The CLIP biosensor reveals the spatiotemporal activity of the Nodal proprotein convertases Furin and Pace4 during embryonic development.
Abstract
Axis formation and allocation of pluripotent progenitor cells to the germ layers are governed by the TGF-β–related Nodal precursor and its secreted proprotein convertases (PCs) Furin and Pace4. However, when and where Furin and Pace4 first become active have not been determined. To study the distribution of PCs, we developed a novel cell surface–targeted fluorescent biosensor (cell surface–linked indicator of proteolysis [CLIP]). Live imaging of CLIP in wild-type and Furin- and Pace4-deficient embryonic stem cells and embryos revealed that Furin and Pace4 are already active at the blastocyst stage in the inner cell mass and can cleave membrane-bound substrate both cell autonomously and nonautonomously. CLIP was also cleaved in the epiblast of implanted embryos, in part by a novel activity in the uterus that is independent of zygotic Furin and Pace4, suggesting a role for maternal PCs during embryonic development. The unprecedented sensitivity and spatial resolution of CLIP opens exciting new possibilities to elucidate PC functions in vivo.
Introduction
In mammals, the proprotein convertase (PC) family comprises nine serine proteases that share a characteristic subtilisin/kexin-like catalytic domain. Among these, Furin, Pace4, Pcsk5, and PC7 are broadly expressed and responsible for cleaving intra- and extracellular precursors of various growth factors, receptors, adhesion molecules, neuropeptides, metalloproteases, viral envelope glycoproteins, and bacterial endotoxins after the minimal dibasic recognition motif RXXR (Thomas, 2002; Seidah et al., 2008). Thus, inhibitors of these proteases are considered to be antiviral or antibacterial agents and to combat tumor progression and invasion (Fugère and Day, 2005; Jiao et al., 2006; Komiyama et al., 2009). However, classical genetic approaches have provided only limited information on the specific roles of individual PCs in normal tissues and during disease, in part because extensive functional overlap among the more widely expressed family members masks their functions. Consequently, the majority of predicted PC substrates remain to be validated in vivo.
Our previous work established that Furin and Pace4 functionally overlap and together activate the TGF-β–related Nodal precursor during early embryogenesis (Constam and Robertson, 1999; Beck et al., 2002). In the mouse embryo, Nodal expression is initiated in the inner cell mass (ICM) and maintained in the epiblast to control the fate of pluripotent progenitor cells and their allocation to the three germ layers during gastrulation (Brennan et al., 2001; Arnold and Robertson, 2009). In contrast, expression of Furin and Pace4 during gastrulation is essential in extraembryonic lineages and limited to the extraembryonic ectoderm (Constam and Robertson, 2000b; Beck et al., 2002). Therefore, to directly cleave proNodal in epiblast cells, secreted forms of Furin and Pace4 would have to act cell nonautonomously. In line with this view, a complex of Nodal with its membrane-bound coreceptor Cripto can be activated in cultured cells by Furin and Pace4 activities that are provided in trans by separately transfected cells (Blanchet et al., 2008). Nevertheless, direct evidence that secreted PC activities can cleave proNodal or any other substrate at the surface of neighboring cells is missing. If PCs can act on neighboring cells, this considerably enlarges the spectrum of possible functions during development and in diseases such as cancer.
To assess when and where endogenous PCs are active in vivo, we here introduce a transgene encoding a cell surface–linked indicator of proteolysis (CLIP) that can image PC activity at high spatial resolution in live cells and tissues. Analysis of CLIP in a Furin;Pace4 double knockout (DKO) background shows that these two proteases are active and responsible for cleaving CLIP in the ICM of preimplantation blastocysts. However, loss of zygotic Furin and Pace4 in DKO mutant blastocysts is partially compensated shortly after implantation in the uterus. This rescue probably involves a maternal source of PCs in the uterus because CLIP was inhibited in Furin;Pace4 DKO embryos that were separated from maternal tissue. Analysis of early Nodal target genes suggests that this novel source of PC activity is sufficient to also initiate Nodal signaling in zygotic Furin;Pace4 double mutants. These findings establish CLIP as a specific and sensitive tool to predict novel PC functions even before overt manifestation of PC mutant phenotypes.
Results
CLIP is a specific and sensitive PC biosensor
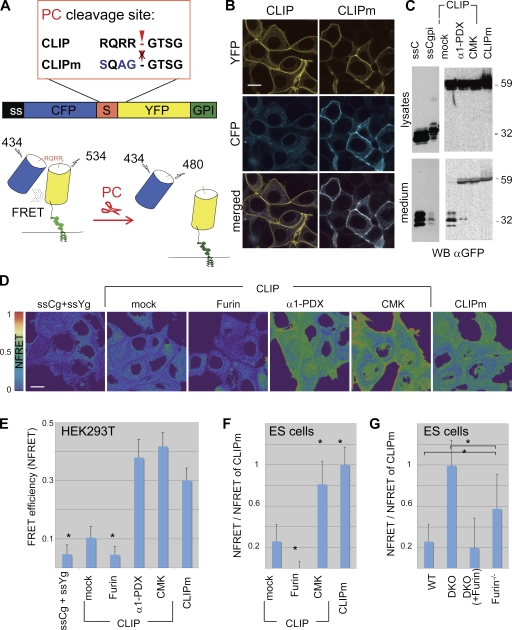
Proteolysis and other protein modifications affecting interatomic distances can be imaged in live cells by monitoring Förster resonance energy transfer (FRET) between suitable pairs of fluorophores (VanEngelenburg and Palmer, 2008). To monitor PC activities, ECFP (hereafter designated CFP) carrying a signal sequence (ss) was joined to citrine (hereafter designated YFP) by the PC recognition motif RQRR so that cleavage should trigger a proportional loss of FRET (Fig. 1 A). As a negative control, YFP was coupled to CFP by the PC-resistant linker sequence SQAG. To also detect extracellular PC activities (Klimpel et al., 1992; Blanchet et al., 2008; Mayer et al., 2008), the resulting fusion proteins CLIP and CLIP mutant (CLIPm) were targeted to the plasma membrane by a glycosylphosphatidylinositol (GPI) signal sequence. Accumulation of YFP combined with loss of CFP thus should mark the cells where processing occurred, provided that shedding of the GPI anchor is limited. Expression of CLIP in transfected HEK293T cells resulted in YFP fluorescence at the plasma membrane, whereas a fragment co-migrating with ssCFP was released into the culture medium (Fig. 1, B and C). In contrast, both YFP and CFP were enriched at the plasma membrane and shed uncleaved into culture medium in cells expressing CLIPm. Cleavage was also blocked if CLIP was cotransfected with α1-PDX, a PC-specific variant of antitrypsin (Jean et al., 1998), or if it was stabilized with the PC inhibitory peptide decanoyl-RVKR-CMK (CMK; Fig. 1, B and C; and Fig. S1 A). Given that basic residues may also be recognized by less specific proteases such as Thrombin, reporter cells expressing CLIP at the cell surface were incubated with recombinant Thrombin. Although Thrombin was significantly active at the highest concentration examined, physiological amounts (<2 U/ml) had no effect (Fig. S1 B). These results suggest that PC activity in HEK293T cells can be specifically detected by monitoring the ratio between CFP and YFP fluorescence of CLIP at the plasma membrane.
