Sds22 defines protein phosphatase 1 location and function at kinetochores and subsequent activity of aurora B in mitosis.
Abstract
We have studied Sds22, a conserved regulator of protein phosphatase 1 (PP1) activity, and determined its role in modulating the activity of aurora B kinase and kinetochore–microtubule interactions. Sds22 is required for proper progression through mitosis and localization of PP1 to mitotic kinetochores. Depletion of Sds22 increases aurora B T-loop phosphorylation and the rate of recovery from monastrol arrest. Phospho–aurora B accumulates at kinetochores in Sds22-depleted cells juxtaposed to critical kinetochore substrates. Sds22 modulates sister kinetochore distance and the interaction between Hec1 and the microtubule lattice and, thus, the activation of the spindle assembly checkpoint. These results demonstrate that Sds22 specifically defines PP1 function and localization in mitosis. Sds22 regulates PP1 targeting to the kinetochore, accumulation of phospho–aurora B, and force generation at the kinetochore–microtubule interface.
Introduction
The kinetochore mediates the attachment of microtubules to mitotic chromosomes and mediates the critical force generation and signaling systems necessary for proper mitotic chromosome segregation. This multiprotein machine consists of at least 80 conserved proteins required for microtubule attachment and dynamics, association with DNA, and the spindle assembly checkpoint (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). Several kinetochore proteins are signaling molecules required for posttranslational modification of kinetochore, chromosome and microtubule proteins that are critical for kinetochore assembly and function. A series of protein kinases including aurora B, Plk1, Bub1, BubR1, and Mps1 have been implicated in critical phosphorylation events at the mitotic kinetochore. In at least some cases, phosphorylation targets are known, and modification sites have been mapped. However, reversibility is a critical component of dynamic signaling pathways, and there is much less known about the protein phosphatases involved in kinetochore assembly and function. Mutations in protein phosphatase 1 (PP1) rescue mutations in worms and yeast aurora B, suggesting that aurora B activity is antagonized by PP1 (Sassoon et al., 1999; Hsu et al., 2000; Emanuele et al., 2008). In Schizosaccharomyces pombe, PP1 (Dis2) mutations cause defects in chromosome segregation (Kinoshita et al., 1990) and interfere with silencing of the spindle assembly checkpoint (Vanoosthuyse and Hardwick, 2009). PP1-α and PP1-γ, two of three human PP1 isoforms, are localized at mitotic kinetochores (Trinkle-Mulcahy et al., 2003, 2006), and targeting of PP1-γ to chromatin by a REPOMan is required for proper chromosome decondensation in anaphase (Vagnarelli et al., 2006). KNL1, a conserved component of the kinetochore, binds and targets PP1-γ to the kinetochore and antagonizes aurora B activity at centromeres, suggesting that multiple pathways exist for targeting PP1 isoforms to different compartments at different times (Liu et al., 2010). Inhibition of PP1 activity during interphase allows activation of aurora B and phosphorylation of chromatin-associated histone H3, suggesting that PP1 inhibits aurora B autoactivation and holds it in an inactive state during interphase (Murnion et al., 2001).
Phosphatase specificity is achieved through association of a catalytic subunit with accessory subunits that can drive localization and thus modulate activity and specificity. In budding yeast, several proteins modulate Glc7p/PP1 function (Pinsky et al., 2006). One of these, Sds22, was originally identified in fission yeast using high copy suppression of S. pombe dis2 (Ohkura and Yanagida, 1991). It is a highly conserved, essential protein that physically interacts with PP1 in yeasts and metazoans (Stone et al., 1993; MacKelvie et al., 1995; Renouf et al., 1995; Dinischiotu et al., 1997). Sds22 binds PP1 using a noncanonical PP1-binding motif, an extended helix (Ceulemans et al., 2002). Sds22 is required for proper mitosis in budding and fission yeasts, suggesting that it interacts with and regulates the function of PP1 during mitosis (Ohkura and Yanagida, 1991; Stone et al., 1993; MacKelvie et al., 1995; Peggie et al., 2002; Pinsky et al., 2006).
In this study, we have directly examined the effect of depleting Sds22 from human cells using RNAi and found effects on cell cycle progression, aurora B regulation, and the interaction between microtubules and kinetochores. Our data suggest that Sds22 plays a critical role in defining the activity of aurora B by regulating the levels of phospho–aurora B at kinetochores.
Results
Sds22 is required for proper chromosome segregation
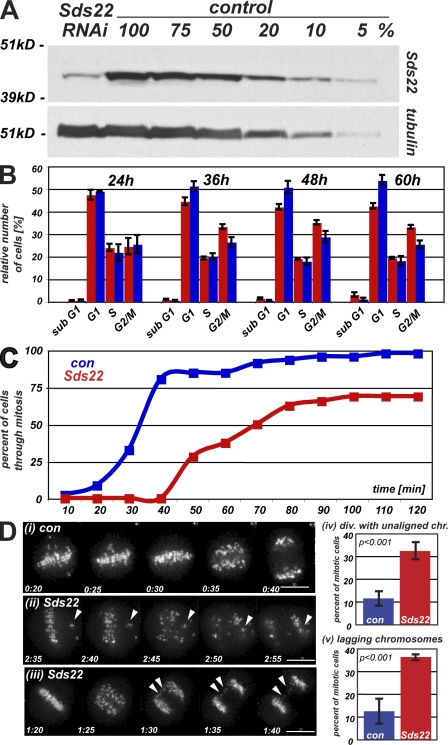
Previous work in budding and fission yeasts has suggested that Sds22 functions in concert with Glc7p or Dis2, the yeast PP1 proteins, most likely to modulate the phosphorylation of chromosomal and/or kinetochore proteins (Peggie et al., 2002; Pinsky et al., 2006). To assess the role of Sds22 in mitotic progression in human cells, we established an RNAi protocol that depleted levels of Sds22 to ≤10% of endogenous levels of proteins (Fig. 1 A). The G2/M population was specifically increased in cells depleted of Sds22 from 36–60 h after initiation of RNAi depletion (Fig. 1 B). To further define this phenotype, we monitored progression through mitosis using differential interference contrast or fluorescence microscopy of HeLa cells stably expressing GFP–CENP-A (Jaqaman et al., 2010) and measured the time from nuclear envelope breakdown (NEB) until anaphase (Videos 1–3). This analysis revealed a delay of between 20 and 50 min in Sds22-depleted cells compared with controls (Fig. 1 C). Notably, all cells showed at least a 20 min delay in mitosis. We also observed an approximately threefold increase in the number of cells entering anaphase before complete alignment of sister kinetochores in Sds22-depleted cells (P < 0.01; Fig. 1 D, ii and iv). Similarly, we observed an approximately threefold increase in the number of cells with visible lagging chromosomes in the spindle midzone during anaphase (Fig. 1 D, iii and v). In separate experiments, we examined (a) segregation of chromosomes using histone-H2B fluorescent protein fusions, (b) mitotic spindle size, bipolarity, and morphology using cells expressing mCherry-tubulin, and (c) spindle pole separation and duplication using metaphase cells expressing centrin-GFP and stained with anti-tubulin and DAPI. These analyses revealed a reproducible increase in spindle pole to pole distance (6.4 ± 2.5%; P < 0.049 by Kolmogorov-Smirnov) in Sds22-depleted cells. These results suggest that Sds22 is required for proper mitotic progression and functions to mediate the proper alignment of chromosomes on the metaphase plate and efficient segregation of chromosomes in anaphase. The phenotypes we observed were consistently weaker than those previously reported for depletion or inhibition of aurora B and the other members of the chromosome passenger complex (Adams et al., 2001; Giet and Glover, 2001; Ditchfield et al., 2003; Hauf et al., 2003); we never observed more than four misaligned pairs in 293 cells followed by time-lapse microscopy.
Figure 1.
Depletion of Sds22 causes defects in mitotic progression. (A) Depletion of Sds22 by siRNAi. 70 µg extracts of logarithmically growing HeLa cells 48 h after transfection with Sds22-specific and control siRNAi were probed with Sds22-specific polyclonal antibody by Western blotting and reprobed with anti-tubulin as a loading control. (B) Cell cycle profile of HeLa cells after Sds22 RNAi. Subpopulation of HeLa in specific cell cycle stages as determined by DNA content analysis (FACS of propidium iodide–stained cells). Cells were transfected with control (blue) or Sds22-specific (red) siRNAi and analyzed at the time points indicated (hours). Results are from four separate experiments. (C) Timing of mitosis in Sds22-depleted and control cells. HeLa (Kyoto) cells stably expressing EGFP–CENP-A were imaged at 5- or 10-min intervals starting 48 h after transfection with Sds22-specific and control RNAi. Time elapsed from NEB to the onset of anaphase was plotted against cumulative percentage of cells having completed mitosis. Data were generated from three independent experiments. Data from two experiments recorded at 5-min intervals were binned to 10 min and combined with one experiment recorded at 10-min intervals. Total number of cells: control, 314; Sds22 RNAi, 293. (D) Mitotic defects in Sds22-depleted cells. Still images of C are shown with time after NEB indicated (hours:minutes). (i) Control cell. (ii and iii) Sds22-depleted cell. Defects are indicated by arrowheads. (iv) Mean percentage of mitotic cells dividing with unaligned chromosomes in C. (v) Mean percentage of mitotic cells dividing with lagging kinetochores in the midzone in C. Error bars indicate SEM. Bars, 10 µm.
