The molecular motors kinesin-1 dynein navigate migrating nuclei around cytoplasmic roadblocks.
Abstract
Kinesin-1 and dynein are recruited to the nuclear envelope by the Caenorhabditis elegans klarsicht/ANC-1/Syne homology (KASH) protein UNC-83 to move nuclei. The mechanisms of how these motors are coordinated to mediate nuclear migration are unknown. Time-lapse differential interference contrast and fluorescence imaging of embryonic hypodermal nuclear migration events were used to characterize the kinetics of nuclear migration and determine microtubule dynamics and polarity. Wild-type nuclei display bidirectional movements during migration and are also able to roll past cytoplasmic granules. unc-83, unc-84, and kinesin-1 mutants have severe nuclear migration defects. Without dynein, nuclear migration initiates normally but lacks bidirectional movement and shows defects in nuclear rolling, implicating dynein in resolution of cytoplasmic roadblocks. Microtubules are highly dynamic during nuclear migration. EB1::green fluorescence protein imaging demonstrates that microtubules are polarized in the direction of nuclear migration. This organization of microtubules fits with our model that kinesin-1 moves nuclei forward and dynein functions to move nuclei backward for short stretches to bypass cellular roadblocks.
Introduction
Microtubule motors kinesin and dynein move many different cargos in the cell. Although typical cargos have an overall direction in their migration, closer time-lapse imaging shows that most cargos move in a bidirectional manner (Welte, 2004; Ross et al., 2008). Individual cargos often contain both plus and minus end–directed motors, but how the relative outputs of kinesin and dynein motors are coordinated remains poorly understood.
The nucleus is the largest intracellular cargo moved by microtubule motors. Moving and anchoring the nucleus to specific intracellular locations is essential for the formation of polarized cells and for the normal development of most eukaryotes. Nuclear positioning is mediated by a nuclear envelope bridge of Sad1p/UNC-84 (SUN) and klarsicht/ANC-1/Syne homology (KASH) proteins that is conserved across eukaryotes (for reviews see Burke and Roux, 2009; Starr, 2009). SUN proteins in the inner nuclear membrane interact with and recruit KASH proteins to the outer nuclear membrane to form the bridge (Padmakumar et al., 2005; McGee et al., 2006; Crisp and Burke, 2008; Ostlund et al., 2009). KASH proteins then mediate interactions between the nuclear envelope and cytoskeleton (Starr and Han, 2002; Malone et al., 2003; Meyerzon et al., 2009; Roux et al., 2009). KASH and SUN proteins are essential for neurological and muscular development in mice (Puckelwartz et al., 2009; Zhang et al., 2009). Furthermore, mutations in KASH and SUN proteins have been shown to cause or to be linked to muscular dystrophies, ataxias, lissencephaly, progeria, and multiple cancers (Vallee and Tsai, 2006; Gros-Louis et al., 2007; Kandert et al., 2007; Zhang et al., 2007; Marmé et al., 2008; Ovarian Cancer Association Consortium, 2010). Moreover, the KASH–SUN nuclear envelope bridges that move nuclei also move chromosomes and ensure homologue pairing in meiosis (Chikashige et al., 2006; Ding et al., 2007; Koszul et al., 2008; Sato et al., 2009).
KASH and SUN proteins were identified by genetic analysis of mutants with misplaced nuclei (Malone et al., 1999; Mosley-Bishop et al., 1999; Starr and Han, 2002). A limitation of the genetic studies is that they focused on terminal phenotypes because of the difficulty to film live nuclear migration events. Many additional studies on KASH and SUN proteins have been performed in mammalian tissue culture cells (Wilhelmsen et al., 2005; Crisp et al., 2006; Haque et al., 2006; Ostlund et al., 2009). A limitation of these studies is that it is difficult to examine nuclear migration events in culture. One exception is in cultured fibroblasts near a wound edge where nuclei migrate to polarize the cell and nuclear migration is associated with cellular migration (Gomes et al., 2005; Levy and Holzbaur, 2008). However, these migrations are short, associated closely with the leading edge of the cell, and the roles of KASH and SUN proteins remain to be determined. Caenorhabditis elegans hypodermal cells offer the unique combination of performing both genetic disruptions and real-time imaging of long-distance nuclear migration events that are genetically separable from leading edge migrations (Spencer et al., 2001; Starr and Han, 2005).
The KASH protein UNC-83 and the SUN protein UNC-84 function during nuclear migration in a variety of C. elegans tissues (Malone et al., 1999; Starr et al., 2001). The SUN domain of UNC-84 directly interacts with the KASH domain of UNC-83 to bridge the nuclear envelope (McGee et al., 2006). We recently elucidated the mechanism of force generation during nuclear migration in the hyp7 embryonic hypodermal precursor cells. The cytoplasmic domain of UNC-83 directly interacts with the KLC-2 light chain of kinesin-1 and two dynein-regulating complexes (Meyerzon et al., 2009; Fridolfsson et al., 2010). Kinesin-1 mutants have severe nuclear migration terminal defects, as assayed by misplaced nuclei in the dorsal cord, similar to unc-83 or unc-84 mutant animals (Meyerzon et al., 2009). In contrast, mutations in nud-2 and bicd-1, two of the dynein regulators identified, have weaker nuclear migration defects and function partially redundantly to each other (Fridolfsson et al., 2010). In mammals, nesprin-4 and Sun1/2 are likely UNC-83 and UNC-84 functional homologues in that they recruit kinesin-1 to the surface of the nucleus to position nuclei (Roux et al., 2009). In this study, we test our model in which UNC-83 functions as a cargo adaptor and coordinates interactions between microtubule motors and the nucleus. Once recruited to the surface of the nucleus, kinesin-1 provides the major forces to move the nucleus, whereas dynein regulates the process (Fridolfsson et al., 2010).
Three nonmutually exclusive models could explain why both plus and minus end–directed motors are recruited to the nucleus. (1) Most cargos of microtubule motors appear to be moving unidirectionally but actually display short runs in both directions. Such bidirectional movement could allow the nucleus to switch microtubule tracks, avoid obstacles, or correct errors (Welte, 2004; Ross et al., 2008). (2) In some examples, kinesin-1 and dynein are interdependent; a mutation in one disrupts movements in both directions (Martin et al., 1999; Shubeita et al., 2008; Ally et al., 2009). (3) Alternatively, dynein could function as a drag on kinesin-1–driven movement as it does in interkinetic nuclear migration (Del Bene et al., 2008). Two obstacles prevent us from distinguishing between these models: we do not know how microtubules are organized during nuclear migration, and past studies were limited by only observing terminal nuclear migration defects. Thus, to elucidate mechanisms of microtubule motor–regulated nuclear migration, we used time-lapse imaging of the nuclear migration events and determined microtubule dynamics and polarity during nuclear migration.
Results
Nuclear migration in the embryonic C. elegans hypodermis
unc-83 and unc-84 are required for nuclear migration in a variety of C. elegans tissues including larval P cells, gonadal distal tip cells, intestinal primordial cells, and embryonic hyp7 precursors (Horvitz and Sulston, 1980; Sulston and Horvitz, 1981; Malone et al., 1999; Starr et al., 2001). The unc-83 and unc-84 hyp7 nuclear migration phenotype is characterized by misplaced nuclei in the larval dorsal cord, but only terminal phenotypes have been examined. Thus, the dynamics of nuclear migration events are unknown. To determine mechanistic roles for unc-83 and unc-84 during nuclear migration, we examined actual nuclear migration events in embryonic hyp7 precursors.