Figure 1.
PC-mediated activation of the cell surface–linked indicator of proteolysis (CLIP). (A) Schematic representation of the CLIP fusion protein containing the PC recognition motif RQRR, which is mutated in CLIPm to block cleavage. (B) Fluorescent imaging of HEK293T cells expressing CLIP or CLIPm. (C) Anti-GFP Western blot analysis of lysates (top) and conditioned medium (bottom) of HEK293T cells transfected with CLIP or CLIPm. ssCFP with or without a GPI signal sequence served as a control. ssCFP is released into conditioned medium in three isoforms migrating at 29, 31, and 34 kD. As expected, their secretion is severely diminished if ssCFP carries a GPI signal. Cleaved ssCFP also accumulates in the medium of cells transfected with CLIP, possibly together with trace amounts of shed YFP-gpi. The PC inhibitors α1-PDX or CMK shift the size of secreted CLIP to that of CLIPm (59 kD). The cleaved YFP-gpi moiety of CLIP at the cell membrane (B, top left) is not sufficiently enriched to be detected in Western blots of cell lysates, reflecting perhaps limited solubility. (D) Heat maps of normalized FRET (NFRET) efficiencies of CLIP and CLIPm in transfected HEK293T cells. NFRET between ssCFP-gpi and cotransfected ssYFP-gpi (background) was minimal. (E) Quantification of NFRET at the cell surface. (F) Relative NFRET efficiencies at the plasma membrane of wild-type ES cells transfected with CLIP or CLIPm. Where indicated, cells were cotransfected with Furin or empty vector (mock) or incubated with PC inhibitor CMK. (G) Relative NFRET of CLIP compared with CLIPm in Furin−/− and Furin−/−;Pace4−/− (DKO) double mutant ES cells cotransfected or not cotransfected with Furin expression vector. Asterisks indicate a significant difference with mock control as determined by t test (P < 0.05). Error bars are means ± SD. Bars, 10 µm.
To quantify PC activity by loss of FRET, FRET values of CLIP and CLIPm were acquired on live cells by sensitized emission analysis (Jares-Erijman and Jovin, 2003; Feige et al., 2005) and normalized to the signal of GPI-anchored YFP, which represents the total amount of biosensor at the plasma membrane. Normalized FRET (NFRET) efficiency was high for CLIPm, reaching on average 30 ± 4% (Figs. 1, D and E and S1 C). A similar NFRET efficiency of 41 ± 4 or 37 ± 6% was observed for CLIP in cells that were treated with CMK or transfected with α1-PDX, respectively. Alternatively, maximal NFRET efficiency was determined by acceptor photobleaching in CMK-treated fixed HEK293T cells. After photobleaching >95% of YFP, CFP fluorescence of stably transfected CLIP increased by 34 ± 4% (Fig. S2 A). Moreover, after addition of CMK to live HEK293T cells, time course experiments showed that NFRET of CLIP increases at an approximately linear rate of 10% per hour, leading to statistically significant changes already within 1 h after treatment (Fig. S2 B). In contrast, NFRET of CLIP in the absence of PC inhibitors only reached 10% and was further reduced to background levels upon cotransfection of Furin, Pace4, or Pcsk5A but not of the unrelated protease Cathepsin B (Figs. 1, D and E and S3 A). Likewise, in mouse embryonic stem (ES) cells, cotransfection of CLIP with Furin diminished NFRET, whereas CMK increased it (Fig. 1 F). Thus, PC activity can be measured by FRET analysis of CLIP at the plasma membrane.
To validate that CLIP is sensitive to variation of endogenous PCs, we measured NFRET in Furin−/−;Pace4−/− DKO ES cells (Constam and Robertson, 2000b; Beck et al., 2002) and in the Furin-deficient LoVo colon carcinoma cell line devoid of Furin activity (Takahashi et al., 1995). In DKO ES cells, NFRET of CLIP was comparable to that of CLIPm, whereas it was reduced below basal levels in rescue experiments in which CLIP was cotransfected with exogenous Furin (Fig. 1 G). Equivalent experiments in LoVo cells showed similar results (Fig. S3 B). Processing of CLIP in LoVo cells was also significantly rescued by cotransfected Pcsk5B or PC7 but not by PC1/3, PC2, or PC4 (Fig. S3 C). Together, these results indicate that (a) CLIP can be cleaved by all of the more widespread PC family members, including Furin, Pace4, Pcsk5, and PC7; (b) among these, the main endogenous PCs active in ES cells are Furin and Pace4; and (c) no other endogenous endopeptidases in these cell lines significantly activate CLIP.
CLIP imaging directly reveals paracrine PCs acting at the cell surface
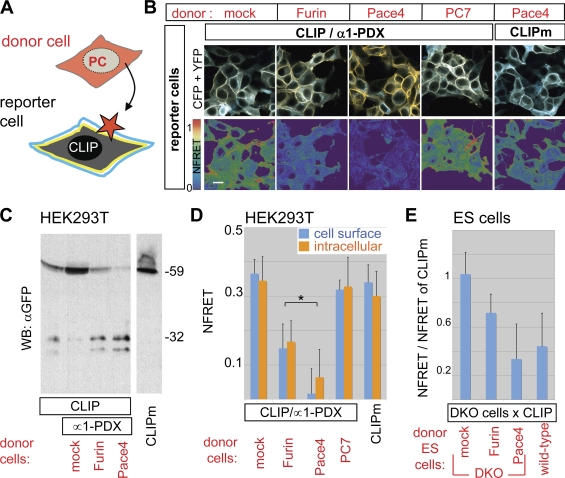
We next tested whether CLIP also detects paracrine PC activities. To block autocrine cleavage, CLIP was cotransfected in HEK293T cells together with α1-PDX. The resulting reporter cells were co-cultured with cells that were separately transfected with PC expression vectors or DsRed fluorescent protein (Fig. 2 A). CLIP did not overlap with DsRed, confirming that it does not pass from reporter to donor cells (Fig. S4 A). Fluorescence imaging, FRET, and immunoblot analysis revealed that both Furin and Pace4 from donor cells cleaved CLIP in reporter cells (Fig. 2, B–D). Similar results were obtained in co-cultures of donor and acceptor cells separated by a 0.4-µm pore sized filter insert, suggesting that cleavage is contact independent (Fig. S4 B). Furin and Pace4 also reduced FRET cells nonautonomously in cytoplasmic compartments (Fig. 2, B and D), suggesting that an intracellular FRET signal mainly emanates from uncleaved CLIP in endosomes. Pace4 was more active in this cell mixing assay than Furin, consistent with observations that it localizes mainly in the extracellular space (Tsuji et al., 2003; Mayer et al., 2008). In contrast, PC7, a PC not shed into the medium, had no effect (Figs. 2, B and D and S4 B), confirming that CLIP processing by cell nonautonomous Furin and Pace4 is specific. Moreover, CLIP processing was also induced in DKO reporter cells mixed with wild-type donor ES cells. In contrast, DKO donor cells had no effect, except if they were transfected with a Furin or Pace4 expression vector (Fig. 2 E). Together, these results establish that CLIP detects paracrine PC activities with high sensitivity and that cell nonautonomous Furin and Pace4 activities can directly cleave a membrane-bound substrate in target cells.