Sds22 functions at kinetochores
Our functional analysis suggests that Sds22 might function at kinetochores, possibly through kinetochore-associated PP1 (Trinkle-Mulcahy et al., 2003). Sds22 is known to interact with PP1 in vivo and in vitro (Ceulemans et al., 2002; Peggie et al., 2002). Human cells express three closely related isoforms of PP1 (PP1-α, PP1-γ, and PP1-β or PP1-δ), and in vitro assays suggest that there is no specificity between Sds22 and any specific PP1 isoform (Ceulemans et al., 2002). Therefore, we generated N- and C-terminal GFP-tagged forms of Sds22 and five cell lines stably expressing either GFP-Sds22 (C103) or Sds22-GFP (D103). Both constructs and all cell lines behaved identically in our experiments.
We first confirmed that GFP-Sds22 binds PP1 in HeLa cell lysates (Fig. S1 A) and analyzed immunoprecipitates from a GFP-Sds22 cell line (D103) by mass spectrometry. GFP-Sds22 immunoprecipitates contained isoform-specific peptides from all three forms of PP1, suggesting that Sds22 binds all forms of PP1 in human cells (Fig. S1 B).
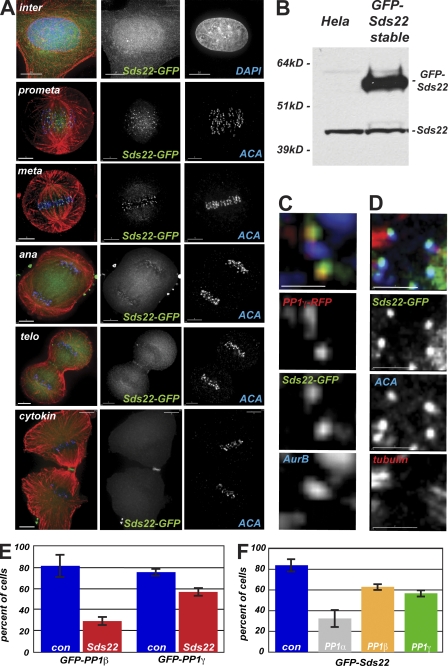
We next compared the localization of Sds22-GFP to other mitotic markers by immunofluorescence either in transiently transfected cells or in five separately isolated stable cell lines. In all cases, we observed a diffuse localization in interphase cells with some concentration in the nucleus (Fig. 2 A). In mitotic cells, we observed a specific concentration of Sds22-GFP near kinetochores starting in prometaphase, continuing through metaphase and anaphase, and disappearing after the completion of cytokinesis (Fig. 2 A). Sds22-GFP was overexpressed relative to endogenous levels in our stable cell lines (Fig. 2 B), but we observed no significant defects in cell cycle progression or mitotic chromosome segregation in these cells. Sds22-GFP colocalized with the known kinetochore components PP1-γ and was easily resolved from aurora B, a component of the chromosome passenger complex that concentrates at the inner centromere at prometaphase and metaphase (Fig. 2, C and D). Turnover of Sds22-GFP at mitotic kinetochores measured by FRAP (see Materials and methods) was rapid (t1/2 = 0.39 ± 0.16 s; compared with turnover in cytoplasm, t1/2 < 0.10 s), suggesting that Sds22 is not a core component of the kinetochore. We also measured turnover of PP1-γ–GFP at mitotic kinetochores using the same technique and found a similar value (t1/2 = 0.88 ± 0.42 s). As these measurements are affected by the very high turnover in the cytoplasm, they are consistent with the idea that Sds22 and PP1 form a complex at the kinetochore. Our antibody generated against recombinantly expressed full-length Sds22 (Fig. 2 B) failed to detect endogenous Sds22 by immunofluorescence but reacted with overexpressed Sds22-GFP at kinetochores (Fig. S1 C).
Figure 2.
Sds22 functions at kinetochores. (A) Intracellular localization of Sds22-GFP. Immunofluorescence of HeLa cells fixed 48 h after transient transfection with GFP-Sds22 plasmid. Overlay shows DAPI or anti–human centromere antibody (ACA; blue), Sds22-GFP (green), and microtubules (red). (B) Stable expression of Sds22-GFP in HeLa cells. Western blot analysis of extracts from untransfected HeLa cells and cell clone D103 stably expressing GFP-Sds22 probed with polyclonal Sds22-specific antibody. (C) Relative localization of Sds22 and PP1 at the kinetochore. Immunofluorescence of HeLa cells transiently transfected with mRFP–PP1-γ and GFP-Sds22 fixed 48 h after transient transfection. Overlay shows mRFP1–PP1-γ (red), Sds22-GFP (green), and aurora B (blue). (D) Relative localization of Sds22 and PP1 at the kinetochore. Immunofluorescence of HeLa cells transiently transfected with GFP-Sds22 plasmid fixed 48 h after transient transfection. Overlay shows Sds22-GFP (green), ACA (blue), and anti-tubulin (red). (E) Dependency of PP1 kinetochore localization on Sds22. Kinetochore localization GFP–PP1-β and GFP–PP1-γ were scored in mitotic spreads of HeLa cells 48 h after transfection with Sds22-specific and control siRNA. Data are from three separate experiments testing localization of either GFP–PP1-β or GFP–PP1-γ. Total number of cells for GFP–PP1-β: control, 47; Sds22 RNAi, 61. Total number of cells for GFP–PP1-γ: control, 162; Sds22 RNAi, 134. P < 0.001. P-values were calculated by Fisher’s exact test. (F) Dependency of Sds22-GFP kinetochore localization on PP1 isoforms. Relative number of cells showing EGFP-Sds22 at kinetochores in mitotic spreads of a stable HeLa cell line (clone D103) 48 h after transfection with control or PP1 isoform–specific RNAi. Data are from five independent experiments. Total number of cells and p-values: control, 93; PP1-α, 40 (P < 0.001); PP1-β, 84 (P = 0.024); PP1-γ, 81 (P = 0.001). Error bars indicate SEM. Bars: (A, top row) 10 µm; (A, rows 2–6) 5 µm; (C and D) 2 µm.
To determine whether Sds22 was required for PP1 localization at kinetochores, we examined the localization of PP1-β and -γ in mitotic chromosome spreads. In both cases, PP1 isoform localization was decreased in cells depleted of Sds22 (Fig. 2 E), indicating that PP1 isoforms require Sds22 for proper kinetochore localization. This dependence is reciprocal, as depletion of any of the individual PP1 isoforms by isoform-specific siRNA reduced the number of mitotic kinetochores in chromosome spreads with concentrated GFP-Sds22 (Fig. 2 F). Together, these data suggest that Sds22 and PP1 isoforms associate at the kinetochore and are mutually required for localization at the mitotic kinetochore.
Sds22 modulates aurora B function at centromeres and kinetochores
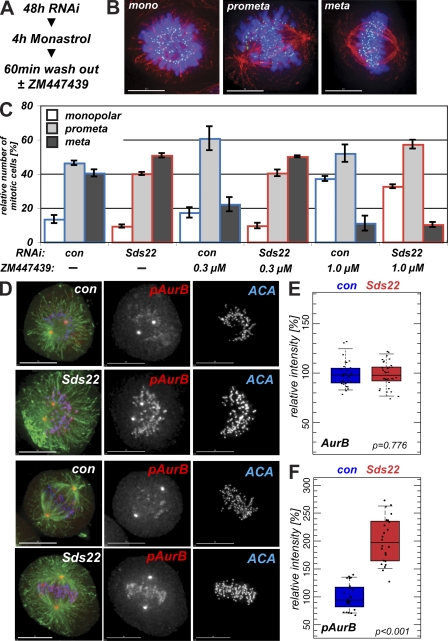
PP1 is thought to antagonize the activation of aurora B by modulating the activation of kinase activity by phosphorylation (Murnion et al., 2001; Rosasco-Nitcher et al., 2008). To determine whether loss of PP1 at kinetochores through Sds22 depletion might increase aurora B activity, we arrested cells in monastrol for 4 h, switched the cells into medium without monastrol, and allowed them to recover for 60 min. We released cells from monastrol arrest into either control media or media containing either 0.3 or 1 µM ZM447439 (ZM), a small molecule inhibitor of aurora kinases (Ditchfield et al., 2003), to assess the dependence of recovery on aurora B kinase activity (Fig. 3 A). After 60 min of recovery, cells were fixed and stained to reveal DNA, microtubules, and kinetochores and scored as being monopolar or in prometaphase or metaphase (Fig. 3 B). Monastrol-treated cells showed a high frequency of monopolar cells and syntelic attachments (Lampson et al., 2004). After release into normal medium, control RNAi cells progressed toward metaphase, whereas control RNAi cells treated with 0.3 ZM progressed much more slowly, and recovery was largely inhibited in cells released into 1 µM ZM (Fig. 3 C). In cells depleted of Sds22 and released into control medium, we observed a higher frequency of metaphase cells than in control cells, which is consistent with an increased level of aurora B activity. Release of Sds22-depleted cells into 0.3 µM ZM (where aurora B is partially inhibited) increased the number of cells in metaphase and generated a distribution of phenotypes very similar to that seen in the absence of ZM. To assess whether alternative recovery pathways are activated in Sds22-depleted cells, we released cells into 1 µM ZM. These cells recovered from monastrol very slowly, and no acceleration of recovery in Sds22-depleted cells was observed. Thus, Sds22 depletion accelerates recovery from monastrol, and this recovery is completely dependent on activity of aurora B.