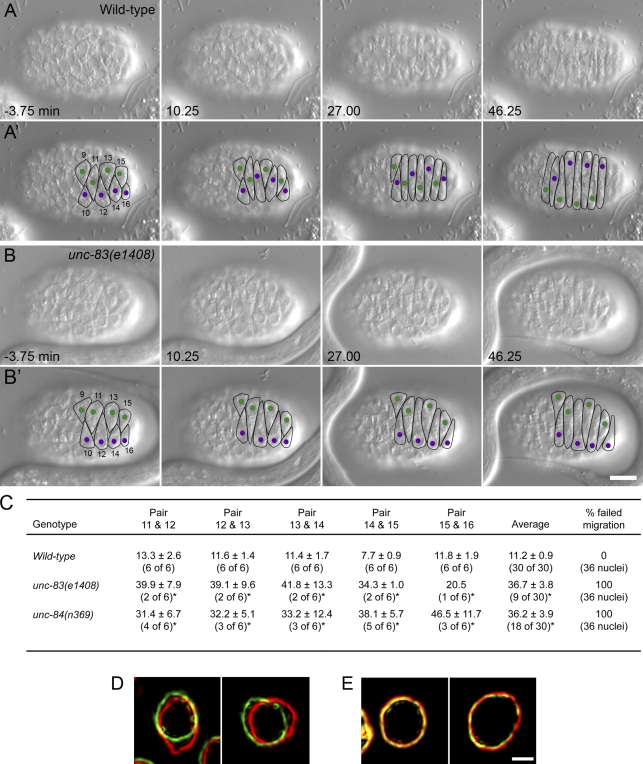
Before examining mutant nuclear migration events, we carefully characterized wild-type nuclear migrations in the dorsal hypodermis of the precomma stage embryo using differential interference contrast (DIC) microscopy, where the cellular boundaries appeared as depressions in the hypodermis and nuclei were easily visualized (Fig. 1 A and Video 1). It has been previously described that normally, two rows of hyp7 precursors align along the dorsal midline. The cells then elongate and intercalate to form a single row of column-shaped epithelial cells spanning the dorsal midline. Intercalation proceeds from anterior to posterior with the exception of the pointer cells that form a wedge shape and delay intercalation. Finally, nuclei migrate across the length of the cell to the opposite lateral side of the embryo (Sulston et al., 1983; Williams-Masson et al., 1998). We followed intercalation and nuclear migration in six cells (hyp7 precursors 11–16, following previous designations; Sulston et al., 1983) posterior to the pointer cells (hyp7 precursors 9 and 10; Fig. 1 A′ and Video 1). Nuclei in adjacent intercalated cells migrated in opposite directions so that nuclei 10, 12, 14, and 16 migrated from left to right (Fig. 1 A′, purple), whereas nuclei 9, 11, 13, and 15 migrated from right to left (Fig. 1 A′, green).
Figure 1.
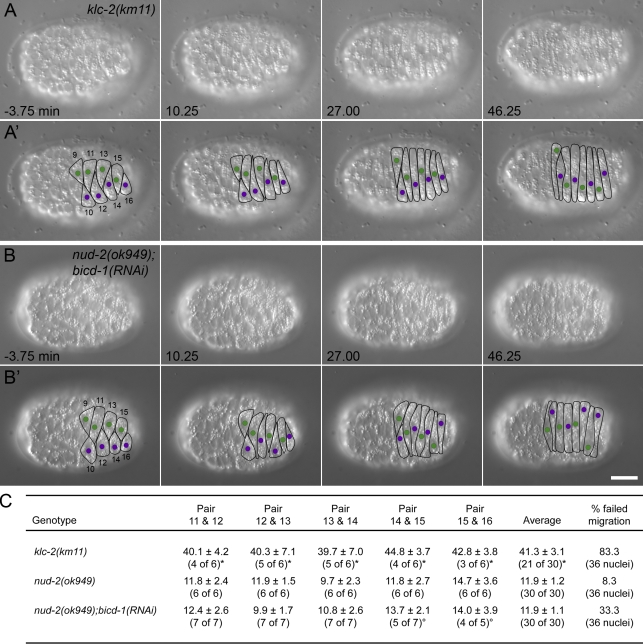
UNC-83 and UNC-84 are required at the onset of nuclear migration. (A and B) DIC images from a representative time-lapse sequence of wild-type (A) and unc-83(e1408) (B) nuclear migration in hyp7 precursors. t = 0 min when the tip of cell 12 reached the opposite seam cell boundary. Dorsal view, anterior is to the left. (A’ and B’) Cell borders are outlined in black, nuclei migrating left to right are purple, and nuclei migrating right to left are green. Hyp7 cells are numbered 9–16, beginning with the posterior pointer cells. Bar, 10 µm. (C) Quantification of side by side nuclear alignment. Ratios represent the number of nuclei that could be scored out of total nuclei. Times are shown in minutes. Many mutant nuclei failed to reach the dorsal midline before filming stopped at least 40 min after intercalation (*). The final column shows the percentage of nuclei that failed to complete migration. (D and E) Lamin::GFP from wild-type (D) and unc-83(e1408) (E) time-lapse imaging. The image of lamin::GFP pseudocolored red was taken 60 s before the image in green. Bar, 1 µm.
Time-lapse DIC microscopy of complete cell intercalation and nuclear migration events in wild-type embryos (up to 80 min of filming per embryo) was performed; images were collected at a rate of one frame every 15 s. For each embryo, t = 0 was defined as when the tip of hyp7 precursor 12 completed intercalation as determined by its growing tip reaching the opposite seam cell boundary. Nuclear migration began in each cell shortly after the tip of that hyp7 precursor reached the opposite seam cell boundary. The mean rate of nuclear migration, from when the nucleus began moving to when it reached the opposite side of the cell, was 13.6 ± 1.6 µm/h (n = 17 nuclei). We quantified nuclear migration by measuring the mean time at which a pair of nuclei in adjacent cells lined up side by side as they were crossing each other during migration (Fig. 1 A′, nuclei 14 and 15 align at 10.25 min). In wild-type embryos, this event occurred at a mean time of 11.2 ± 0.9 min (n = 30 pairs; Fig. 1 C). We also quantified nuclear migration events by measuring the mean distance traveled within 10 min of the completion of intercalation, which was ∼3.3 µm (Table I).
Table I.
Nuclear migration defects at the start of migration
| Genotype | Defectsa |
| µm | |
| Wild type | 3.3 ± 0.2 (36) |
| unc-83(e1408) | 0.05 ± 0.05 (21) |
| unc-84(n369) | 0.06 ± 0.03 (20) |
| klc-2(km11) | 0.3 ± 0.1 (26) |
| nud-2(ok949) | 3.2 ± 0.3 (36) |
| nud-2(ok949);bicd-1(RNAi) | 3.3 ± 0.3 (36) |
| unc-83(e1408);klc-2::KASH | 3.1 ± 0.2 (20) |
| Start migration (20 of 42)b | |
| unc-83(e1408);klc-2::KASH | 0.05 ± 0.03 (22) |
| No migration (22 of 42)b | |
The mean distance ± SEM nuclei migrated in the first 10 min after the completion of intercalation of that cell. The sample size in number of nuclei is in parentheses.