Figure 2.
CLIP detects paracrine PC activity. (A) Strategy to detect cell nonautonomous PC activities. (B) Fluorescent images and heat maps of NFRET in HEK293T cells exposed to donor cells expressing the indicated constructs. Bar, 10 µm. (C) Anti-GFP immunoblotting of conditioned medium from the co-cultures such as those shown in B. (D) Cell nonautonomous PC activity quantified by FRET analysis of CLIP at the surface (blue bars) or in the cytoplasm (orange bars) of the reporter cells shown in B. (E) FRET efficiency of CLIP in Furin−/−;Pace4−/− ES cells (DKO) co-cultured with wild-type ES cells or with separately transfected DKO cells expressing Furin, Pace4, or empty vector. The asterisk indicates a significant difference as determined by t test (P < 0.05). Error bars are means ± SD.
Furin and Pace4 activities overlap in the mouse ICM
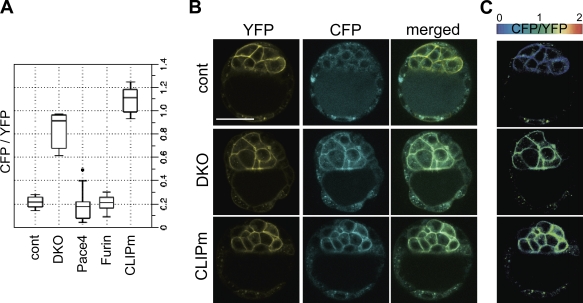
To test whether CLIP can detect PC activities in a physiological context, we generated transgenic mouse embryos expressing CLIP or CLIPm with the hybrid promoter of chicken β-actin and a human cytomegalovirus (CMV) enhancer of the CX-EGFP transgene (Table I; Hadjantonakis et al., 1998). In particular, because deletion of Furin and Pace4 blocked CLIP processing in ES cells, we asked whether they are also active in ES cell progenitors in the ICM. Among all blastocysts examined from Pace4 and Furin single heterozygous intercrosses, only one showed a CFP/YFP ratio >0.4 (n = 21; Fig. 3 A). In contrast, 6 of 28 (22%) CLIP-positive blastocysts from Furin+/−;Pace4+/− intercrosses resembled CLIPm blastocysts, showing bright CFP fluorescence at the surface of ICM cells (Fig. 3, A–C). This corresponds to the expected Mendelian frequency of DKO embryos because Furin and Pace4 are linked on chromosome 7. The CFP/YFP ratios were uniform throughout a given ICM (Fig. 3 C), reaching mean values of 0.84 ± 0.13 in DKO blastocysts (n = 6) compared with 0.21 ± 0.04 in control littermates (n = 7; Fig. 3 A). These results establish that processing of CLIP in the ICM, like in ES cells, depends on redundant activities of Furin and Pace4.
Table I.
Germline transmission of CLIP and CLIPm transgenes
| Founders | Germline transmission | Expression (E5.5) | Expression in adult tissue |
| CLIP (12) | 10/12 | 6/9 | 3/8 |
| CLIPm (9) | 6/7 | 4/5 | 2/5 |
| Total (21) | 16/19 (84%) | 10/14 (71%) | 5/13 (38%) |
Transgenic lines positive for germline transmission were tested for transgene expression at E5.5 and in adult tissues. Expression was considered positive when detectable by an epifluorescent M205FA stereomicroscope. Because of premature loss, three lines could not be analyzed.
Figure 3.
CLIP imaging reveals the presence of redundant Furin and Pace4 activities in the ICM of murine blastocysts. (A) Box plot of cell surface CFP/YFP ratios of E3.5 CLIP embryos from Furin and Pace4 single or double heterozygous intercrosses. CLIPm transgenic embryos are shown for comparison. The dot indicates a CFP/YFP ratio >0.4. (B) Blastocysts from Furin+/−;Pace4+/− intercrosses without (top row) or with (middle row) CFP at the surface of ICM cells. A CLIPm transgenic is shown for comparison (bottom row). (C) Heat maps of CFP/YFP ratios in the ICM of the blastocysts in A. (B and C) Error bars are means ± SD. Bar, 50 µm.
The epiblast of implanted embryos has access to a PC activity other than zygotic Furin and Pace4
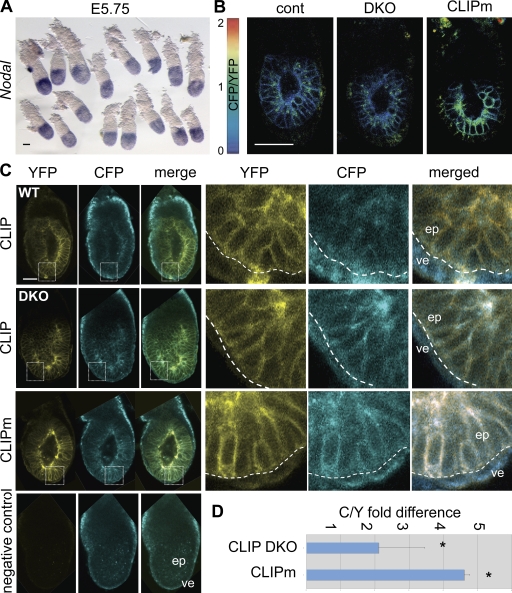
Known substrates of Furin and Pace4 include the TGF-β–related Nodal precursor (Constam and Robertson, 1999), which is expressed in the ICM and in its derivatives, the epiblast and surrounding visceral endoderm (Collignon et al., 1996; Brennan et al., 2002; Mesnard et al., 2006). Targeted mutations disrupting the entire Nodal locus or, specifically, the PC cleavage motif established that processed Nodal is already essential at stage E4.5–5.5 to up-regulate Nodal expression in an autoinductive feedback loop and to induce additional target genes that pattern the epiblast and visceral endoderm (Brennan et al., 2001; Ben-Haim et al., 2006; Mesnard et al., 2006). However, in DKO embryos lacking Furin and Pace4, Nodal and its targets remained normally expressed until E5.5 (Fig. 4 A and Table II). To address this discrepancy, litters obtained from Furin+/−;Pace4+/− intercrosses were also imaged for CLIP processing. CFP fluorescence at the surface of epiblast cells was not significantly increased in any of these embryos (n = 21; Fig. 4 B). These results demonstrate that zygotic Furin and Pace4 are not the only convertases acting on the epiblast once the conceptus has implanted in the uterus.
Figure 4.