Figure 3.
Sds22 depletion affects aurora B activity. (A) Experimental protocol. (B) Monopolar, prometaphase, and metaphase data from monastrol recovery. Examples of cells at the three points of recovery counted in C. Overlay shows DAPI (blue), ACA (green), and microtubules (red). (C) Effect of Sds22 depletion on the recovery from monastrol arrest. 48 h after transfection of HeLa cells with control or Sds22-specific RNAi, duplexes were treated for 4 h with monastrol and released into fresh medium or medium containing 0.3 or 1 µM ZM. After 1 h, cells were fixed, stained (A), and mitotic cells were scored for their progression through mitosis. Total number of cells: 0 µM control, 347; 0 µM Sds22, 1,056; 0.3 µM control, 1,336; 0.3 µM Sds22, 1,230; 1.0 µM control, 1,261; 1.0 µM Sds22, 1,360. (D) Aurora B T232 phosphorylation depends on Sds22. HeLa cells were fixed and immunostained with anti–phospho-T232 antibody 48 h after transfection with control (con) or Sds22-specific duplexes. Overlay shows tubulin (green), anti–phospho-T232 aurora B (red), and ACA (blue). The spindle pole staining is not sensitive to inhibition with 1 µM ZM and is likely spurious and the result of cross-reaction with phospho–aurora A (Fuller et al., 2008). (E and F) Effect of Sds22 depletion on total aurora B and T232 aurora B phosphorylation in metaphase. Quantification of metaphase cells fixed and stained with aurora B (E) and anti-phospho–aurora B T232 (F) antibodies in fixed HeLa cells 48 h after transfection with control or Sds22-specific duplexes (see Materials and methods). Intensities were normalized relative to the general kinetochore marker ACA. Box plots show mean (middle line), top and bottom quartiles of the data as top and bottom of the box, and whiskers as the extent of 90% of the data. P-values were calculated using a Kolmogorov-Smirnov test. (E) Total number of cells: control RNAi, 30; Sds22 RNAi, 34. (F) Total number of cells: control RNAi, 24; Sds22 RNAi, 24. Error bars show SEM from three independent experiments. Bars, 10 µm.
Activation of aurora B kinase occurs through the phosphorylation of T232 in the kinase activation loop, a modification that can be detected with anti–phospho-T232 aurora B antibody (Yasui et al., 2004; Sessa et al., 2005; Rosasco-Nitcher et al., 2008; Wang et al., 2008). We examined the levels of T232 phosphorylation after depletion of Sds22 and observed increased staining with anti–phospho-T232 aurora B, whereas levels of total aurora B were unchanged (Fig. 3, D–F). We repeated our monastrol recovery and ZM treatment protocol and again observed increased anti–phospho-T232 staining in cells released into control or 0.3 µM ZM medium (Fig. S2). All detectable anti–phospho-T232 staining was lost in cells released into 1 µM ZM, which is consistent with a complete inhibition of aurora B function (Fig. S2). We conclude that Sds22 depletion results in an increase in aurora B kinase activity and the rate at which downstream effects of aurora B activity occur.
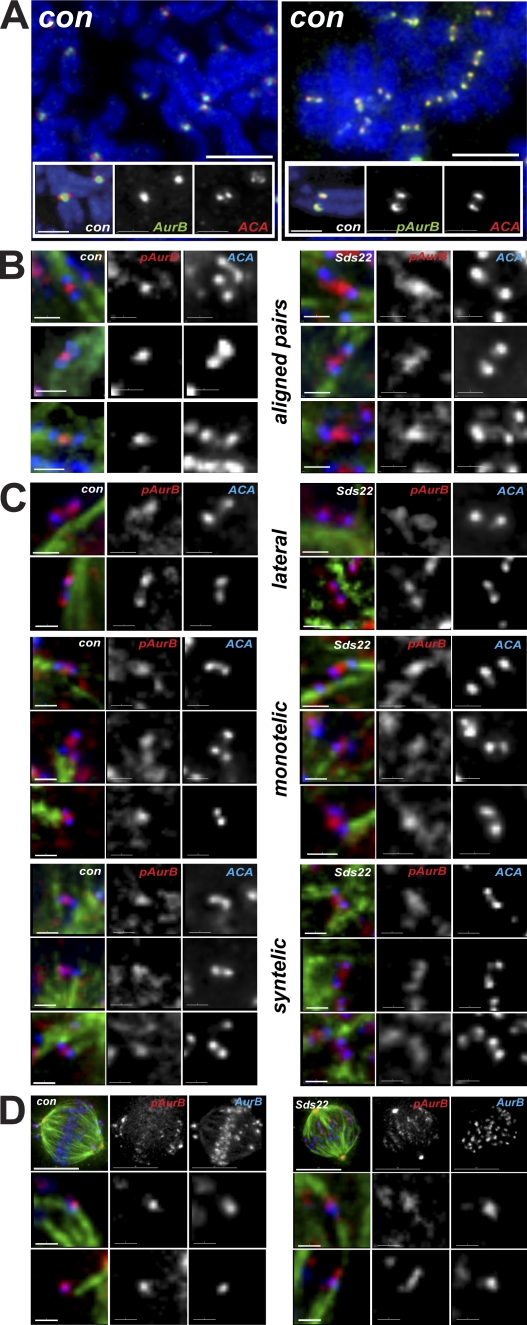
Dynamic localization of phospho–aurora B at centromeres and kinetochores
To determine how increased amounts of phospho-T232 aurora B might drive recovery from monastrol, we arrested cells with nocodazole and examined the localization of aurora B in chromosome spreads using immunofluorescence. Aurora B was localized at centromeres as previously described, but phospho-T232 aurora B was localized to distal to centromeres, near kinetochores (Fig. 4 A). To confirm this result, we examined the localization of phospho-T232 aurora B during monastrol recovery in whole cells and its dependence on Sds22. In control cells, phospho-T232 aurora B concentrated between bioriented kinetochores that were well separated and attached to microtubule ends (Fig. 4 B). In Sds22-depleted cells, phospho-T232 aurora B localized along the extended centromere, but still appeared, at the limit of the resolution of the imaging, to be distinct from the inner kinetochore. Mono- and syntelically attached kinetochore pairs in control cells also had phospho-T232 aurora B concentrated at the centromeres, and again, cells depleted of Sds22 consistently showed a broader localization, with some examples of phospho–aurora B localizing at kinetochores (Fig. 4 C). In laterally attached pairs, we reproducibly observed phospho-T232 aurora B proximal to kinetochores in all cells, but this effect was most pronounced in Sds22-depleted cells. Finally, direct comparison of bulk aurora B and phospho-T232 aurora B revealed similar trends: concentrated colocalization of bulk and phospho-T232 aurora B at centromeres of control cells and extension of phospho-T232 aurora B along the centromere in Sds22-depleted cells (Fig. 4 D).
Figure 4.
Localization of phospho–T232 aurora B is determined by Sds22. (A) Localization of aurora B and phospho-T232 aurora B in chromosome spreads after arrest with nocodazole. Phospho-T232 aurora B is concentrated at kinetochores, whereas bulk aurora B remains concentrated at centromeres. Overlay shows DAPI (blue), aurora B or anti–phospho-T232 aurora B (green), and ACA (red). (B) Localization of phospho-T232 aurora B in bioriented chromosomes in fixed intact cells. Sds22 depletion causes phospho-T232 aurora B to spread toward kinetochores. Overlay shows ACA (blue), tubulin (green), and anti–phospho-T232 aurora B (red). (C) Localization of phospho-T232 aurora B in laterally and monooriented chromosomes in fixed intact cells. Phospho-T232 aurora B appears on kinetochores in control and Sds22-depleted cells. Overlay shows ACA (blue), tubulin (green), and anti–phospho-T232 aurora B (red). (D) Colocalization of phospho-T232 aurora B and total aurora B in fixed intact cells. Sds22 depletion causes phospho-T232 aurora B to spread toward kinetochores. Overlay shows phospho-T232 aurora B (red), aurora B (blue), and tubulin (green). Bars: (A, top) 5 µm; (A, insets) 2 µm; (B, C, and D [bottom]) 1 µm; (D, top) 10 µm.
We next determined whether changes in phospho–aurora B localization were caused by significant changes in the localization of bulk aurora B. Aurora B localization in unaligned pairs was, as expected, concentrated in the inner centromere, and we never observed a substantial change in the localization of total aurora B in Sds22-depleted cells (Fig. S3 A). We next compared the localization of aurora B and the inner kinetochore marker anti-centromere antibody (ACA) by 3D structured illumination (3DSIM; Gustafsson, 2000; Schermelleh et al., 2008) using an OMX microscope. In control and Sds22-depleted cells, aurora B was still largely localized between sister kinetochores, although some kinetochore pairs in Sds22-depleted cells had concentrations of aurora B kinase near the kinetochore (Fig. S3 B). We examined the same cell by 3D deconvolution on a conventional microscope, conventional imaging on OMX, and 3DSIM on OMX. This analysis showed that the subtle differences in aurora B localization were visible by conventional microscopy but made much clearer in 3DSIM, suggesting that the detailed localization we observed were not artifacts of the imaging or processing methods. We conclude that Sds22 depletion does not significantly change the distribution of aurora B on centromeres but increases the phosphorylation of a small portion of aurora B proximal to kinetochores.