42 nuclei were filmed in the unc-83(e1408);klc-2::KASH strain. 20 of them began nuclear migration at a normal time, and the rest failed to migrate.
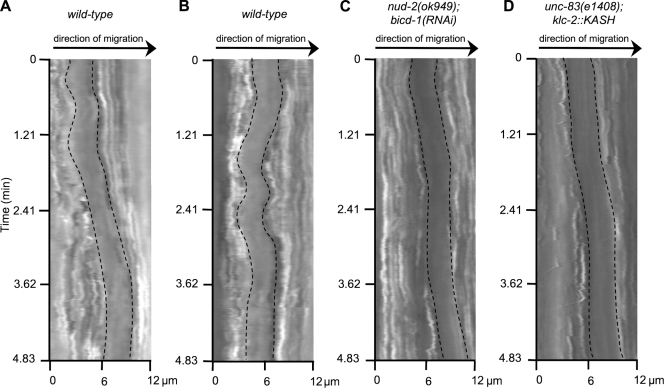
Higher resolution time-lapse imaging and kymographs of moving nuclei compiled at a rate of five frames per second showed that seven of nine nuclei had at least one movement >1 µm in the direction opposite of migration (Fig. 2 B and Fig. S1 A). These kymographs also indicated that the nucleus had periods of no or very slow movement interrupted by periods of faster movements (Fig. 2 A). During faster periods of movement, nuclei moved with a mean peak velocity of 122.4 ± 8.8 µm/h (n = 5 nuclei), ∼10 times faster than the mean forward velocity observed. Time-lapse fluorescent imaging of lamin::GFP provided an additional method of quantifying nuclear movements. During nuclear migration, 63.2% (n = 38) of nuclei in wild-type embryos displayed deformations of the nuclear lamina >0.5 µm in 60 s (Fig. 1 D).
Figure 2.
Nuclei make relatively fast movements and bidirectional movements. Kymographs of nuclei made from DIC time-lapse imaging at 200-ms intervals. Edges of the nucleus are outlined. (A) Example of brief, faster nuclear movement in wild type. (B) Example of wild-type nuclear movements backward and forward during migration. (C) Example of normal forward migration, including a rapid run forward, of a nud-2(ok949);bicd-1(RNAi) nucleus. (D) Example of a rapid forward run of an unc-83(e1408);klc-2::KASH nucleus. The x axis is the distance traveled in micrometers. The forward direction of nuclear migration is left to right.
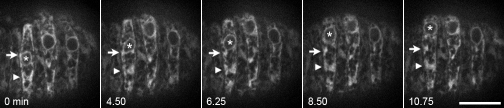
The nucleus is the largest organelle in the cell, and its diameter is nearly the width of the elongated hyp7 precursor cell. Furthermore, the cell is full of cytoplasmic granules, ER, and other organelles. We predict that as organelles accumulate in front of migrating nuclei, the resistance against forward nuclear migration will increase. Releasing this pressure to allow the nucleus to move forward should be accompanied by reorganization of the cellular architecture. Considering the ER is physically connected to the nucleus, how its organization is affected by nuclear migration was of particular interest. Time-lapse imaging of an SP-12::GFP marker of the ER expressed in hypodermal cells revealed that as the nucleus migrates, large pieces of the ER remained stable, but sections closer to the nucleus were briefly pulled along (Fig. 3 and Video 2).
Figure 3.
ER morphology during nuclear migration. Images of SP-12::GFP from a time-lapse sequence. Large sections of the ER behind the nucleus (*) are stretched but otherwise remain fairly stationary during migration (arrowheads). ER closer to the nucleus is briefly pulled along before detaching (arrows). Anterior is to the left, nuclear migration is up. Bar, 10 µm.
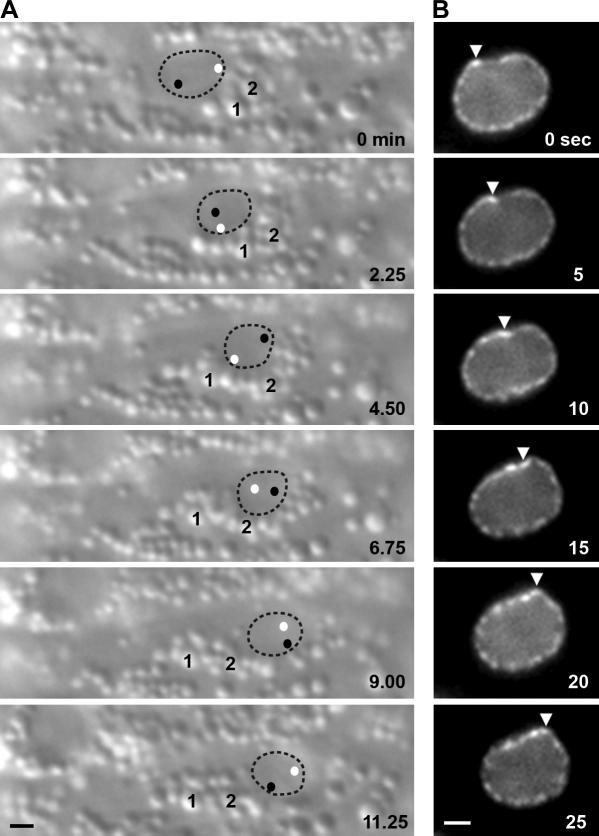
More drastic rearrangements of the cellular architecture were observed by DIC time-lapse imaging, which allowed the observation of rolling nuclei, sometimes >360° (Fig. 4 A and Video 3). As nuclei migrated, an accumulation of cytoplasmic granules formed in front of the nucleus. Often the accumulation of granules compressed as the nucleus attempted to move past the blockage (Fig. 4 A; and Video 3, first second). Eventually, the blockage was resolved, and the nucleus moved past the granules. Sometimes the granules flowed past the sides of or over the top of the nucleus, but no obvious shifting of the nucleus was observed (unpublished data). However, in 16.7% of wild-type hyp7 precursors, nuclei rolled to pass the blockage of granules (Table II, Fig. 4 A, and Video 3). When nuclear rolling was observed, it was quite fast, rotating a mean of nearly 30 angular degrees per minute (Table II). We captured one example of a rapid movement of the nuclear envelope that could lead to nuclear rolling by filming lamin::GFP (Fig. 4 B and Video 4). We propose that rolling is a mechanism for nuclei to pass accumulations of cytoplasmic granules during migration through narrow cells.
Figure 4.
Actively migrating nuclei roll. (A) DIC images from a time-lapse sequence of nuclear rolling. Nuclear migration is to the right, and rolling is clockwise. The nucleus is outlined; black and white dots represent the position of nucleoli within the nucleus. Two cytoplasmic vesicles (1 and 2) are marked. (B) Images of lamin::GFP from a time-lapse sequence. Rolling is led by a deformation in the lamina (arrowheads). Nuclear migration is to the left, and rolling is clockwise. Bar, 1 µm.
Table II.