Zygotic Furin;Pace4 DKO mutants show alternative PC activity at E5.5. (A) Whole mount in situ hybridization analysis of Nodal mRNA in embryos from Furin+/−;Pace4+/− intercrosses at stage E5.5–5.75. (B) Heat maps of CFP/YFP ratios in DKO and wild-type CLIP embryos and in CLIPm controls at E5.5. (C) From top to bottom, analysis of CFP and YFP signals in E6.5 wild type and DKO expressing CLIP, CLIPm, and nontransgenic embryos. Insets show the mean relative intensities of YFP (yellow) and CFP (blue) at the surface of epiblast cells. Dashed outlines indicate the boundary between the epiblast (ep) and visceral endoderm (ve). Bars, 50 µm. (D) CFP/YFP ratios were measured at the cell surface of E6.5 CLIP embryos from Furin+/−;Pace4+/− intercrosses or E6.5 CLIPm embryos. The fold difference was calculated relative to the mean CFP/YFP ratio of wild-type embryos. The asterisk indicates a significant difference as determined by t test (P < 0.05). Error bars are means ± SD.
Table II.
In situ hybridization analysis of E5.5 embryos from Furin+/−;Pace4+/− intercrosses
| Marker | Number of embryos | Normal expression | Reduced expression | Loss of expression |
| Nodal | 61 | 57 | 4 | 0 |
| Fgf8 | 16 | 13 | 3 | 0 |
| Lefty-1/-2 | 14 | 14 | 0 | 0 |
| Lim1 | 9 | 9 | 0 | 0 |
| Fgf5 | 27 | 22 | 3 | 2 |
| Foxd3 | 18 | 18 | 0 | 0 |
| Cer-l | 21 | 21 | 0 | 0 |
| Oct4 | 20 | 20 | 0 | 0 |
E5.5 embryos from Furin+/−;Pace4+/− intercrosses were stained by whole mount in situ hybridization for markers that are down-regulated at this stage in Nodal mutants (Ben-Haim et al., 2006; Mesnard et al., 2006).
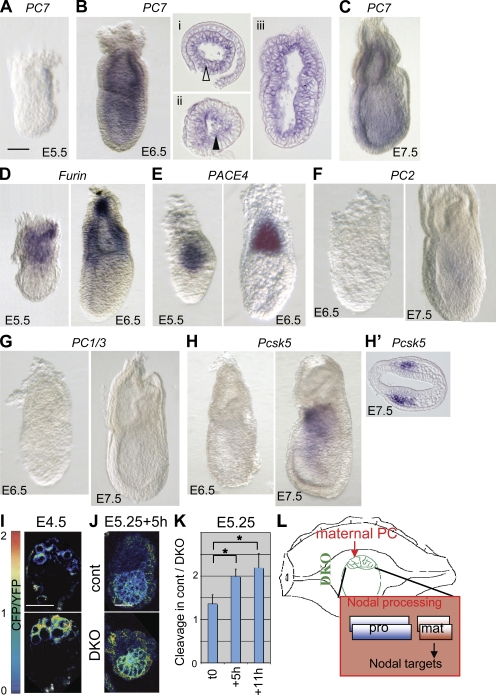
Zygotic Furin and Pace4 are essential to maintain Nodal signaling during gastrulation (Constam and Robertson, 2000b; Beck et al., 2002). At this stage (E6.5), the CFP/YFP ratio at the surface of epiblast cells on average increased twofold in DKO mutants compared with wild type (Fig. 4, C and D). By comparison, CLIPm transgenic epiblast cells showed a fivefold mean increase of CFP/YFP, concurring with the notion that processing is PC specific (Fig. 4, C and D). Thus, impaired CLIP processing correlates with the onset of the DKO mutant phenotype despite the presence of some residual overlapping cleavage activity.
Analysis of CLIP in the ICM/epiblast lineage between E3.5 and E5.5 implicates the uterus as a likely source of maternal PC activity
Given that processing of CLIP was blocked in LoVo and PC-deficient ES cells and in cells transfected with the PC-specific inhibitor α1-PDX, a role for endogenous proteases other than PCs seems unlikely. Moreover, among the PCs, only Furin and Pace4 are significantly expressed after implantation until gastrulation, as determined by whole mount in situ hybridization staining (Fig. 5, A–H). Pcsk5, like PC2 and PC4, is already down-regulated at the morula stage (St Germain et al., 2005). To narrow the time when CLIP can be processed independently of zygotic Furin and Pace4, DKO embryos were also analyzed at stage E4.5. As predicted by a Mendelian distribution, 5 out of 29 newly implanted blastocysts from Furin+/−;Pace4+/− intercrosses retained strong CFP fluorescence at the cell surface (Fig. 5 I), suggesting that zygotic Furin and Pace4 are responsible to cleave CLIP until implantation. Concurring with this result, CLIP processing was also inhibited in DKO blastocysts that were isolated at E3.5 and developed for 24 h in culture (Fig. S5, A and B). Therefore, to distinguish whether the PC-like activity detected in E5.5 DKO embryos is induced de novo or provided by maternal tissues, E5.25 embryos were cultured ex vivo, thereby removing any further maternal contribution. Interestingly, unlike in control littermates, in DKO embryos (n = 8/8) the CFP/YFP ratio of CLIP increased on average by 50% within 5–7 h (Fig. 5, J and K). This result demonstrates that processing within the epiblast of DKO embryos depends, at least in part, on the uterine environment (Fig. 5 L).
Figure 5.
Evidence for maternal PC activity in E5.5 embryos. (A–C) Zygotic expression of PCs before and during gastrulation. Although PC7 mRNA is not detectable at E5.5–5.75, even after >3 d of staining (A), it is clearly induced at E6.25–6.5 in embryonic and extraembryonic lineages (B). Transverse (i and ii) and sagittal (iii) sections show that PC7 mRNA is enriched near the apical surface of epiblast cells (closed arrowhead), whereas it is uniformly distributed in the extraembryonic ectoderm (open arrowhead). (C) PC7 remains broadly expressed at E7.5. (D and E) Furin (D) and Pace4 mRNAs (E) are expressed in the extraembryonic ectoderm both before and during gastrulation as described in Beck et al. (2002) and Mesnard et al. (2006). (F–H) Neither PC2 (F), PC1/3 (G), nor Pcsk5 transcripts (H) were detected until E6.5 even after prolonged staining for several days. Pcsk5 expression initiated between E7.25 and E7.5, specifically in anterior mesoderm (H’). Embryos are viewed laterally, and anterior is to the left. (I) Heat maps of CFP/YFP ratios in implanted E4.5 DKO and wild-type blastocysts. (J) Heat maps of CFP/YFP ratios in E5.25 DKO and wild-type embryos cultured for 5 h in vitro. Bars, 50 µm. (K) Relative CFP/YFP ratios of DKO versus wild-type embryos at 0, 5, and 11 h of in vitro culture. The asterisk indicates a significant difference as determined by t test (P < 0.05). (L) Model for the transient activation of Nodal in ICM cells of implanted DKO blastocysts by maternal PC activities from the deciduum, a tissue producing high levels of Pcsk5 (Constam and Robertson, 1999; Essalmani et al., 2008). Pro and mat indicate pro and mature domains of Nodal, respectively. Error bars are means ± SD.