Sds22 regulates phosphorylation of aurora B substrates
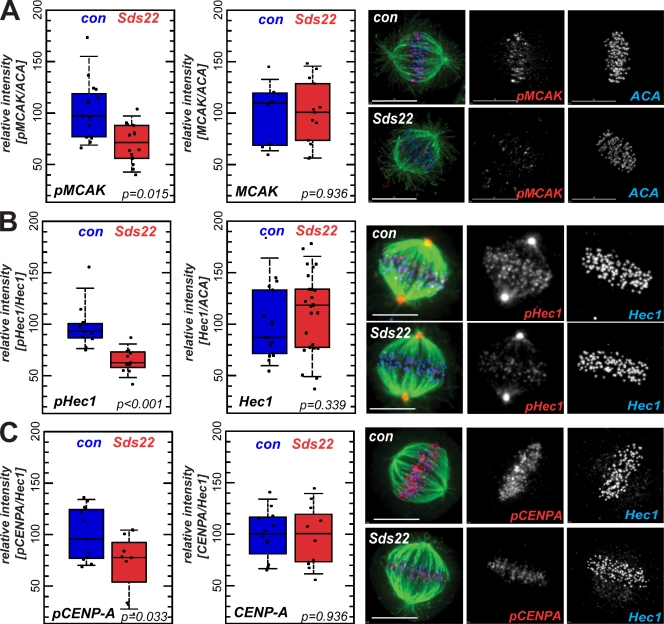
We next tested a series of antibodies raised to known aurora B phosphoepitopes on MCAK, Hec1, and CENP-A as reporters of aurora B activity toward kinetochore substrates. Surprisingly, in all cases, we observed a substantial decrease in phosphorylation of aurora B substrates in Sds22-depleted cells but no change in the total amount of each substrate (Fig. 5, A–C). We individually depleted PP1 isoforms and observed a similar decrease in CENP-A phosphorylation as in cells depleted of Sds22 (Fig. S4). We also confirmed that loss of MCAK and Hec1 phosphorylation occurred throughout M phase, during and after chromosome biorientation and alignment on the metaphase plate, and that Sds22 depletion caused loss of phosphorylation on a second site on Hec1 (Fig. S5, A–C). These data suggest that Sds22/PP1 modulates the phosphorylation of aurora B substrates on kinetochores. Paradoxically, depletion of kinetochore-associated Sds22/PP1 increases aurora B activity at kinetochores (Figs. 3 and 4) but reduces the level of phosphorylation of MCAK, Hec1, and CENP-A.
Figure 5.
Effect of Sds22 depletion on substrate phosphorylation and centromere binding of aurora B. (A–C) Representative images and quantification of metaphase cells fixed and stained with anti–phospho-Ser92 MCAK and total MCAK (A), anti–phospho-Hec1 and total Hec1 (B), and anti-phospho–CENP-A and total CENP-A (C) antibodies in fixed HeLa cells 48 h after transfection with control (con) or Sds22-specific duplexes (see Materials and methods). Intensities were normalized relative to general kinetochore markers (Hec1 and ACA as indicated). Box plots and statistical analysis are as in Fig. 3 (E and F). Bars, 10 µm.
Sds22 modulates generation of force at the kinetochore
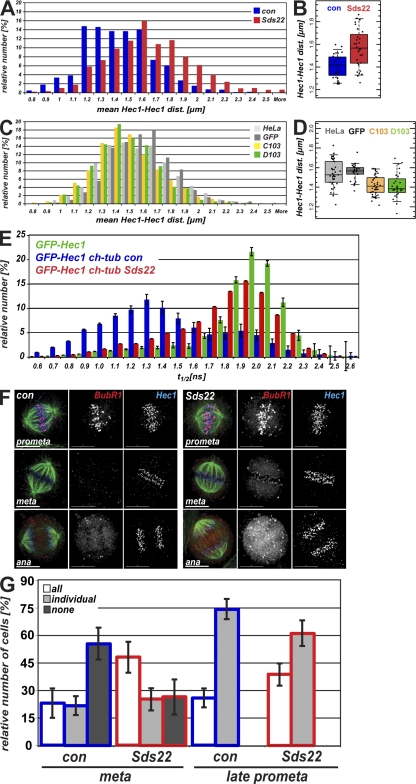
Previous work has shown that dephosphorylated MCAK concentrates at kinetochores, where it may contribute to force generation by driving microtubule depolymerization (Andrews et al., 2004). In addition, dephosphorylated Hec1, a microtubule-binding component of kinetochores, has an increased affinity for the microtubule lattice and increases the distance between sister kinetochores (Cheeseman et al., 2006; DeLuca et al., 2006). Based on our results in Fig. 5 (A and B), we predicted that in Sds22-depleted cells, decreased phosphorylation of MCAK and Hec1 would increase force generation, kinetochore affinity, and distance between sister kinetochores. Consistent with this, Sds22-depleted metaphase cells showed a notable increase in distances between sister kinetochores (Fig. 6 A). Most importantly, using a frequency histogram, we observed a distribution of interkinetochore distances between 1.2 and 1.6 µm, which likely reflect the sampling of a dynamic, oscillating system at fixed time points. Depletion of Sds22 by RNAi caused the sister kinetochore distance to shift to a single population centered around 1.6 µm. An increase in sister kinetochore distance after Sds22 depletion was also observed plotting the mean distances in each cell (Fig. 6 B) and in living CENP-A–GFP-expressing HeLa cells using automatic sister kinetochore identification and tracking (Fig. S5 D; Jaqaman et al., 2010). We next measured the distance between sister kinetochores in two stable cell lines that express GFP-Sds22 at levels five times endogenous levels (Fig. 2 B) and compared these distances with those measured in normal HeLa cells or a HeLa cell line that stably expresses GFP. Both the normal and GFP-expressing HeLa lines contained two populations of sister kinetochore pairs distances centered around 1.4 and 1.6 µm, and a Kolmogorov-Smirnov test revealed little difference between the two distributions (Fig. 6 C). In contrast, overexpression of Sds22-GFP (C103) or GFP-Sds22 (D103) shifted the distribution of sister kinetochores to smaller distances, the opposite effect of that observed in cells depleted of Sds22 (Fig. 6, A and C). Control and Sds22-depleted cells treated with nocodazole had interkinetochore distances that were not statistically distinguishable (control RNAi, 0.786 ± 0.006 µm [n = 66 cells]; Sds22 RNAi, 0.811 ± 0.006 µm [n = 64 cells]; P < 0.05). Together with the absence of effect on Hec1 and CENP-A targeting (Fig. 5, B and C), these data suggest that Sds22 deletion does not substantially change the architecture of the kinetochore, although more subtle changes might occur, possibly mediated by changes in phosphorylation of CENP-A. Nonetheless, our data demonstrate that Sds22 modulates the distance between sister kinetochores, most likely by regulating the phosphorylation of important regulators of microtubule binding and dynamics like MCAK and Hec1 by aurora B.
Figure 6.
Effects of Sds22 on sister kinetochore dis-tances and interactions between microtubules and kinetochores. (A–D) Histograms showing distribution of interkinetochore distances (A and C) and interkinetochore distances per cell (B and D). (A and B) Sister kinetochore distances after Sds22 depletion. Hec1–Hec1 distances of sister pairs were measured in HeLa cells fixed 48 h after transfection with control siRNA or Sds22-specific siRNA (C and D) Hec1–Hec1 distances of sister pairs in untransfected HeLa cells or stable cell lines expressing GFP alone or GFP-Sds22 (C103 and D103). Data from two independent experiments are shown. Total number of cells: control siRNA, 28; Sds22 RNAi, 33; normal HeLa, 37; HeLa GFP, 30; C103, 42; D103, 28. (B) Kolmogorov-Smirnov test of significance: control siRNA and Sds22-depleted cells, P < 6 × 10−5. (D) Kolmogorov-Smirnov test of significance: normal HeLa and HeLa GFP, P < 0.05; HeLa GFP and C103, P < 6 × 10−6; HeLa GFP and D103, P < 1 × 10−5; C103 and D103, P < 0.9. (E) Effect of Sds22 on the microtubule–kinetochore attachment. Fluorescence lifetime measurement of FRET between EGFP-Hec1 and mCherry-tubulin is shown. Fluorescence lifetime of EGFP was determined in HeLa cells 48 h after transfection with either EGFP-Hec1 (green bars) or cotransfection with EGFP-Hec1 and mCherry-tubulin together with either control (blue bars) or Sds22-specific (red bars) RNAi duplexes. (F) Recruitment of BubR1 in Sds22-depleted cells. HeLa cells were fixed and immunostained 48 h after transfection with control or Sds22-specific RNAi duplexes. Bars, 10 µm. (G) Effect of Sds22 depletion in localization of BubR1. HeLa cells in F that showed a defined plate with aligned chromosomes were subdivided into cells where all chromosomes had clearly aligned to the metaphase plate as opposed to cells with one or more unaligned chromosomes. Both categories were scored on whether all, individual, or no kinetochores at all stained positive for BubR1. Data from four independent experiments are shown. Total number of cells: control RNAi, 153; Sds22 RNAi, 99. P-values between control and Sds22 RNAi were calculated by χ2 test: aligned kinetochores, P = 0.0004; unaligned kinetochores, P = 0.1172. Error bars indicate mean ± SEM.