Quantification of nuclear migration and rolling
| Migration quantification | Wild type | unc-83(e1408) | unc-84(n369) | klc-2(km11) | nud-2(ok949) | nud-2(ok949); bicd-1(RNAi) | unc-83(e1408); klc-2::KASH |
| Successful migration | |||||||
| Migration (%) | 100 | 0 | 0 | 16.7 | 91.7 | 66.7 | 31.0 |
| Roll (%) | 16.7 | NA | NA | 16.7 | 12.1 | 8.3 | 0 |
| Speed of rollinga | 29.4 ± 3.0 | NA | NA | 34.3 | 21.1 ± 5.0 | 21.9 ± 2.9 | NA |
| Partial migrationb | |||||||
| Migration (%) | 0 | 0 | 0 | 0 | 8.3 | 33.3 | 16.7 |
| Roll (%) | NA | NA | NA | NA | 0 | 0 | 0 |
| Failed migrationb | |||||||
| Migration (%) | 0 | 100 | 100 | 83.3 | 0 | 0 | 52.4 |
For each indicated genotype, 36 nuclei were examined, except for unc-83(e1408);klc-2::KASH, in which 42 nuclei were examined. NA, not applicable.
Of the nuclei that rolled, the mean speed of rolling in angular degrees per minute ± SEM is shown.
Nuclei that partially migrate initiate movement at the normal time and speed but fail to complete their migrations. Typically, such a nucleus migrates about halfway across the cell before stopping or returning to its original position. Failed nuclear migrations do not move at all until much later in development. Nuclei that failed to migrate could not be scored for rolling.
unc-83 and unc-84 are required from the onset of nuclear migration
In unc-83 or unc-84 mutant embryos, nuclei fail to migrate, and most nuclei are found mispositioned in the dorsal cord of L1 larvae instead of the lateral surface (Horvitz and Sulston, 1980; Sulston and Horvitz, 1981; Malone et al., 1999; Starr et al., 2001). Before this study, the mechanisms and timing of how the nuclei end up in the dorsal cord were unknown. Time-lapse DIC imaging revealed that unc-83(e1408) or unc-84(n369) mutant nuclei had no active migrations and remained on the side of the embryo they began on long after the tip of the cell reached the opposite seam cell boundary (Fig. 1, B and B′; Video 5; and not depicted). In unc-83(e1408) embryos, 21 of 30 pairs of nuclei failed to align, even after 40 min (Fig. 1 C). The nine pairs that did align took a mean of 36.7 ± 3.8 min (as compared with 11.2 ± 0.9 min in wild type). All nuclei failed to initiate nuclear migration on time (Table I) and failed to migrate past the dorsal midline. Similarly, in unc-84(n369) embryos, nuclei failed to initiate nuclear migration at the normal time (Table I), and those that did eventually move (18 of 30 pairs) aligned with their opposing partner late, at a mean of 36.2 ± 3.9 min (Fig. 1 C). Previous measures of the terminal nuclear migration phenotype found that 85–90% of hyp7 nuclei were misplaced in the dorsal cord of unc-83 or unc-84 mutant L1 larvae (Malone et al., 1999; Starr et al., 2001). Live imaging showed that this variability was caused by nuclei that remained on the lateral side that they started on. The pointer cells’ nuclei in particular, which were positioned more laterally to begin with and normally migrated later than the others, often remained in their lateral starting positions in unc-83 and unc-84 mutant embryos. Therefore, 100% of the unc-83 and unc-84 mutant nuclei had nuclear migration defects.
There was no significant difference in the initiation or timing of nuclear movements between unc-83 and unc-84 mutant embryos. The difference in the number of unc-83 versus unc-84 nuclei that crossed paths late (Fig. 1 C) was likely because of the fact that many crossings occurred near the end of our filming, which was frequently cut short when the embryo rotated orientations within the eggshell. Alternatively, nuclear anchorage, which requires unc-84 but not unc-83 (Starr and Han, 2002), could antagonize the late nuclear movements observed in unc-83 mutant embryos. In contrast to wild type, nuclei failed to roll in unc-83 and unc-84 embryos, suggesting that nuclear rolling was mediated by unc-83 and unc-84–dependent forces acting on nuclei during migration (Table II). Consistent with a lack of forces in unc-83 mutant embryos, time-lapse imaging of lamin::GFP for 2-min periods indicated that 0% (n = 35) of nuclei showed deformations or movements of the nuclear envelope >0.5 µm over 60 s (Fig. 1 E).
Nuclei fail to migrate properly in kinesin-1 mutants
UNC-83 recruits kinesin-1 to the cytoplasmic surface of the nuclear envelope to mediate nuclear migration (Meyerzon et al., 2009). Mutations in either the kinesin-1 heavy chain unc-116 or the light chain klc-2 cause severe hyp7 nuclear migration defects as assayed by the abnormal presence of hyp7 nuclei in the dorsal cords of L1 larvae (Meyerzon et al., 2009). In the hypomorphic klc-2(km11) mutant background, 52% of the nuclei end up in the dorsal cord, as compared with 0% in wild type and 84% in unc-83(e1408) null (Meyerzon et al., 2009). Time-lapse imaging was used to characterize nuclear movements in klc-2(km11) embryos. Consistent with the hypothesis that kinesin-1 provides the major force to move the nucleus, nuclei failed to initiate migration upon the completion of intercalation (Table I). A majority of the nuclei moved to the dorsal midline very late, similar to unc-83(e1408) or unc-84(n369) embryos (Fig. 5, A and A′; and Video 6). In the 30 pairs of nuclei filmed from six embryos, 21 pairs met their partner nucleus from the opposite side in an adjacent cell at a mean of 41.3 ± 3.1 min after the completion of intercalation, 30 min later than wild type (compare Fig. 5 C with Fig. 1 C). The other nine pairs failed to migrate to the midline while we were able to film. Occasionally (6 of the 36 nuclei analyzed from six embryos), and well after the completion of intercalation, nuclei migrated past the dorsal midline to the opposite side of the embryo (Fig. 5 A′, nucleus 16). Because klc-2(km11) is a partial loss of function, presumably there is enough KLC-2 activity for nuclear migration but not enough for it to occur at the proper time. These events explain why the terminal klc-2(km11) phenotype is slightly less severe than unc-83(e1408). Of the nuclei that were not actively migrating, there was no occurrence of nuclear rolling. However, 16.7% of the few nuclei that did migrate to the opposite side of the embryo rolled (Table II).
Figure 5.
Nuclear migration in kinesin-1 and dynein-regulatory mutant hyp7 precursors. (A and B) DIC images from a representative time-lapse sequence of klc-2(km11) (A) and nud-2(ok949);bicd-1(RNAi) (B) nuclear migration events. t = 0 min when the tip of cell 12 reached the opposite seam cell boundary. Anterior is to the left. (A’ and B’) Cell borders are outlined in black, nuclei migrating left to right are purple, and nuclei migrating right to left are green. Hyp7 cells are numbered 9–16, beginning with the posterior pointer cells. Bar, 10 µm. (C) Quantification of side by side nuclear alignment. Ratios represent the number of nuclei that could be scored out of total nuclei. Times are shown in minutes. Some nuclei failed to reach the dorsal midline before filming stopped at least 40 min after intercalation (*), and others could not be seen (°). The final column shows the percentage of nuclei that failed to complete migration.