CLIP imaging in adult tissues
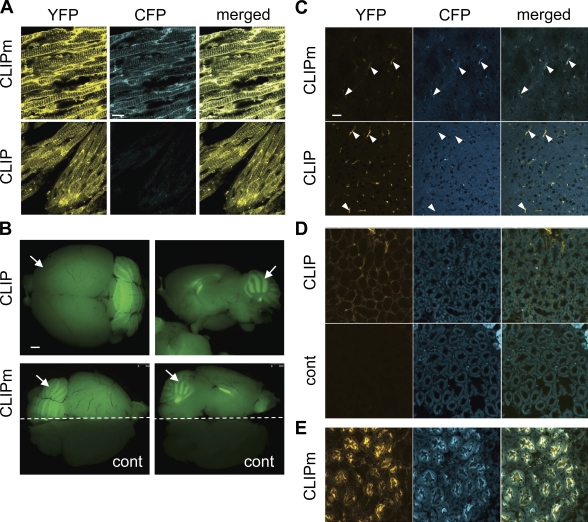
To test whether PC activity can also be monitored in adults, cryosections of fixed tissues from CLIP and CLIPm transgenics were analyzed by confocal microscopy. Strong fluorescence of both CFP and YFP was detected in CLIPm transgenic tissues expressing high levels of β-actin, such as cardiac muscle (Fig. 6 A). In contrast, CFP was reduced to background levels in the corresponding tissues expressing CLIP. Significant ubiquitous fluorescence was also seen in the brain, gut, kidney, and skin. However, relative expression levels of CLIP and CLIPm were not uniform among individual lines and among different tissues (Figs. 6, B–E and S5, B and C).
Figure 6.
PC activities in adult tissues of CLIP transgenic mice. (A) CFP and YFP fluorescence of CLIP or CLIPm in cryosections of cardiac striated muscle cells. CLIP mice, unlike CLIPm controls, lose CFP from the surface. (B) Biosensor expression was tested by GFP epifluorescence imaging of live brains of CLIP (top), CLIPm, and nontransgenic control mice (bottom). Only one half of the CLIPm brain is shown together with one half of a nontransgenic one. Both CLIP and CLIPm are expressed ubiquitously, with peak levels in the cerebellum (arrows). Left, dorsal views; right, lateral views. (C) In cryosections of fixed brain cortex, YFP and CFP signal intensities are barely detectable, except in capillaries (arrowheads). (D) Analysis of YFP and CFP fluorescence of CLIP in interstitial mesenchyme of kidney medulla. CFP signal intensity is reduced to the background levels observed in nontransgenic controls (cont), indicating that CLIP is efficiently processed. (E) Kidney medulla of a CLIPm transgenic line showing expression in ductal cells only. Bars: (A and C–E) 20 µm; (B) 1 mm.
Discussion
In this paper, we introduce CLIP, an affordable and convenient biosensor to image PC activities in live cells and whole tissues. Our analysis of wild-type and Furin and Pace4 single and DKO mutant blastocysts reveal that zygotic Furin and Pace4 are already active and responsible for activating CLIP in the ICM both before and during implantation (E3.5–4.5). Unlike the cleavage mutant negative control CLIPm, CLIP was also efficiently processed in the epiblast of wild-type E5.5–6.5 embryos, thus confirming that this tissue is exposed to PC activities, even though it does not endogenously express any known PC. Moreover, processing in the epiblast of zygotic DKO mutant was impaired at E6.5, coinciding with the loss of Nodal activity. This finding is consistent with a direct role for Furin and Pace4 in Nodal precursor processing (Beck et al., 2002) and with the model that Nodal matures at the plasma membrane in a complex with Cripto (Blanchet et al., 2008; Constam, 2009b). By contrast, at E5.5, deletion of zygotic Furin and Pace4 inhibited neither processing of CLIP nor the induction of Nodal and its target genes. This was unexpected because, on a different, inbred genetic background, zygotic DKO mutants phenocopy NodalNr/Nr embryos, which only express a cleavage-deficient mutant Nodal precursor (Beck et al., 2002; Ben-Haim et al., 2006). PC7 is not involved because it can neither cleave proNodal ex vivo nor rescue Nodal signaling in DKO mutants at E6.5 (Beck et al., 2002). Before E6.5, implanted embryos also do not express detectable amounts of PC7 or Pcsk5 (Fig. 5, A–H). Therefore, the residual processing of Nodal and CLIP in DKO embryos at E5.5 is most likely mediated by a maternal PC from the uterus (Fig. 5 L). Consistent with this model, the CFP fluorescence of CLIP rapidly increased in E5.25 DKO mutants that were cultured outside the uterus until E5.5–5.75 but not in cultured control littermates. Thus, although zygotic Furin/Pace4 activities clearly contribute to CLIP processing in the epiblast, their loss is efficiently compensated at E5.5 in utero, most likely by one or several maternal PCs. A likely candidate for the maternal activity is Pcsk5, which is not expressed in embryonic lineages until late gastrulation (Essalmani et al., 2008; Szumska et al., 2008) but is abundantly present in the E4.5 deciduum (Essalmani et al., 2008) and able to cleave Nodal (Beck et al., 2002).
The sensitivity and specificity of CLIP were also stringently validated in DKO mutant ES cells and by quantitative FRET measurements. Furthermore, confirming earlier predictions (Beck et al., 2002; Blanchet et al., 2008), our analysis of CLIP in mixed cell cultures provides direct evidence that Furin and Pace4 can cell nonautonomously cleave a membrane-bound protein. In this assay, CLIP also detected robust cell nonautonomous activity of Pcsk5 (Fig. S4 B), which binds to heparan sulfate proteoglycans through a cysteine-rich region that is conserved in Pace4 (Nour et al., 2005; Mayer et al., 2008). In cultured cells, activation of Nodal at the membrane is directed to lipid rafts by the GPI-anchored coreceptor Cripto, which also binds Furin and Pace4 (Blanchet et al., 2008). Cripto binds the Furin P domain that is conserved in all PCs (Blanchet et al., 2008). However, this interaction is clearly not essential to engage PCs at the plasma membrane because CLIP can be efficiently cleaved at the surface of HEK293T cells, which express neither Cripto nor the related protein Cryptic (Minchiotti et al., 2000; Yan et al., 2002). Thus, soluble PCs may be enriched at the plasma membrane by heparan sulfate proteoglycans or other receptors, whereas Cripto serves to localize processing to specific membrane domains (Constam, 2009a).
To label cells exposed to autocrine or paracrine PC activities, CLIP was tethered to the plasma membrane like Cripto by a GPI anchor. This approach differs from earlier studies in tissue culture and in tumor xenografts monitoring intracellular PC activities by the release of alkaline phosphatase from a Golgi-resident fusion protein into the culture medium (Coppola et al., 2007) or by adding caged bioluminescent substrates (Dragulescu-Andrasi et al., 2009). PC activities have also been detected at the surface of transfected cells by the internalization of a tagged ligand of processed anthrax protective antigen (Hobson et al., 2006). However, none of these smart systems provided information on endogenous PC activities in vivo or on their individual relative contributions to the processing of membrane-bound substrates.
