H, Krogan NJ, Karp RM, et al. Genome-wide association data reveal a global map of genetic interactions among protein complexes. PLoS Genet. 2009;5:e1000782. doi: 10.1371/journal.pgen.1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oberdorf R, Kortemme T. Complex topology rather than complex membership is a determinant of protein dosage sensitivity. Mol Syst Biol. 2009;5:253. doi: 10.1038/msb.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venancio TM, Balaji S, Aravind L. High-confidence mapping of chemical compounds and protein complexes reveals novel aspects of chemical stress response in yeast. Mol Biosyst. 2010;6:165–171. doi: 10.1039/b911821g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barutcuoglu Z, Schapire RE, Troyanskaya OG. Hierarchical multi-label prediction of gene function. Bioinformatics. 2006;22:830–836. doi: 10.1093/bioinformatics/btk048. [DOI] [PubMed] [Google Scholar]
- 88.Tian W, Zhang LV, Taşan M, Gibbons FD, King OD, Park J, et al. Combining guilt-by-association and guilt-by-profiling to predict Saccharomyces cerevisiae gene function. Genome Biol. 2008;9 Suppl 1:S7. doi: 10.1186/gb-2008-9-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gray RS, Abitua PB, Wlodarczyk BJ, Szabo-Rogers HL, Blanchard O, Lee I, et al. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat Cell Biol. 2009;11:1225–1232. doi: 10.1038/ncb1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ostlund G, Lindskog M, Sonnhammer ELL. Network-based identification of novel cancer genes. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900227-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, et al. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–1106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 94.Hidalgo CA, Blumm N, Barabási A, Christakis NA. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leontief, Wassily W. The structure of American economy, 1919–1929. Cambridge, Mass: Harvard University Press; 1941. [Google Scholar]
- 96.Page L, Brin S, Motwani R, Winograd T. The PageRank citation ranking: bringing order to the Web. Stanford InfoLab. 1999 [Google Scholar]
- 97.Birkland A, Yona G. BIOZON: a hub of heterogeneous biological data. Nucleic Acids Res. 2006;34:D235–D242. doi: 10.1093/nar/gkj153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Voevodski K, Teng S, Xia Y. Spectral affinity in protein networks. BMC Syst Biol. 2009;3:112. doi: 10.1186/1752-0509-3-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chipman KC, Singh AK. Predicting genetic interactions with random walks on biological networks. BMC Bioinformatics. 2009;10:17. doi: 10.1186/1471-2105-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Voevodski K, Teng S, Xia Y. Finding local communities in protein networks. BMC Bioinform. 2009;10:297. doi: 10.1186/1471-2105-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang L, Xu L, He L. A CitationRank algorithm inheriting Google technology designed to highlight genes responsible for serious adverse drug reaction. Bioinformatics. 2009;25:2244–2250. doi: 10.1093/bioinformatics/btp369. [DOI] [PubMed] [Google Scholar]
- 104.Morrison JL, Breitling R, Higham DJ, Gilbert DR. GeneRank: using search engine technology for the analysis of microarray experiments. BMC Bioinform. 2005;6:233. doi: 10.1186/1471-2105-6-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]