Most nuclei migrate with normal dynamics in dynein-regulatory mutant backgrounds
Besides recruiting kinesin-1 to the outer surface of the nucleus, UNC-83 also recruits dynein (Fridolfsson et al., 2010). UNC-83 interacts with two dynein-regulatory complexes, one consisting of the NudE and Lis1 homologues NUD-2 and LIS-1 and the other consisting of the Bicaudal D, egalitarian, and LC8 dynein light chain homologues BICD-1, EGAL-1, and DLC-1 (Fridolfsson et al., 2010). In our model, dynein functions as a regulator of nuclear migration, ensuring a more efficient migration by allowing for bidirectional transport. In the nud-2(ok949)–null mutant, ∼1% of the nuclei end up in the dorsal cord (Fridolfsson et al., 2010). Consistent with this weak nud-2(ok949) terminal nuclear migration defect, nearly all hyp7 nuclei migrated normally when observed by DIC time-lapse imaging (Table I and Video 7). Similar to wild-type nuclear migration, nud-2(ok949) mutant nuclei crossed their adjacent neighbors at ∼11.9 ± 1.2 min (n = 30 pairs) after intercalation (Fig. 5 C). The mean rate of nuclear migration was ∼13.2 ± 1.9 µm/h (n = 15 nuclei), and nuclei were able to reach the same peak velocity for short runs as in wild type. In three of the six embryos that were analyzed, a single nucleus started to migrate at the appropriate time but failed to complete its migration (Video 7, nucleus 11). Such failed nuclear migration events are likely what cause the terminal nud-2(ok949) phenotype, where occasionally a single nucleus is observed in the dorsal cord, an event that never occurs in wild type (Fridolfsson et al., 2010). Of the nuclei that migrated properly, 12.1% displayed rolling, but rolling was not observed in the cases where nuclear migration failed (Table II).
nud-2(ok949) hyp7 cells represent only a partial disruption of dynein function at the nuclear envelope because the BICD-1–EGAL-1–DLC-1 complex is still functioning (Fridolfsson et al., 2010). Therefore, we filmed nuclear migration in nud-2(ok949);bicd-1(RNAi) embryos, in which ∼14% of nuclei end up in the dorsal cord (Fridolfsson et al., 2010). Consistent with this stronger nuclear migration defect, more nud-2(ok949);bicd-1(RNAi) nuclei failed to migrate when observed by DIC time-lapse imaging as compared with nud-2(ok949) alone (Fig. 5, B and B′; and Video 8). 33.3% of nud-2(ok949);bicd-1(RNAi) nuclei failed to reach the opposite side of the embryo (Table II). Despite an increase in migration failure, nud-2(ok949);bicd-1(RNAi) mutant nuclei started to migrate normally (Table I). The mean and peak velocities of successful nuclear migrations were similar to wild type. High resolution kymographs of migration just after intercalation showed that nud-2(ok949);bicd-1(RNAi) mutant nuclei alternated between slow and swift forward movements (Fig. 2 C). However, nud-2(ok949);bicd-1(RNAi) mutant nuclei did not display bidirectional movements at this time. No movements >1 µm in the direction opposite of migration were observed in a total of 43.5 min of imaging of nine nuclei for 4.8 min just after intercalation (as compared with seven of nine wild-type nuclei that did move backward; Fig. S1 B). Normal initiation of migration of nud-2(ok949);bicd-1(RNAi) mutant nuclei allowed them to cross their adjacent neighbors at a normal time of 11.9 ± 1.1 min (n = 30 pairs) after intercalation (Fig. 5 C). Thus, all nud-2(ok949);bicd-1(RNAi) nuclei initiated migration properly, and the majority of nuclei were able to complete their migration normally. However, 12 out of 36 nuclei failed to complete their migration and either stalled at the midline or returned to their starting position. Of nuclei that migrated normally, 8.3% displayed rolling; nuclear rolling was never seen in the nuclei that failed to migrate (Table II).
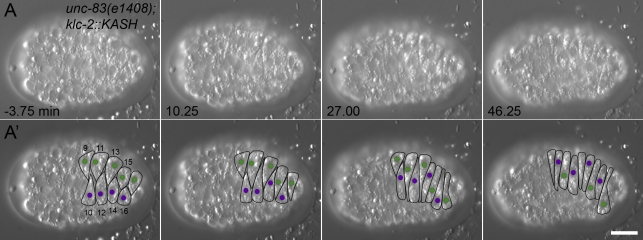
Some nuclei rolled in the nud-2(ok949);bicd-1(RNAi) mutant embryos, suggesting that not all dynein-regulatory activity was disrupted. To further examine the role of dynein regulators and anything else that might be binding to the cytoplasmic domain of UNC-83, we filmed nuclear migration in an unc-83(e1408)–null background expressing a transgene encoding a KLC-2::KASH hybrid. KLC-2 was fused to the C-terminal 290 residues of UNC-83, creating a protein that was targeted to the nuclear envelope and could partially rescue nuclear migration defects independent of UNC-83 (Meyerzon et al., 2009). KLC-2::KASH included the transmembrane and KASH domains of UNC-83 but was missing most of the DLC-1– and NUD-2–binding domains (Fridolfsson et al., 2010). DIC time-lapse imaging of unc-83(e1408);klc-2::KASH embryos indicated that ∼50% of nuclei initiated migration normally (Fig. 6, Table I, and Video 9). High resolution kymographs of migration just after intercalation showed that unc-83(e1408);klc-2::KASH nuclei did not display bidirectional movements (Fig. 2 D). No movements >1 µm in the direction opposite of migration were observed in a total of 43.5 min of imaging of nine nuclei for 4.8 min just after intercalation (Fig. S1 C). Additionally, 7 out of 20 nuclei that initiated migration normally failed to complete their migration and either stalled at the midline or returned to their starting position. None of the nuclei that initiated migration normally displayed rolling (Table II). Because NUD-2 and BICD-1 were likely not recruited to the nuclear envelope, it is possible that their loss alone caused the lack of nuclear rolling and backward movement. However, their known dynein-dependent functions make it likely that backward movements and rolling are mediated through dynein (Kardon and Vale, 2009).
Figure 6.
Nuclear migration in unc-83(e1408);klc-2::KASH hyp7 precursors. (A) DIC images from a representative time-lapse sequence of unc-83(e1408);klc-2::KASH nuclear migration events. t = 0 min when the tip of cell 12 reached the opposite seam cell boundary. Anterior is to the left. (A’) Cell borders are outlined in black, nuclei migrating left to right are purple, and nuclei migrating right to left are green. Hyp7 cells are numbered 9–16, beginning with the posterior pointer cells. Bar, 10 µm.
Microtubules dynamically reorganize during intercalation and nuclear migration
To fully understand the mechanisms of how kinesin-1 and dynein function together to move nuclei, it was necessary to determine the organization of the microtubule cytoskeleton during nuclear migration. Microtubules are necessary for intercalation and nuclear migration because nocodazole treatment results in the failure of both (Williams-Masson et al., 1998). The organization of microtubules during intercalation has previously been described by images of fixed embryos; before intercalation, microtubules appear as a meshwork, but by the time intercalation is complete, microtubules form long, thick bundles that align parallel to the long axis of the cell (Williams-Masson et al., 1998). Based on these images, a model has been proposed in which microtubules reorganize during intercalation into stable, polarized bundles with their plus ends at the new tip of the cell (Williams-Masson et al., 1998; Meyerzon et al., 2009). Such an organization of microtubules would be ideal for kinesin-1 at the surface of the nucleus to move nuclei (Meyerzon et al., 2009).