Earlier efforts to image proteases using FRET probes have focused on matrix metalloproteases (MMPs). A synthetic FRET probe was sufficiently sensitive ex vivo to detect endogenous MMP12 in isolated macrophages, a rich source of MMPs (Cobos-Correa et al., 2009). However, spatial resolution at the tissue level has not been analyzed, and synthetic probes also have the disadvantage that they require costly chemistry and specialized expertise. As an alternative, FRET has been monitored using a YFP–CFP fusion that can be partially cleaved by recombinant soluble MMPs in vitro (Yang et al., 2007). However, targeting of this probe to the cell surface by the transmembrane domain of platelet-derived growth factor receptor was inefficient, and MMP inhibitors had no effect on FRET until after prolonged treatment for at least 72 h (Yang et al., 2007), implying that both the sensitivity and specificity of cleavable YFP–CFP fusion proteins might be insufficient to image cell surface proteases in vivo. In contrast, our results obtained with CLIP establish that a fusion of widely used fluorescent proteins is suitable to image secreted proteases with unmatched sensitivity and spatial resolution. On the other hand, although our reporter can image the onset of enzyme activation, the time required for its own synthesis might be rate limiting to detect more rapid changes in PC activity. CLIP also detected PC activities in adult tissues, even though its expression levels were heterogeneous across different tissues. More uniform expression might be achieved in the future by inserting the transgene into a defined locus. To our knowledge, CLIP is the first biosensor to detect specific proteolytic activities in the in vivo physiological context of a transgenic mouse model.
Materials and methods
Plasmids and expression vectors
CLIP and CLIPm expression vectors were generated in pcDNA3.1 (Invitrogen). In brief, cDNA fragments comprising the signal sequence of lactase-phlorizin hydrolase, ECFP, a linker region, citrine, and the gpi attachment signal of lymphocyte function–associated antigen 3 (Keller et al., 2001) were amplified by PCR and ligated in frame. For convenient exchange, the linker sequences were flanked by unique EcoRI and Cla1 restriction sites. Linkers comprising the PC consensus cleavage site RQRR or the cleavage-resistant sequence SQAG were obtained by annealing the oligonucleotide 5′-AATTCTCGTCCTCAGAGTTGAGCGCTCGACAACGACGCGGCACAAGCGGCAGAT-3′ with 5′-CGATGCTGCCGCTTGTGCCGCGTCGTTGTCGAGCGCTCAACTCTGAGGACGAG-3′, and 5′-AATTCTCGTCCTCAACCGGTATCCCTGTTTCACTCGGCAGCGGAAGCGGCAGCAT-3′ with 5′-CATGCTGCCGCTTGTGCCGCCGGCTTGAGAACCGGTCAACTCTGAGGACGAG-3′, respectively. To derive ssCFP-gpi and ssYFP-gpi, the ECFP-linker-YFP sequence of CLIP was replaced by ECFP or YFP, respectively. Corresponding transgenes containing the CMV/chick β-actin hybrid promoter, the first intron of β-actin, and a rabbit β-globin 3′ untranslated region were generated by replacing the EcoRI-EGFP cDNA fragment of the CX-EGFP plasmid (Hadjantonakis et al., 1998) with the open reading frames of CLIP or CLIPm. After linearization using HindIII or XmnI, respectively, the resulting transgenes (4,380 bp) were purified using gel extraction columns (QIAquick PCR purification kit; QIAGEN) and resuspended at 20 ng/µl in 10 mM Tris-HCl, pH 7.5, containing 0.1 mM EDTA for pronuclear injection in mouse oocytes. PC expression vectors contained a CMV promoter directing the expression of full-length mouse proteins from the plasmid pRc/CMV (Invitrogen) or pCS2+. Expression of the α1-PDX cDNA was under the control of the pRc/CMV vector (gift of G. Thomas, Oregon Health and Science University, Portland, OR).
Mice
Transgenic mice were generated at the Swiss Federal Institute of Technology Lausanne core facility by pronuclear injection of linearized CLIP or CLIPm into zygotes of FVB and NMRI mice (Harlan and Janvier). 12 and 9 founders, respectively, and their offspring were genotyped by 31 PCR amplification cycles at the annealing temperature of 63°C using the CLIP-specific primer 5′-GAGTTGAGCGCTCGACAACGACG-3′ or the CLIPm-specific primer 5′-CTCAGAGTTGACCGGTTCTCAAGC-3′ and the YFP-specific primer 5′-GAAGCACATCAGGCCGTAGCCG-3′. Cell surface fluorescence of the biosensor was monitored in embryos at the stages indicated. Furin and Pace4 single and double heterozygotes of a C57BL/6 inbred background (Roebroek et al., 1998; Constam and Robertson, 2000a; Beck et al., 2002) were serially backcrossed to an NMRI outbred background for more than eight generations.
Whole embryo culture
Whole embryo in vitro culture was performed as described in Beck et al. (2002) but in the absence of STO fibroblast feeders. In brief, embryos were recovered at 5.25 d after coitum. Reichert’s membrane was mechanically removed using fine needles. Dissected embryos were transferred to filter inserts (pore size of 12 µm; Millipore) and incubated in OptiMEM I (Invitrogen) supplemented with 15% knockout replacement serum, 1% gentamycin, and glutamine sulfate in the presence of 5% CO2- and water-saturated atmosphere.
Whole mount in situ hybridization
Whole mount in situ hybridization was performed as described previously using digoxigenin-labeled antisense probes (Varlet et al., 1997; Beck et al., 2002). PC7 mRNA was detected by an antisense probe spanning nucleotides 1,606–2,407 under GenBank/EMBL/DDBJ accession no. U48830. The Pcsk5 probe comprised nucleotides 740–2,086 (GenBank accession no. D12619), PC2 nucleotides 1,145–1,588 (available at GenBank under accession no. NM_008792), and PC1/3 nucleotides 862–1,305 (GenBank accession no. NM_013628). Antidigoxigenin antibodies conjugated to alkaline phosphatase and the substrate BM purple were obtained from Roche. Color reactions were developed until saturation at RT or maximally for 3 d at 4°C.
Cell transfection and Western blot analysis
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum, 100 µg/ml gentamycin, and 2 mM glutamine. Furin knockout and Furin;Pace4 DKO ES cells (Constam and Robertson, 2000b; Beck et al., 2002) were maintained on irradiated STO fibroblasts in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum, 100 µg/ml gentamycin, 2 mM glutamine, and 0.1 mM β-mercaptoethanol. LoVo cells were cultured in Ham’s F12 medium (Sigma-Aldrich) containing 10% fetal calf serum, 100 µg/ml gentamycin, and 2 mM glutamine. For transient transfection, HEK293T and LoVo cells were plated on 24-well plates at a density of 5 × 104 cells/well. The next day, the cells were incubated with plasmids (0.5 µg/well) in Lipofectamine 2000 CD Reagent (Invitrogen) during 4–6 h. Thereafter, the medium was replaced by OptiMEM containing 0.25% KnockOut Serum Replacement (Invitrogen). After 24 h, conditioned medium was harvested, and cells were lysed in Laemmli buffer containing 5% β-mercaptoethanol. Expression and processing of CLIP were monitored by chemiluminescent immunoblotting of the ECFP and YFP moieties using monoclonal anti-GFP antibody (Sigma-Aldrich) and HRP-conjugated anti–mouse secondary antibody (GE Healthcare). To monitor processing of biosensors by secreted PC activities, donor cells were washed with PBS 6 h after transfection, trypsinized, and added at a 1:1 ratio to reporter cells that were separately transfected with CLIP. Medium was conditioned during the next 24 h. ES cells were transfected and analyzed identically, except that they were plated on gelatin-coated 24-well dishes at a density of 2.5 × 105 cells/well.