To further dissect the interaction of kinetochores and microtubules, we next assessed the stability of microtubules in Sds22-depleted cells after cooling cells on ice and observed no detectable effect on microtubule attachment (unpublished data), indicating that Sds22 does not significantly destabilize microtubule–kinetochore attachments. This is consistent with our results from live cell imaging (Fig. 1, C and D) showing that Sds22 depletion causes a mild arrest in mitosis where most chromosomes successfully migrate to the metaphase plate. However, a cold-stable stability assay might not detect subtle changes in kinetochore–microtubule interaction. To determine whether Sds22 depletion directly affected kinetochore–microtubule interactions, we coexpressed GFP-Hec1 and mCherry-tubulin fusions in living HeLa cells and measured Förster resonance energy transfer (FRET) between GFP and mCherry using time-correlated single photon counting fluorescence lifetime microscopy Wouters et al., 2001; Duncan et al., 2004). This assay measures the interaction between Hec1, a critical microtubule interaction component, and the surface of the microtubule. Hec1 is known to be a microtubule-binding protein whose affinity for the microtubule lattice is modulated by phosphorylation, most likely by aurora B (Cheeseman et al., 2006; DeLuca et al., 2006). The fluorescence lifetime (t1/2) of cells expressing GFP-Hec1 alone was distributed around 2,000 ps, similar to the fluorescence lifetime of native GFP (Fig. 6 E; Pepperkok et al., 1999). Coexpression of mCherry-tubulin with GFP-Hec1 produced a significant shift in the distribution of GFP t1/2, centered at ∼1,300 ps, with a minor population remaining at ∼1,900 ps. The substantially shorter t1/2 is indicative of FRET between GFP and mCherry. Depletion of Sds22 in the same cells restored the longer lifetime, suggesting a loss of FRET and, thus, a change in the relative arrangement of Hec1 and microtubules in Sds22-depleted cells.
We are not able to distinguish a change in fluorophore orientation from a simple change in relative distance between Hec1 and the microtubule lattice in our FRET assay; however, a change in kinetochore–microtubule interactions might explain the mitotic delay observed in Fig. 1 C. We note that Sds22 depletion causes 33% of cells to divide with misaligned chromosomes but that all cells suffer a mitotic delay (Fig. 1 D). Thus, although most Sds22-depleted cells can align their chromosomes, they still delay in metaphase. Our FRET analysis is consistent with this and suggests that the majority of kinetochores in living metaphase cells experience a change in proximity of Hec1 and tubulin after Sds22 depletion (Fig. 6 E). Together, these data indicate that Sds22 depletion perturbs the interactions of most kinetochores with microtubules.
Sds22 depletion engages the spindle checkpoint
Perturbation of kinetochore–microtubule interactions can engage the spindle checkpoint. Therefore, we measured the levels of the mitotic checkpoint protein BubR1 on prometaphase and metaphase kinetochores in control and Sds22-depleted cells. Prometaphase cells had many kinetochores with high levels of BubR1, and there was no detectable difference in BubR1 levels between control and Sds22-depleted cells (Fig. 6 F). However, in metaphase Sds22-depleted cells, we observed low levels of BubR1 localizing at all kinetochores, suggesting that interactions between kinetochores and microtubules were partially perturbed in these cells. We counted the frequency of this phenotype and cells with bright BubR1 staining at a subset of kinetochores, as is typical of cells with a few unaligned chromosomes (Fig. 6 G). In late prometaphase, we observed no significant difference in the distribution of these phenotypes. However, in cells where all chromosomes were aligned at the metaphase plate, we observed a significant increase in cells where all kinetochores had low, detectable BubR1 staining. As these aligned kinetochores show changes in microtubule interaction (Fig. 6 E) and checkpoint engagement (Fig. 6 G), our data suggest that Sds22 depletion systematically perturbs the interaction of microtubules and kinetochores, most likely by continuous overactivation of aurora B through prometaphase and metaphase. There is substantial evidence suggesting that aurora B directly regulates the components of the spindle assembly checkpoint (Morrow et al., 2005; King et al., 2007) and that PP1 is required to reverse checkpoint response (Vanoosthuyse and Hardwick, 2009). Although we cannot rule out contributions from these signaling pathways, our data show that Sds22 depletion likely engages the checkpoint through perturbation of the microtubule–kinetochore interface.
Discussion
To understand the localized regulation of aurora B at the mitotic centromere, we have explored the function of Sds22, a known modulator of PP1 activity during mitosis (Peggie et al., 2002). We find that Sds22 modulates aurora B function at centromeres and kinetochores and thus defines timely progression through mitosis. Sds22 binds PP1 and is required for correct localization of PP1 to the kinetochore. Sds22/PP1 regulates the level of aurora B activity and rate of correction of erroneous kinetochore–microtubule attachments, likely by modulating the amount of activated phospho-T232 aurora B localized at the kinetochore. Phospho-T232 is elevated in prometaphase and metaphase in Sds22-depleted cells, showing that Sds22 plays a critical role in balancing aurora B activity in unaligned and aligned sister kinetochores. Sds22 depletion causes a loss of phosphorylation on Hec1 and MCAK and an increase in interkinetochore distance in aligned sister kinetochores. We conclude that Sds22, through its binding of PP1 and regulation of aurora B phosphorylation, regulates the interaction of kinetochores and microtubules and thus the engagement of the spindle assembly checkpoint and progression through mitosis.
As loss of Sds22/PP1 from the kinetochore results in significant increase of phospho–aurora B, our results suggest that Sds22/PP1 antagonizes the autophosphorylation of aurora B at centromeres and kinetochores, most likely by dephosphorylating aurora B on T232. Sds22/PP1 might gain access to centromeric phospho–aurora B through its high turnover at the kinetochore. In the absence of Sds22/PP1, the amount of phospho–aurora B increases, leading to increased aurora B activity at centromeres and kinetochores. Functionally, we observe the consequences of aurora B activation as increased recovery from monastrol and perturbation of the kinetochore–microtubule interface in aligned sister kinetochore pairs.
Aurora B phosphorylates a large number of centromere and kinetochore proteins, making it difficult to determine whether changes in interkinetochore distance are the result of direct effects on Hec1 and MCAK or the result of other changes in the centromere or kinetochores. However, we observe no change in kinetochore targeting of CENP-A or Hec1 (Fig. 5) and no significant change in interkinetochore distances in nocodazole, suggesting that the fundamental components of the centromere and kinetochore are not substantially altered in Sds22-depleted cells. Although more detailed studies will be needed, the simplest current model is that Sds22 depletion changes the phosphorylation state of Hec1 and MCAK and thus alters force generation across sister kinetochore pairs.
Spatiotemporal regulation of aurora B
Previous results have highlighted changes in aurora B levels throughout the cell cycle (Kimura et al., 1998). In our study, we have demonstrated a much finer level of control of aurora B in which the specific phosphorylation and activity of aurora B are determined by both the attachment state of individual kinetochore pairs and the localization of Sds22/PP1 to the kinetochore. Before biorientation has been established, we detect a small population of aurora B at the kinetochore that is active through phosphorylation on the kinase T-loop. This population is reduced or not detected in aligned, bioriented kinetochore pairs (Fig. 4). This dynamic regulation of aurora B suggests that the activities of aurora B and PP1 are finely balanced across kinetochore pairs and may respond dynamically to changes in distance between sister kinetochores.
Recently, a FRET-based aurora B probe has been used to demonstrate a gradient of aurora B activity at the centromere and kinetochore and to suggest that the separation between aurora B at the centromere and kinetochore-associated substrates defines the phosphorylation state of aurora B targets (Liu et al., 2009). Our results are consistent with this idea and suggest that Sds22/PP1 may contribute to defining the gradient of aurora B activity. However, we have demonstrated that Sds22/PP1 also determines the appearance of an additional pool of activated aurora B at the kinetochore, where it is presumably near substrates at the kinetochore and microtubule end. Thus, especially before biorientation, activated aurora B is proximal to its substrates and well positioned to modulate microtubule–kinetochore interactions.
Force generation and error correction
The phenotypes we have measured are the result of Sds22-dependent changes in aurora B kinase activity and, thus, reflect changes in the level of aurora B phosphorylation of many centromere and kinetochore-associated substrates (Cheeseman et al., 2002). Notably, Sds22 depletion causes a loss of phosphorylation of Hec1 and MCAK and, as predicted (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Cheeseman et al., 2006; DeLuca et al., 2006), increased sister kinetochore distance (Figs. 5 and 6). However, Sds22 depletion also increases aurora B phosphoactivation, accelerates recovery from monastrol, and modulates interactions between microtubules and kinetochores (Figs. 3 and 6). Our results are certainly consistent with evidence that MCAK and Hec1 are critical downstream substrates for aurora B. However, we suggest that MCAK and Hec1 have critical roles in force generation at the kinetochore–microtubule interface but that other kinetochore components are involved with correction of improper microtubule–kinetochore attachments during biorientation. Our data further suggest that separate pathways involving aurora B and Sds22/PP1 mediate force generation and error correction at the kinetochore. Modulation of aurora B activity through Sds22/PP1 can be used to identify and characterize these other components.