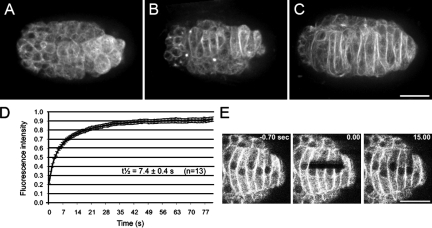
To directly test this hypothesis, we used live imaging of GFP::β-tubulin expressed specifically in the hypodermis. Our time-lapse imaging showed that the microtubule cytoskeleton underwent a dramatic reorganization during intercalation. The microtubules were arranged as a meshwork before intercalation, but by the time intercalation completed, microtubules were arranged in bundles parallel to the long axis of the cell (Fig. 7, A–C; and Video 10). We tested whether microtubule bundles were stabilized during nuclear migration using FRAP. In embryos expressing GFP::β-tubulin, an area across the width of several cells during nuclear migration was photobleached. The microtubules remained highly dynamic, as indicated by a half-time of recovery of only 7.4 ± 0.4 s (n = 13 embryos; Fig. 7, D and E). This recovery rate is slower than the diffusion time of free tubulin dimers and presumably reflects microtubule polymer turnover (Salmon et al., 1984). Similar rates for microtubule dynamics have been observed in the mitotic spindle (Cheerambathur et al., 2007). These data cannot eliminate the possibility that there is a small population of stable microtubules, although no acetylated tubulin, a hallmark of stabilized microtubules used by kinein-1 (Reed et al., 2006), was observed in these cells (unpublished data).
Figure 7.
Microtubules are dynamic in hyp7 precursors. (A–C) Representative images of GFP::β-tubulin from time-lapse sequences during intercalation and nuclear migration. Anterior is to the left. (A) Microtubules are a meshwork early in intercalation. (B) As cells intercalate, microtubules reorganize into parallel bundles. (C) During nuclear migration, most microtubules are in long bundles that appear dense at cell–cell boundaries. (D) FRAP curves from GFP::β-tubulin embryos. (E) Images of GFP::β-tubulin from a representative FRAP experiment. Bars, 10 µm.
Microtubules polarize in the direction of nuclear migration
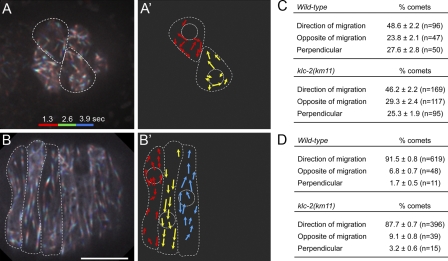
The model of microtubules in hyp7 precursors at the time of nuclear migration predicts that the parallel microtubule bundles are polarized so that plus ends are in the direction of nuclear migration. This orientation would be consistent with our findings that kinesin-1, a plus end–directed motor, is providing the major force to move the nucleus (Meyerzon et al., 2009). To observe microtubule polarity during intercalation and nuclear migration, EBP-1::GFP was expressed in hypodermal cells. EBP-1 is the C. elegans homologue of mammalian EB1 (Motegi et al., 2006), which binds the plus ends of growing microtubules in a comet-like pattern (Morrison et al., 1998). In the early stages of intercalation, microtubules were partially polarized with 48.6 ± 2.2% of EBP-1::GFP comets growing toward the intercalating, dorsal tip of the cell (n = 96 comets; Fig. 8, A, A′, and C; and Video 11). The other half of the comets were split between growing toward the old, lateral end of the cell (23.8 ± 2.1%; n = 47) or perpendicularly across the cell (27.6 ± 2.8%; n = 50). Analysis of EBP-1::GFP in embryos with actively migrating nuclei indicated that the microtubules had polarized so that 91.5 ± 0.8% (n = 619 comets) of the EBP-1 grew in the direction of migration (Fig. 8, B, B′, and D; and Video 12), 6.8 ± 0.7% (n = 48) grew in the opposite direction of migration, and 1.7 ± 0.5% (n = 11) grew perpendicularly across the cell. Early in intercalation, the comets grew at a mean speed of 0.40 ± 0.01 µm/s (n = 70), and during nuclear migration, they grew at the same rate (0.40 ± 0.01 µm/s; n = 70). This indicated that although microtubules reorganized during intercalation, their growth rates remained constant throughout the process.
Figure 8.
Microtubules polarize in the direction of nuclear migration. (A and B) Images representing 3.9 s of EBP-1::GFP time-lapse sequences. The first 1.3 s are colored red, the next 1.3 s are colored green, and the last 1.3 s are colored blue to show directionality. Cell borders are outlined in white. Anterior is to the left. (A) Cells early in intercalation. (B) Cells with migrating nuclei. The nucleus in the left-most cell is migrating up, and direction alternates in every cell to the posterior. (A’ and B’) Cartoon representing direction of microtubule growth based on data in A and B. Nuclei and cell boundaries are outlined in white. (C and D) Quantification of the direction of microtubule growth by visualization of EBP-1::GFP comets early in intercalation (C) and during nuclear migration (D) for wild-type and klc-2(km11). Bars, 10 µm.
To test whether disrupting kinesin-1 blocked nuclear migration by disrupting the organization of microtubules, we examined EBP-1::GFP polarity in klc-2(km11) mutant embryos. Microtubules were normally organized before and after intercalation in klc-2(km11) embryos (Fig. 8, C and D). The velocity of these comets was the same as in wild type. These data are consistent with a role for kinesin-1 in physically pulling the nucleus during nuclear migration rather than a role for kinesin-1 in organizing microtubules.
During hyp7 nuclear migration, the centrosome remains attached to the nucleus by an UNC-83–independent mechanism (Starr et al., 2001). To better characterize the role of the centrosome during nuclear migration in hyp7 precursors, we fixed embryos at the time of nuclear migration and immunolocalized centrosomes using the IFA antibody (Leung et al., 1999). The centrosome was positioned randomly around the migrating nucleus. Division of the nucleus into four equal quadrants indicated that the centrosome was in the front of the nucleus 23.4% of the time, the back of the nucleus 29.8% of the time, and on the sides of the nucleus 46.8% of the time (n = 47; Fig. S2). Therefore, centrosome position did not correlate with the direction of nuclear migration.
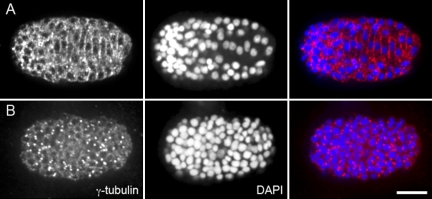
To further characterize the organization of the microtubule network, we examined the localization of γ-tubulin, which nucleates microtubules and is usually localized to the centrosome or cytoplasmic complexes (Stearns and Kirschner, 1994; Murphy and Stearns, 1996). An antibody against C. elegans γ-tubulin (Ow et al., 2008) was used for immunolocalization experiments in embryos fixed just before or at the time of nuclear migration. Before cell intercalation, γ-tubulin localized strongly to centrosomes (Fig. 9 B). However, at the time of intercalation, γ-tubulin redistributed and was broadly localized to the membrane between cells (Fig. 9 A). Similarly, γ-tubulin localizes to the membrane in the C. elegans gonad, where it functions to nucleate microtubules required for nuclear positioning (Zhou et al., 2009). Localization of γ-tubulin to the cell–cell boundaries could create the noncentrosomal parallel microtubule arrays observed in these hyp7 precursors. Alternatively, γ-tubulin may be localized in puncta along the length of microtubules to allow nucleation from the sides of existing polymers. Cortical microtubule arrays established by this mechanism have been identified in plants (Joshi and Palevitz, 1996).