Imaging
For confocal imaging, cells were plated on glass-bottom 24-well plates and maintained in the buffered OptiMEM medium. To avoid evaporation and temperature variation, the plate was sealed using parafilm and maintained at 37°C throughout the imaging process. Image acquisition was achieved on a confocal microscope (TCS SP II; Leica) at the following settings: CFP channel, 458-nm excitation and 462–510-nm emission; FRET channel, 458-nm excitation and 518–580-nm emission; and YFP channel, 514-nm excitation and 518–580-nm emission. Scanning was done sequentially between lines at 400 Hz with a PlanApo 63× 1.4 NA oil objective (Leica). Heat maps of C/Y ratios were generated by Imaris 5.3 software (Bitplane) using a Matlab script. A mask was created for YFP signals above a critical threshold to highlight cell surface staining. Each CFP/YFP ratio was then attributed a proportional value between 1 (CFP/YFP = 0; blue) and 256 (CFP/YFP = 2; red). Values >2 were considered as artifactual (white). Areas not included in the mask were attributed a black color. GFP imaging was performed under an epifluorescent stereomicroscope (M205FA; Leica) using the GFP2 filter set (480/40-nm excitement filter, 505-nm long pass dichromatic beam splitter, and 510-nm long pass emission filter).
FRET analysis
To measure FRET efficiency of CLIP and CLIPm in live transfected HEK293T or mouse ES cells, confocal microscopy images were processed using the commercial TCS SP II sensitized emission wizard or the ImageJ (National Institutes of Health) pixFRET plug-in (Feige et al., 2005). Correction factors α, β, γ, and δ were established on cells transfected with either ssCFP-gpi or ssYFP-gpi plasmids as described in van Rheenen et al. (2004). Because CFP may be lost upon cleavage and therefore cannot be used to estimate the ratio of cleaved versus uncleaved biosensor, FRET efficiency (NFRET) was calculated by normalizing the corrected FRET signal to the corrected YFP signal representing the total amount of biosensor at the plasma membrane. Owing to this normalization, NFRET tends to be slightly over- or underestimated in regions where YFP signals are extremely low or high, respectively. Therefore, NFRET data were acquired in regions of comparable intermediate YFP fluorescence intensity values. For quantitative analysis, mean NFRET values were calculated by defining regions of interest (ROIs) at the cell surface (Fig. S1 C) of 15 reporter cells over three different fields of cells imaged for each specific condition.
Online supplemental material
Fig. S1 shows the specificity of CFP and YFP fluorescence of CLIP and CLIPm at the cell surface. Fig. S2 shows maximal value and dynamics of FRET recovery upon inhibition of CLIP processing. Fig. S3 shows the analysis of CLIP specificity. Fig. S4 shows validation that paracrine PC activity does not require cell contact between donor and reporter cells. Fig. S5 shows analysis of PC activity in cultured DKO blastocysts and in adult skin and gut. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005026/DC1.
Acknowledgments
We are grateful to Dr. G. Thomas for the gift of α1-PDX expression vector, to A. Griffa and J.-C. Sarria for imaging support, and to Drs. P. Gönczy, A. Grapin-Botton, and C. Fuerer for critical reading of the manuscript.
This work was supported by the Swiss National Science Foundation (grant 3100A0-118080) and by the European Union Sixth Framework Programme Specific Targeted Research Projects (LSHM-CT-2006-037831).
Footnotes
Abbreviations used in this paper:
- CLIP
- cell surface–linked indicator of proteolysis
- CLIPm
- CLIP mutant
- CMV
- human cytomegalovirus
- DKO
- double knockout
- ES
- embryonic stem
- FRET
- Förster resonance energy transfer
- GPI
- glycosylphosphatidylinositol
- ICM
- inner cell mass
- MMP
- matrix metalloprotease
- NFRET
- normalized FRET
- PC
- proprotein convertase
- ROI
- region of interest
References
- Arnold S.J., Robertson E.J. 2009. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10:91–103 10.1038/nrm2618 [DOI] [PubMed] [Google Scholar]
- Beck S., Le Good J.A., Guzman M., Ben Haim N., Roy K., Beermann F., Constam D.B. 2002. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 4:981–985 10.1038/ncb890 [DOI] [PubMed] [Google Scholar]
- Ben-Haim N., Lu C., Guzman-Ayala M., Pescatore L., Mesnard D., Bischofberger M., Naef F., Robertson E.J., Constam D.B. 2006. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell. 11:313–323 10.1016/j.devcel.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Blanchet M.-H., Le Good J.A., Mesnard D., Oorschot V., Baflast S., Minchiotti G., Klumperman J., Constam D.B. 2008. Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J. 27:2580–2591 10.1038/emboj.2008.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J., Lu C.C., Norris D.P., Rodriguez T.A., Beddington R.S., Robertson E.J. 2001. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 411:965–969 10.1038/35082103 [DOI] [PubMed] [Google Scholar]
- Brennan J., Norris D.P., Robertson E.J. 2002. Nodal activity in the node governs left-right asymmetry. Genes Dev. 16:2339–2344 10.1101/gad.1016202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos-Correa A., Trojanek J.B., Diemer S., Mall M.A., Schultz C. 2009. Membrane-bound FRET probe visualizes MMP12 activity in pulmonary inflammation. Nat. Chem. Biol. 5:628–630 10.1038/nchembio.196 [DOI] [PubMed] [Google Scholar]
- Collignon J., Varlet I., Robertson E.J. 1996. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 381:155–158 10.1038/381155a0 [DOI] [PubMed] [Google Scholar]
- Constam D.B. 2009a. Riding shotgun: a dual role for the epidermal growth factor-Cripto/FRL-1/Cryptic protein Cripto in Nodal trafficking. Traffic. 10:783–791 10.1111/j.1600-0854.2009.00874.x [DOI] [PubMed] [Google Scholar]
- Constam D.B. 2009b. Running the gauntlet: an overview of the modalities of travel employed by the putative morphogen Nodal. Curr. Opin. Genet. Dev. 19:302–307 10.1016/j.gde.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Constam D.B., Robertson E.J. 1999. Regulation of bone morphogenetic protein activity by pro domains and proprotein convertases. J. Cell Biol. 144:139–149 10.1083/jcb.144.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam D.B., Robertson E.J. 2000a. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 14:1146–1155 [PMC free article] [PubMed] [Google Scholar]
- Constam D.B., Robertson E.J. 2000b. Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development. 127:245–254 [DOI] [PubMed] [Google Scholar]