Function of PP1 at kinetochores
PP1 is a well-established regulator of aurora B activity either through competitive dephosphorylation of aurora B substrates or direct regulation of aurora B activity itself (Murnion et al., 2001; Yasui et al., 2004; Rosasco-Nitcher et al., 2008). In human cells, as in yeast, Sds22 appears to have a fairly specific effect on PP1 function in mitosis, as we did not observe pleiotropic effects on cell growth or viability after Sds22 depletion (Fig. 1). There is a reciprocal dependence for the localization of PP1 and Sds22 to kinetochores, and this localization is critical to maintain the proper level of aurora B activity and the correct concentration of phospho–aurora B at the kinetochore. We have no direct data suggesting that Sds22-bound PP1 dephosphorylates aurora B substrates. In fact, loss of Sds22 actually decreases phosphorylation of all kinetochore-associated aurora B substrates we have tested. We do not believe that this occurs through delocalization of kinetochore-bound PP1, as this represents <5% of the total PP1 in the cell (Trinkle-Mulcahy et al., 2003). Other kinetochore-associated phosphatases, e.g., PP2A, may fulfill this role. Moreover, many other PP1-binding proteins are known, and one or more of these might mediate the dephosphorylation of kinetochore proteins by PP1 (Pinsky et al., 2006). KNL1, a key mediator of kinetochore function, binds and localizes PP1-γ to kinetochores and also antagonizes aurora B activity detected with an aurora B–specific FRET sensor, although its role in modulating endogenous aurora B substrates is not known (Liu et al., 2010). Given the critical requirement for KNL1 in kinetochore microtubule attachment (Desai et al., 2003; Cheeseman et al., 2008), this interaction may play a critical role in error correction. The interaction between KNL1 and PP1 may explain why despite near-complete depletion of Sds22, we only ever observed partial depletion of PP1 from kinetochores (Fig. 2).
The aurora B/Sds22/PP1 regulatory circuit
Both aurora B and PP1-γ turn over rapidly at mitotic kinetochores (Murata-Hori and Wang, 2002; Trinkle-Mulcahy et al., 2003). The dynamic association of these critical regulators with their respective binding sites suggests that the turnover of these enzymes may be a component of the circuitry that defines the final phosphorylation states of their target substrates. The level of phosphorylation of any aurora B substrate in the kinetochore would be set by the distance from the centromere and kinetochore, the pools of aurora B located at the inner centromere and kinetochore and the PP1 located at that kinetochore, and the binding affinities of each enzyme and, thus, the gradient of kinase and phosphatase activities. Our data cannot determine whether free diffusion might set up this gradient of enzymatic activity or whether the chromatin that links the kinetochore and centromere may provide a medium that governs the movement of these competing enzymes. A critical avenue for the future is the determination of the physical and enzymatic properties of all of the components of this regulatory circuit.
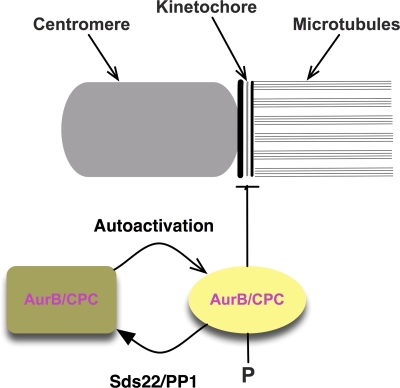
Fig. 7 summarizes our data and suggests a model for the effects of Sds22 in aurora B function at centromeres and kinetochores. Normally, the levels of phospho–aurora B at kinetochores are set by a competition between autoactivation by aurora B and deactivation by PP1. Removal of Sds22/PP1 changes this balance and removes the activity that antagonizes aurora B activation. Thus, Sds22/PP1 defines the amount of phospho–aurora B and, through substrates that must still be identified, the stability of the interaction between the microtubule and kinetochore. This same pathway is used before and after establishment of biorientation. In maloriented sister pairs, centromeres and kinetochores are close to one another, allowing autoactivation of aurora B and binding to the kinetochore. Although we have no evidence for how this occurs, this most likely involves inhibition of Sds22/PP1 activity by phosphorylation of PP1 by aurora B or other kinetochore-associated kinases. Indeed, PP1 has previously been shown to be inhibited by CDK1/cyclin B–mediated phosphorylation in S. pombe (Yamano et al., 1994).
Figure 7.
A model for regulation of aurora B by Sds22 and PP1. The drawing shows a schematic model of the activation of aurora B kinase activity and its regulation by Sds22/PP1. Aurora B activates through autophosphorylation of phospho-T232 at the both the centromere and kinetochore. Phospho–aurora B can destabilize interactions between kinetochores and microtubules through phosphorylation of unknown targets. Sds22/PP1 dephosphorylates phospho–aurora B and, thus, indirectly functions to stabilize kinetochore–microtubule interactions. Centromere-associated aurora B may inhibit Sds22/PP1 when distance between centromere and kinetochore are low, e.g., in syntelic or merotelic attachments.
Our data suggest that activated, kinetochore-associated aurora B stimulates destabilization of microtubule attachments. Once biorientation is established, the distance between aurora B at the centromere and Sds22/PP1 at the kinetochore increases, relieving the inhibition of PP1 by aurora B and allowing Sds22/PP1 to dephosphorylate kinetochore-associated phospho–aurora B and, thus, protecting established kinetochore-associated microtubules from depolymerization. This model predicts that loss of Sds22/PP1 should cause perturbations in microtubule–kinetochore interactions, as we have observed (Fig. 6). However, this simple circuit is incomplete, as it does not explain how in Sds22-depleted cells, phosphorylation of MCAK and Hec1 decrease, and it suggests that other factors that control phosphatase activity must be involved.
Materials and methods
Tissue culture, siRNA, FACS, and fluorescent protein fusions
HeLa S3 cells were grown in DME supplemented with 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen) at 37°C with 5% CO2 in a humidified incubator. PP1-γ–EGFP-expressing stable cell line (Trinkle-Mulcahy et al., 2003) was maintained at 300 µg/ml G418, and GFP-Sds22 cell lines were maintained at 1 µg/ml puromycin. Stable HeLa (Kyoto) EGFP–CENP-A cell line (Jaqaman et al., 2010) was maintained in 500 ng/ml puromycin.
To increase the yield of mitotic cells, cells were collected by mitotic shake off after 18 h in medium containing 330 nM nocodazole or 10 h after release from double-thymidine block. For thymidine block, cells were grown for 18 h in medium containing 2 mM thymidine, washed twice, and released into fresh medium; after 8 h, 2 mM thymidine was added for another 18 h before being released into fresh medium. Finally, after 10 h, mitotic cells were collected by mitotic shake off.
For experiments with monastrol, cells were incubated for 4 h in 100 µM monastrol (Sigma-Aldrich), washed, and released into equilibrated medium. ZM was purchased from Tocris Bioscience and used at the specified concentrations.
To measure G2/M or sub-G1 fractions, cells were collected, washed in PBS, and fixed/permeabilized in 70% ethanol at −20°C for >30 min before staining with 50 µg/ml propidium iodide in PBS containing 50 µg/ml RNase A and 0.1% Triton X-100. The cell profile was recorded on a flow cytometer (FACSCalibur; BD) using CellQuest Pro software (BD). Data analysis was performed using Flowjo software (Tree Star, Inc.). Clusters of two or more cells were excluded from analysis by gating.
N- and C-terminal fusion proteins of human Sds22 (GenBank accession no. Z50749) with and without a 5× Gly-Ala linker were expressed from a bicistronic vector and selected with 1.5 µg/ml puromycin. Five or more clones of each fusion (C103: Sds22-GFP, D103 GFP-5×GlyAla-Sds22) were selected and analyzed. Transient transfectants (200 ng DNA per 6-wells; Effectene; QIAGEN) were analyzed 48 h after transfection.
Double-stranded Sds22 siRNAi (5′-CUCUCAAAGGAGAUUGUCGUUCAUCC-3′) and medium GC control (Stealth RNAi; Invitrogen) were introduced by oligofectamine (Invitrogen) or alternatively using HiPerFect (QIAGEN) according to the manufacturers’ instructions. Typically, 9 µmol siRNAi was used per 6-well plate; unless stated otherwise, cells were analyzed 48 h after transfection. PP1-specific RNAi duplexes (PP1-α, 5′-CCGCAATTCCGCAAAGCCAA-3′; PP1-β, 5′-TACGAGGATGTCGTCCAGGAA-3′; and PP1-γ, 5′-AACATCGACAGCATTATCCAA-3′) were obtained from QIAGEN. Lipofectamine 2000 was used for cotransfections of mCherry-tubulin and EGFP-Hec1 (provided by D.A. Compton [Dartmouth Medical School, Hanover, NH]) with siRNAs in fluorescence lifetime imaging microscopy (FLIM) FRET experiments.