Figure 9.
γ-Tubulin localizes at cell–cell boundaries. (A and B) Immunostaining of anti–γ-tubulin in fixed wild-type embryos (left), costained with DAPI to visualize nuclei (middle), and merged (right). Dorsal view, anterior is to the left. (A) During nuclear migration, γ-tubulin localizes to hyp7 cell–cell boundaries. (B) In earlier embryos, γ-tubulin localizes primarily to centrosomes. Bar, 10 µm.
Discussion
We previously proposed a model for hyp7 nuclear migration based on the genetic analysis of terminal nuclear migration defects. The model posits that UNC-83 recruits both kinesin-1 and dynein to the outer nuclear membrane and transfers forces generated by the motors though UNC-84 to the nuclear lamina. In this model, kinesin-1 provides the major forces to move the nucleus, whereas dynein has a regulatory role in the process (Meyerzon et al., 2009; Fridolfsson et al., 2010). To test our model, live imaging of nuclear migration in C. elegans hyp7 precursors was used to characterize the dynamics of nuclear movement and to analyze microtubule organization. Our findings led to a better understanding of the mechanisms of how microtubule motors mediate nuclear migration.
UNC-83 and UNC-84 are required for active nuclear migration
Nuclei normally migrate across the length of C. elegans embryonic hyp7 precursors from one lateral side of the embryo to the other (Sulston et al., 1983). In unc-83 or unc-84 mutant embryos, nuclear migration is disrupted, resulting in nuclei wrongly positioned in the middle of the cell at the dorsal midline (Malone et al., 1999; Starr et al., 2001). Our DIC time-lapse imaging allowed us, for the first time, to study the dynamics of nuclear movements in unc-83 and unc-84 mutant embryos and to compare them with wild-type nuclear migrations.
In wild-type embryos, hyp7 precursor nuclei began to migrate as soon as intercalation completed and passed the midline of the cell ∼11 min later (Fig. 1). Nuclear migration was not a smooth continuous process. For brief periods, wild-type nuclei moved forward relatively fast. Additionally, they often backed up, showing bidirectional movements. Filming of lamin::GFP in wild-type hyp7 precursors showed that during nuclear migration, the nuclear envelope underwent many deformations. Presumably, motors pulling on the nuclear envelope caused these deformations. Consistent with this hypothesis, deformations of the nuclear envelope were absent in unc-83 mutant embryos (Fig. 1). The discontinuous bidirectional movements of nuclei were similar to many other cargos transported by microtubule motors. These data support the first model discussed in the Introduction in which kinesin-1 and dynein function during nuclear migration to mediate bidirectional movements.
The timing of when unc-83 or unc-84 mutant nuclei move to the dorsal midline was significantly slower (at least 25 min slower and sometimes much longer; Fig. 1) than in wild-type embryos. The late nuclear movements in mutant embryos coincided with the morphogenesis of hyp7 precursors, which elongated into long, thin sheets, limiting the space for nuclei. At approximately the same time, body wall muscle cells that are initially positioned laterally begin to migrate dorsally (Hresko et al., 1994). The body wall muscles squeeze the hypodermal cells and establish linkages through the hypodermis to the cuticle (Francis and Waterston, 1991). We propose that migration of the underlying body wall muscle cells and/or morphogenesis of the hyp7 precursors pushes hyp7 nuclei to the dorsal cord in unc-83 and unc-84 mutant embryos.
Microtubules during nuclear migration are dynamic and polarized
Before this study, our working model was that nuclear migration is mediated by microtubule motors recruited to the surface of the nucleus by UNC-83 (Meyerzon et al., 2009; Fridolfsson et al., 2010). The model predicted that nuclei migrate along a stable, polarized bundle of microtubules. However, the model relied on fixed images of microtubules taken during hyp7 precursor intercalation and nuclear migration (Williams-Masson et al., 1998). Imaging our GFP::β-tubulin lines confirmed that before intercalation, microtubules were organized as a meshwork. As the cells elongated, microtubules formed long parallel bundles that were denser at the cell–cell boundaries. Surprisingly, we found that the microtubule bundles at the time of nuclear migration were very dynamic (Fig. 7), suggesting that the polarity of the cells is not maintained by stable microtubules formed at the time of intercalation. Instead, our EBP-1::GFP videos showed that microtubule growth was polar; most microtubules grew in the direction of nuclear migration (Fig. 8). Such a polar microtubule network is ideally organized for nuclei to move via forces generated by kinesin-1 at the surface of the nucleus.
In many organisms, nuclear migration events are led by the centrosome, which usually remains closely attached to the nucleus (for review see Starr, 2009). For example, in migrating neurons, the centrosome polarizes to the front of the nucleus and leads nuclear migration (Tsai et al., 2007). However, hyp7 nuclei migrate in a centrosome-independent manner. We found that centrosomes remained attached to the nucleus during migration but were randomly positioned with respect to the direction of migration (Fig. S2). This is consistent with our findings that γ-tubulin was broadly localized (Fig. 9), leading to a model in which most microtubules nucleate far from the centrosome during nuclear migration. Thus, before nuclear migration, the microtubule network transformed from a meshwork to dynamic, centrosome-independent bundles of polarized microtubules. A similar switch in microtubule organization occurs during mammalian epidermal differentiation (Lechler and Fuchs, 2007; Meng et al., 2008). Furthermore, the linear arrays in mammalian epithelial cells are polarized in a way that has been hypothesized to allow nesprin-4– and kinesin-1–mediated positioning of the nucleus (Roux et al., 2009).
Dynein is required for nuclear migration past roadblocks
There are many examples of opposing motors being recruited to the same cargo, but the mechanisms of how kinesin-1 and dynein activities are coordinated to move the cargo are mostly unknown. Models to explain how microtubule motors of opposite directionality function together to move a single cargo include the bidirectional model, which would allow cargo to pass obstacles, change microtubule tracks, or correct errors (Welte, 2004; Ross et al., 2008); the interdependent model, in which the disruption of one motor affects movement in both directions (Martin et al., 1999; Shubeita et al., 2008; Ally et al., 2009); and the drag model, in which dynein acts to slow kinesin-1–mediated transport (Del Bene et al., 2008). We could not film nuclear migration in dynein heavy chain dhc-1 mutant embryos. DHC-1 is required for early embryonic development (Gönczy et al., 1999), making it difficult to directly test the role of dynein in hyp7 nuclear migration (Fridolfsson et al., 2010). Instead, we focused on the phenotypes of nud-2 and bicd-1 mutants that complete early development normally (Fridolfsson et al., 2010). NUD-2 and BICD-1, the NudE and Bicaudal D homologues, are not known to function outside of dynein regulation (Kardon and Vale, 2009) and, therefore, make the best approximation of a dynein disruption that we could study in vivo and use to discriminate between these three models. We predicted that if the bidirectional model were true, dynein disruption would affect backward movements; in the interdependent model, dynein disruption should block all movement, and in the drag model, dynein disruption should speed up nuclear movements. We showed that neither the initiation nor the rate of nuclear migration was affected by the disruption of dynein, but backward movements were (Figs. 2, 5, and S1; and Table I). Thus, our data favor the first model, in which dynein is used for brief backward movements to allow kinesin-1–driven nuclear migration to move past roadblocks in the cell.