- Coppola J.M., Hamilton C.A., Bhojani M.S., Larsen M.J., Ross B.D., Rehemtulla A. 2007. Identification of inhibitors using a cell-based assay for monitoring Golgi-resident protease activity. Anal. Biochem. 364:19–29 10.1016/j.ab.2007.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragulescu-Andrasi A., Liang G., Rao J. 2009. In Vivo Bioluminescence Imaging of Furin Activity in Breast Cancer Cells Using Bioluminogenic Substrates. Bioconjug. Chem. 20:1660–1666 10.1021/bc9002508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essalmani R., Zaid A., Marcinkiewicz J., Chamberland A., Pasquato A., Seidah N.G., Prat A. 2008. In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc. Natl. Acad. Sci. USA. 105:5750–5755 10.1073/pnas.0709428105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J.N., Sage D., Wahli W., Desvergne B., Gelman L. 2005. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 68:51–58 10.1002/jemt.20215 [DOI] [PubMed] [Google Scholar]
- Fugère M., Day R. 2005. Cutting back on pro-protein convertases: the latest approaches to pharmacological inhibition. Trends Pharmacol. Sci. 26:294–301 10.1016/j.tips.2005.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis A.K., Gertsenstein M., Ikawa M., Okabe M., Nagy A. 1998. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech. Dev. 76:79–90 10.1016/S0925-4773(98)00093-8 [DOI] [PubMed] [Google Scholar]
- Hobson J.P., Liu S., Rønø B., Leppla S.H., Bugge T.H. 2006. Imaging specific cell-surface proteolytic activity in single living cells. Nat. Methods. 3:259–261 10.1038/nmeth862 [DOI] [PubMed] [Google Scholar]
- Jares-Erijman E.A., Jovin T.M. 2003. FRET imaging. Nat. Biotechnol. 21:1387–1395 10.1038/nbt896 [DOI] [PubMed] [Google Scholar]
- Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A.J., Thomas G. 1998. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc. Natl. Acad. Sci. USA. 95:7293–7298 10.1073/pnas.95.13.7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao G.S., Cregar L., Wang J., Millis S.Z., Tang C., O’Malley S., Johnson A.T., Sareth S., Larson J., Thomas G. 2006. Synthetic small molecule furin inhibitors derived from 2,5-dideoxystreptamine. Proc. Natl. Acad. Sci. USA. 103:19707–19712 10.1073/pnas.0606555104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P., Toomre D., Díaz E., White J., Simons K. 2001. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3:140–149 10.1038/35055042 [DOI] [PubMed] [Google Scholar]
- Klimpel K.R., Molloy S.S., Thomas G., Leppla S.H. 1992. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc. Natl. Acad. Sci. USA. 89:10277–10281 10.1073/pnas.89.21.10277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T., Coppola J.M., Larsen M.J., van Dort M.E., Ross B.D., Day R., Rehemtulla A., Fuller R.S. 2009. Inhibition of furin/proprotein convertase-catalyzed surface and intracellular processing by small molecules. J. Biol. Chem. 284:15729–15738 10.1074/jbc.M901540200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer G., Hamelin J., Asselin M.C., Pasquato A., Marcinkiewicz E., Tang M., Tabibzadeh S., Seidah N.G. 2008. The regulated cell surface zymogen activation of the proprotein convertase PC5A directs the processing of its secretory substrates. J. Biol. Chem. 283:2373–2384 10.1074/jbc.M708763200 [DOI] [PubMed] [Google Scholar]
- Mesnard D., Guzman-Ayala M., Constam D.B. 2006. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development. 133:2497–2505 [DOI] [PubMed] [Google Scholar]
- Minchiotti G., Parisi S., Liguori G., Signore M., Lania G., Adamson E.D., Lago C.T., Persico M.G. 2000. Membrane-anchorage of Cripto protein by glycosylphosphatidylinositol and its distribution during early mouse development. Mech. Dev. 90:133–142 10.1016/S0925-4773(99)00235-X [DOI] [PubMed] [Google Scholar]
- Nour N., Mayer G., Mort J.S., Salvas A., Mbikay M., Morrison C.J., Overall C.M., Seidah N.G. 2005. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol. Biol. Cell. 16:5215–5226 10.1091/mbc.E05-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroek A.J.M., Umans L., Pauli I.G.L., Robertson E.J., van Leuven F., Van de Ven W.J.M., Constam D.B. 1998. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 125:4863–4876 [DOI] [PubMed] [Google Scholar]
- Seidah N.G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. 2008. The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol. 40:1111–1125 10.1016/j.biocel.2008.01.030 [DOI] [PubMed] [Google Scholar]
- St Germain C., Croissandeau G., Mayne J., Baltz J.M., Chrétien M., Mbikay M. 2005. Expression and transient nuclear translocation of proprotein convertase 1 (PC1) during mouse preimplantation embryonic development. Mol. Reprod. Dev. 72:483–493 10.1002/mrd.20271 [DOI] [PubMed] [Google Scholar]
- Szumska D., Pieles G., Essalmani R., Bilski M., Mesnard D., Kaur K., Franklyn A., El Omari K., Jefferis J., Bentham J., et al. 2008. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 22:1465–1477 10.1101/gad.479408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Nakagawa T., Kasai K., Banno T., Duguay S.J., Van de Ven W.J., Murakami K., Nakayama K. 1995. A second mutant allele of furin in the processing-incompetent cell line, LoVo. Evidence for involvement of the homo B domain in autocatalytic activation. J. Biol. Chem. 270:26565–26569 10.1074/jbc.270.44.26565 [DOI] [PubMed] [Google Scholar]
- Thomas G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753–766 10.1038/nrm934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A., Sakurai K., Kiyokage E., Yamazaki T., Koide S., Toida K., Ishimura K., Matsuda Y. 2003. Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix. Potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim. Biophys. Acta. 1645:95–104 [DOI] [PubMed] [Google Scholar]
- VanEngelenburg S.B., Palmer A.E. 2008. Fluorescent biosensors of protein function. Curr. Opin. Chem. Biol. 12:60–65 10.1016/j.cbpa.2008.01.020 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Langeslag M., Jalink K. 2004. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys. J. 86:2517–2529 10.1016/S0006-3495(04)74307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet I., Collignon J., Robertson E.J. 1997. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development. 124:1033–1044 [DOI] [PubMed] [Google Scholar]
- Yan Y.T., Liu J.J., Luo Y., E C., Haltiwanger R.S., Abate-Shen C., Shen M.M. 2002. Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol. Cell. Biol. 22:4439–4449 10.1128/MCB.22.13.4439-4449.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang Z., Lin J., Lu J., Liu B.F., Zeng S., Luo Q. 2007. Detection of MMP activity in living cells by a genetically encoded surface-displayed FRET sensor. Biochim. Biophys. Acta. 1773:400–407 10.1016/j.bbamcr.2006.11.002 [DOI] [PubMed] [Google Scholar]