Sample preparation for proteomic analysis
Cells were lysed in hypotonic buffer (20 mM Tris acetate, pH 7.5, 1 mM EDTA, 1 mM EGTA, 10 mM Na β-glycerophosphate, 5 mM Na pyrophosphate, 1mM Na orthovanadate, 50 mM NaF, 0.1% 2-ME, 0.27 M sucrose, 1% Triton X-100, and 1 µM microcystein), incubated on ice for 15 min, and spun at 1,000 g for 10 min.
GFP fusion proteins were immunoprecipitated with mouse anti-GFP IgG (Roche), prebound, and dimethyl pimelimidate linked to protein G–Sepharose (Bio-Rad Laboratories) or with GFP-Trap (ChromaTek). Beads were preblocked with 0.02% insulin and washed three times with lysis buffer and 0.01% Triton X-100.
Immunoprecipitate eluates were separated on 1D PAGE gel and stained (SimplyBlue; Invitrogen), and protein bands were excised, chopped into ∼1 × 1–mm pieces, and destained at RT (2 × 30 min in 10% acetonitrile (ACN)/25 mM triethylammonium bicarbonate buffer [TEAB], pH 8.5). After 10-min dehydration in 100% ACN, proteins in gel were reduced by incubation in 10 mM DTT/TEAB for 60 min at 37°C and alkylated in 50 mM iodoacetic acid/TEAB for 30 min in the dark at RT. For trypsin treatment, gel pellets were dehydrated in 100% ACN and rehydrated on ice (for 1 h) with TEAB buffer containing 10 ng trypsin; excess trypsin was removed, and digestion was performed overnight at 37°C in 50 µl TEAB.
Digested peptides were extracted with 2 × 100 µl 20% ACN/5% formic acid (FA) and once with 100% ACN. Combined extracts were evaporated down to ∼20 µl. The final solution was diluted to 100 µl with 3% ACN/0.25% TFA and subjected to cleaning and concentrating procedure on homemade C18 tips. Peptides were eluted from C18 tips in 15 µl 70% ACN/0.1% FA, diluted with 0.1% FA, and concentrated again by evaporation. Liquid chromatography mass spectrometry analysis was performed at Proteomics Facility of the Wellcome Trust Biocentre (University of Dundee, Dundee, Scotland, UK) using a mass spectrometer (Orbitrap; Thermo Fisher Scientific). Combined peak lists were searched against the National Center for Biotechnology Information database using the Mascot program (version 2.2; Matrix Science).
Immunostaining
For immunofluorescence, cells grown on coverslips were fixed after washing once in 37°C PBS by incubation in 3.7% formaldehyde/PBS, pH 6.8, two times for 5 min at 37°C. Cells were permeabilized in PBS/0.1% Triton X-100 for 10 min at 37°C, blocked (2% BSA in TBS/0.1% Triton X-100 and 0.1% normal donkey serum) for 40 min at RT, incubated with primary antibodies for 1 h, washed (TBS/0.1% Triton X-100), and incubated with secondary antibodies for 45 min. If required, cells were stained with 0.1 µg/ml DAPI. After a final set of washes, cells were mounted in p-phenylenediamin/glycerol homemade mounting medium (0.5% p-phenylenediamine, 20 mM Tris, pH 8.8, and 90% glycerol). For phospho-specific antibodies, heat-inactivated blocking solution (30 min at 55°C) and wash and fixing solutions were supplemented with 500 nM microcystin and 80 nM okadaic acid. For pHec1 antibody, cells were permeabilized for 2 min in PHEM buffer and 0.2% Triton X-100 and fixed in PHEM and 3.7% PFA.
For aqueous chromosome spreads, cells were grown on coverslips in the presence of 330 nM nocodazole for 18 h, swelled in 60% water and 40% medium for 7 min, and spun down onto coverslips for 4 min at 1,000 g before being processed for immunofluorescence.
Anti-AIM1 monoclonal antibody (BD) was used at 1:200 dilution. Affinity-purified anti–phospho-MCAK (Andrews et al., 2004) and Sds22 polyclonal antibodies were diluted to 1 µg/ml. Anti-phospho–Ser55-Hec1 antibody was used at a 1:15,000 dilution and anti-phospho–Ser44-Hec1 was used at 1:3,000. Rat anti–α-tubulin (AbD Serotec) and mouse α-Hec1 (Abcam) were used at a 1:500 dilution. Human ACA (provided by S. Marshall and Tayside Tissue Bank, University of Dundee), anti-phospho–CENP-A (BD), sheep anti-BubR1 (provided by S. Taylor, University of Manchester, Manchester, England, UK), and anti–phospho-T232 aurora B (Rockland Scientific) were diluted at 1:1,000. Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. (FITC, Texas red, TRITC, or Cy5) or Invitrogen (Alexa Fluor 488 and 568). Polyclonal sheep anti-PP1 antibody was provided by P.T. Cohen (University of Dundee).
To generate Sds22-specific antibodies, full-length human Sds22 was expressed as a GST fusion in bacteria, purified on a glutathione column, cleaved from the GST tag, and injected into rabbits. Antisera were purified over recombinant full-length MBP-Sds22 bound to an amylose column.
Microscopy
Fixed imaging was performed on a microscope (DeltaVision Core; Applied Precision) built around a stand (IX70; Olympus) with a 100× 1.4 NA lens and a camera (CoolSNAP HQ; Photometrics; Andrews et al., 2004; Porter et al., 2007). Fixed cells on No. 1.5 coverslips were mounted in 0.5% p-phenylenediamine in 90% glycerol. Optical sections were recorded every 0.2 µm. 3D datasets were deconvolved using constrained iterative restoration (Swedlow et al., 1997; Wallace et al., 2001) as implemented in SoftWoRx software (Applied Precision). Hec1–Hec1 distances for each condition were measured in SoftWoRx for clearly distinguishable metaphase centromere pairs lying along the pole to pole axis using individual optical sections from 3D datasets.
Image intensity analysis for fixed image data was performed in OMERO software (Swedlow et al., 2009). A region covering all of the kinetochores at mitotic plate was defined, and intensities through all sections were summed and divided by the area of the region of interest for each channel. An intensity value per area for the background obtained in a similar fashion was subtracted, and relative intensities were expressed as a ratio relative to a general kinetochore marker (ACA or Hec1).
For analysis of cell cycle progression, HeLa cells expressing CENP-A–GFP were imaged in 35-mm glass-bottom dishes (Microwell; MatTek) or cover glass (Labtek) starting 48 h after transfection. Datasets (512 × 512 pixels with 2 × 2 binning, 0.05-s exposure, and five z sections spaced by 0.5 µm) were acquired every 5 min on a microscope (DeltaVision Core) fitted with a 37°C environmental chamber (Solent Scientific) with a 40× 1.3 NA objective and a camera (CoolSNAP HQ). Datasets were deconvolved, and time courses were presented as maximum intensity projections of deconvolved 3D datasets.
Live cell imaging of HeLa cells expressing CENP-A–GFP for measurement of sister pair distances was performed exactly as previously described (Jaqaman et al., 2010).
FRET measurements by FLIM were performed on a confocal laser-scanning microscope (Radiance 2100MP; Bio-Rad Laboratories) on a stand (TE2000; Nikon) using a 60×/1.4 NA Plan Apo oil immersion lens (Nikon) equipped with a titanium sapphire laser (Chameleon 1; Coherent) providing femtosecond pulses at a 90-MHz repetition rate. Light shielding and environmental control were achieved using a matt black environmental chamber that surrounded the microscope stage and stand (Solent Scientific) maintaining cells at 37°C and limiting stray light from entering the detectors. Presence of both GFP and mCherry was confirmed using the confocal light path with the 488-nm argon ion and 543-nm HeNe laser lines. Two photon excitation for FLIM was performed at 880 nm with a 600 fps scan speed at 512 × 512 resolution for 60 s, and fluorescent light was collected on a nondescanned detector (5783P; Hamamatsu Photonics) using a 670-nm long-pass dichroic mirror and a 528/50-nm band-pass emission filter, such that only GFP was excited and collected. Time correlated single photon counting was performed using a photon-counting card (SPC830; Becker & Hickl), and subsequent analysis was performed with SPCImage (Becker & Hickl).
3DSIM was performed on a microscope system (OMX version 2; Applied Precision) equipped with 405, 488, and 593 solid-state lasers. Images were acquired using a UPlanS Apochromat 100× 1.4 NA oil immersion objective lens and back-illuminated 512 × 512 electron microscopy charge-coupled device cameras (Cascade II; Photometrics). Samples were illuminated by a coherent scrambled laser light source that had passed through a diffraction grating, thus generating interference of light orders in the image plane to create a 3D sinusoidal pattern with lateral stripes ∼0.2 µm apart. The pattern was shifted laterally through five phases and three angular rotations of 60° for each z section. Optical sections were separated by 0.125 µm. Exposure times were typically between 200 and 500 ms, and the power of each laser was adjusted to achieve optimal intensities of between 2,000 and 4,000 counts in a raw image of 16-bit dynamic range at the lowest laser power possible to minimize photobleaching. Each frame acquisition was separated by a 300-ms pause. Multichannel imaging was achieved through sequential acquisition of wavelengths by separate cameras.
Raw 3DSIM images were processed and reconstructed (Gustafsson, 2000; Schermelleh et al., 2008). The channels were aligned in the image plane and around the optical axis using predetermined shifts as measured using a target lens and the SoftwoRx alignment tool.
Online supplemental material
Fig. S1 shows biochemical characterization of the interaction between Sds22-GFP and PP1 isoforms and the localization of Sds22-GFP on kinetochores detected with anti-Sds22. Fig. S2 shows the dose-dependent inhibition of aurora B phosphorylation at T232 by ZM. Fig. S3 shows the high resolution localization of total aurora B in control and Sds22-depleted cells using 3DSIM microscopy on an OMX microscope. Fig. S4 shows loss of CENP-A phosphorylation in fixed cells after depletion of individual PP1 isoforms. Fig. S5 shows the effects of Sds22 depletion on phosphorylation of known aurora B substrates and sister interkinetochore distances in living cells. Videos 1–3 show time-lapse imaging of GFP–CENP-A HeLa cells as shown in Fig.1 D and maximum intensity projections from 3D time-lapse images. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912046/DC1.
Acknowledgments
We thank Ana Amaro and Patrick Meraldi for the gift of the CENP-A–GFP HeLa cell line, Laura Trinkle-Mulcahy for the gift of the PP1-γ expression plasmids, Sara Marshall and the Tayside Tissue Bank for human anti-centromere antibodies, Marek Gierlinski (Dundee Data Analysis Group) for advice on statistical analysis, and David Lleres for advice on processing of FLIM images. We are grateful to all members of the Swedlow laboratory, Tomo Tanaka, and Mike Stark for helpful discussions and critical comments on the manuscript.
This work was funded by grants to J.R. Swedlow (067433) and by the Wellcome Trust Centre for Gene Regulation and Expression (grant 083524). Use of the OMX microscope was supported by the Scottish University Life Sciences Alliance.
Footnotes
Abbreviations used in this paper:
- 3DSIM
- 3D structured illumination
- ACA
- anti-centromere antibody
- ACN
- acetonitrile
- FA
- formic acid
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- Förster resonance energy transfer
- NEB
- nuclear envelope breakdown
- PP1
- protein phosphatase 1
- TEAB
- triethylammonium bicarbonate buffer
- ZM
- ZM447439
References
- Adams R.R., Maiato H., Earnshaw W.C., Carmena M. 2001. Essential roles of Drosophila inner centromere protein (Incenp) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880 10.1083/jcb.153.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Ceulemans H., Vulsteke V., De Maeyer M., Tatchell K., Stalmans W., Bollen M. 2002. Binding of the concave surface of the Sds22 superhelix to the alpha 4/alpha 5/alpha 6-triangle of protein phosphatase-1. J. Biol. Chem. 277:47331–47337 10.1074/jbc.M206838200 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinischiotu A., Beullens M., Stalmans W., Bollen M. 1997. Identification of sds22 as an inhibitory subunit of protein phosphatase-1 in rat liver nuclei. FEBS Lett. 402:141–144 10.1016/S0014-5793(96)01514-1 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R.R., Bergmann A., Cousin M.A., Apps D.K., Shipston M.J. 2004. Multi-dimensional time-correlated single photon counting (TCSPC) fluorescence lifetime imaging microscopy (FLIM) to detect FRET in cells. J. Microsc. 215:1–12 10.1111/j.0022-2720.2004.01343.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S., Stukenberg P.T. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S.E., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Glover D.M. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G. 2000. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198:82–87 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.-Y., Sun Z.-W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Jaqaman K., King E.M., Amaro A.C., Winter J.R., Dorn J.F., Elliott H.L., McHedlishvili N., McClelland S.E., Porter I.M., Posch M., et al. 2010. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 188:665–679 10.1083/jcb.200909005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Matsuda Y., Yoshioka T., Sumi N., Okano Y. 1998. Identification and characterization of STK12/Aik2: a human gene related to aurora of Drosophila and yeast IPL1. Cytogenet. Cell Genet. 82:147–152 10.1159/000015089 [DOI] [PubMed] [Google Scholar]
- King E.M., Rachidi N., Morrice N., Hardwick K.G., Stark M.J. 2007. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 21:1163–1168 10.1101/gad.431507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N., Ohkura H., Yanagida M. 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell. 63:405–415 10.1016/0092-8674(90)90173-C [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKelvie S.H., Andrews P.D., Stark M.J. 1995. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol. Cell. Biol. 15:3777–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C.J., Tighe A., Johnson V.L., Scott M.I., Ditchfield C., Taylor S.S. 2005. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 118:3639–3652 10.1242/jcs.02487 [DOI] [PubMed] [Google Scholar]
- Murata-Hori M., Wang Y.L. 2002. Both midzone and astral microtubules are involved in the delivery of cytokinesis signals: insights from the mobility of aurora B. J. Cell Biol. 159:45–53 10.1083/jcb.200207014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnion M.E., Adams R.R., Callister D.M., Allis C.D., Earnshaw W.C., Swedlow J.R. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656–26665 10.1074/jbc.M102288200 [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T.J. 2004. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 15:2895–2906 10.1091/mbc.E04-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Yanagida M. 1991. S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell. 64:149–157 10.1016/0092-8674(91)90216-L [DOI] [PubMed] [Google Scholar]
- Peggie M.W., MacKelvie S.H., Bloecher A., Knatko E.V., Tatchell K., Stark M.J. 2002. Essential functions of Sds22p in chromosome stability and nuclear localization of PP1. J. Cell Sci. 115:195–206 [DOI] [PubMed] [Google Scholar]
- Pepperkok R., Squire A., Geley S., Bastiaens P.I. 1999. Simultaneous detection of multiple green fluorescent proteins in live cells by fluorescence lifetime imaging microscopy. Curr. Biol. 9:269–272 10.1016/S0960-9822(99)80117-1 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kotwaliwale C.V., Tatsutani S.Y., Breed C.A., Biggins S. 2006. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26:2648–2660 10.1128/MCB.26.7.2648-2660.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter I.M., McClelland S.E., Khoudoli G.A., Hunter C.J., Andersen J.S., McAinsh A.D., Blow J.J., Swedlow J.R. 2007. Bod1, a novel kinetochore protein required for chromosome biorientation. J. Cell Biol. 179:187–197 10.1083/jcb.200704098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renouf S., Beullens M., Wera S., Van Eynde A., Sikela J., Stalmans W., Bollen M. 1995. Molecular cloning of a human polypeptide related to yeast sds22, a regulator of protein phosphatase-1. FEBS Lett. 375:75–78 10.1016/0014-5793(95)01180-M [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., Stukenberg P.T. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon I., Severin F.F., Andrews P.D., Taba M.R., Kaplan K.B., Ashford A.J., Stark M.J., Sorger P.K., Hyman A.A. 1999. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 13:545–555 10.1101/gad.13.5.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G.L., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., Musacchio A. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Stone E.M., Yamano H., Kinoshita N., Yanagida M. 1993. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr. Biol. 3:13–26 10.1016/0960-9822(93)90140-J [DOI] [PubMed] [Google Scholar]
- Swedlow J.R., Sedat J.W., Agard D.A. 1997. Deconvolution in optical microscopy. Deconvolution of Images and Spectra. Second edition Jansson P.A., editor Academic Press, San Diego: 284–309 [Google Scholar]
- Swedlow J.R., Goldberg I.G., Eliceiri K.W.; OME Consortium 2009. Bioimage informatics for experimental biology. Annu. Rev. Biophys. 38:327–346 10.1146/annurev.biophys.050708.133641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andrews P.D., Wickramasinghe S., Sleeman J., Prescott A., Lam Y.W., Lyon C., Swedlow J.R., Lamond A.I. 2003. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol. Biol. Cell. 14:107–117 10.1091/mbc.E02-07-0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andersen J., Lam Y.W., Moorhead G., Mann M., Lamond A.I. 2006. Repo-Man recruits PP1γ to chromatin and is essential for cell viability. J. Cell Biol. 172:679–692 10.1083/jcb.200508154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Hudson D.F., Ribeiro S.A., Trinkle-Mulcahy L., Spence J.M., Lai F., Farr C.J., Lamond A.I., Earnshaw W.C. 2006. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 8:1133–1142 10.1038/ncb1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K.G. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace W., Schaefer L.H., Swedlow J.R. 2001. A workingperson’s guide to deconvolution in light microscopy. Biotechniques. 31:1076–1078: 1080: 1082 passim [DOI] [PubMed] [Google Scholar]
- Wang W., Stukenberg P.T., Brautigan D.L. 2008. Phosphatase inhibitor-2 balances protein phosphatase 1 and aurora B kinase for chromosome segregation and cytokinesis in human retinal epithelial cells. Mol. Biol. Cell. 19:4852–4862 10.1091/mbc.E08-05-0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters F.S., Verveer P.J., Bastiaens P.I. 2001. Imaging biochemistry inside cells. Trends Cell Biol. 11:203–211 10.1016/S0962-8924(01)01982-1 [DOI] [PubMed] [Google Scholar]
- Yamano H., Ishii K., Yanagida M. 1994. Phosphorylation of dis2 protein phosphatase at the C-terminal cdc2 consensus and its potential role in cell cycle regulation. EMBO J. 13:5310–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., Furukawa K., Takahashi T., Izawa I., Inagaki M. 2004. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 279:12997–13003 10.1074/jbc.M311128200 [DOI] [PubMed] [Google Scholar]