More specifically, our data suggest that dynein functions to ensure efficient nuclear migration by facilitating nuclear movement past blockages of cytoplasmic granules and other organelles that kinesin-1 cannot resolve on its own. During hyp7 nuclear migration, the nucleus is nearly as large as the width of the cell. Our DIC videos showed that as the nucleus migrates, granules and other organelles accumulate in front of the nucleus. Simply plowing forward is likely not the most efficient way to move nuclei; the nucleus must allow other organelles to pass. We propose that dynein resolves the largest accumulations of organelles in front of nuclei. Dynein briefly backs up or otherwise shifts the nucleus to allow organelles to pass. In the most severe cases, dynein rolls nuclei past the blockage of granules (Fig. 4). Aggregates of organelles only completely disrupt the nuclear migration in a minority of events; most of the time, kinesin-1 is sufficient to move past granules. However, 16.7% of nuclei in wild-type embryos roll (Table II), which we propose is a mechanism to release the build up of organelles in front of the migrating nucleus. This model explains why dynein mutants only block a small percentage of nuclear migrations (Fridolfsson et al., 2010; this study). In support of this model, we found a strong correlation between failure to migrate properly and lack of rolling. In nud-2(ok949) and nud-2(ok949);bicd-1(RNAi) mutants, nuclei began to migrate normally but often failed to complete the migration. Those that failed to complete migration never rolled (Table II). We sometimes observed nuclei that appeared to migrate normally to the midline but then rapidly returned to their starting point (Video 7, nucleus 11). Often, immediately preceding the backward movement of such nuclei, the accumulation of granules in front of the nucleus is visibly compressed by the nucleus attempting to move forward (Video 7, left-marked nucleus in seconds 11–14). Therefore, we propose that the backward movements of nud-2(ok949) and nud-2(ok949);bicd-1(RNAi) mutant nuclei after they started normal-looking migrations were caused by a failure to resolve cellular blockages.
It is likely that BICD-1 and NUD-2 function through dynein at the nuclear envelope (Kardon and Vale, 2009; Fridolfsson et al., 2010). Dynein has previously been shown to play a role in nuclear rotation in 3T3 fibroblasts, although the rotation events in tissue culture cells are considerably slower (angular velocities of 2–5°/min; Paddock and Albrecht-Buehler, 1986a,b; Levy and Holzbaur, 2008) than the ones described in this study (∼30°/min; Table II). We propose that dynein causes the nucleus to roll by actively pulling it backward or holding one side in place while kinesin-1 continues to pull the other side forward. One film of lamin::GFP showed a spot on the nuclear envelope rapidly move opposite to the direction of migration, which appeared to cause the nucleus to roll (Fig. 4 B). Based on the polarity of microtubules, dynein present on the nuclear envelope would be ideally positioned to cause such a movement and the subsequent rolling of the nucleus. Alternatively, disruptions of klc-2, nud-2, or bicd-1 could lead to global trafficking defects that contribute to the nuclear migration defect. However, klc-2(km11) mutant embryos had no detectable effect on the organization of microtubules (Fig. 8). Furthermore, BICD-1 and NUD-2 are only required for a subset of dynein’s phenotypes (Fridolfsson et al., 2010). Because the phenotypes of klc-2, nud-2, or bicd-1 mutants mimic unc-83 nuclear migration phenotypes, it is most likely that the motor mutant defects are caused by a loss of motor activity on the cytoplasmic face of the nucleus.
In conclusion, our data suggest that UNC-83 functions to mediate nuclear migration by recruiting both kinesin-1 and dynein to the nuclear envelope. Kinesin-1 then provides the major forces to move nuclei along a dynamic, polarized microtubule network. Dynein is required in a subset of nuclear migrations. Our working model is that dynein mediates backward movements and/or rolling to allow the nucleus to bypass roadblocks in the cell.
Materials and methods
C. elegans strains
C. elegans were cultured using standard conditions, and N2 was used as wild type (Brenner, 1974). The unc-83(e1408), klc-2(km11), and nud-2(ok949) alleles were described previously (Starr et al., 2001; Sakamoto et al., 2005; Fridolfsson et al., 2010). The AJM-1::GFP strain was used to identify adherens junctions (Raich et al., 1999). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. Transgenic lines expressing GFP markers or KLC-2::KASH hybrid proteins were created by standard DNA microinjection techniques using odr-1::rfp or sur-5::gfp as a transformation marker (Mello et al., 1991; Yochem et al., 1998; Sagasti et al., 2001) and were generated by injecting the following constructs into N2: pSL268 to make strain UD291 that expresses GFP::β-tubulin, pSL501 to make strain UD299 that expresses EB1::GFP, pSL521 to make strain UD324 that expresses lamin::GFP, and pSL516 to make strain UD320 that expresses an ER GFP marker. Injection of pSL501 (EB-1::GFP) into klc-1(km11) generated strain UD313. Injection of pSL440 (KLC-2::KASH; Meyerzon et al., 2009) into unc-83(e1408) generated strain UD303. UD324 (lamin::GFP) was crossed into unc-83(e1408) to create UD329. bicd-1(RNAi) was performed as previously described (Fridolfsson et al., 2010). Imaging of bicd-1(RNAi) embryos began 48 h after injection of dsRNA.
Molecular cloning of plasmids for GFP fusion proteins
Constructs used to express GFP fusion proteins in the hypodermis were generated as follows. The lbp-1 promoter, 1,056 bp upstream of the predicted ATG, was cut out of pKK1 (provided by J. Plenefisch, University of Toledo, Toledo, OH; Plenefisch et al., 2000) with HindIII and XbaI and inserted into the corresponding sites of pPD49.26 or pPD97.55 to make pSL246 and pSL500, respectively (Fire laboratory vector kit; Addgene). To express GFP::β-tubulin, gfp::tbb-2 was amplified by PCR from pJH4.66 (provided by G. Seydoux, Johns Hopkins University, Baltimore, MD; Strome et al., 2001) and cloned into the NheI site of pSL246 to create pSL268. A full-length cDNA for ebp-1 (yk807f9; provided by Y. Kohara, National Institute of Genetics, Shizuoka, Japan) was amplified by PCR and inserted into the XbaI and KpnI sites of pSL500 to make pSL501. A full-length cDNA of lmn-1 (yk97a11) was amplified by PCR and inserted into the KpnI site of pSL500 to make pSL521. The ER marker was generated by amplifying SP-12 (C34B2.10; provided by M. Rolls, Penn State University, University Park, PA; Rolls et al., 2002) genomic DNA and inserting it into the XbaI and KpnI sites of pSL500 to make pSL516.
Microscopy and image analysis
Embryos were dissected from gravid hermaphrodites and mounted on a 2% agar pad in M9 solution. Time-lapse microscopy of nuclear migration was observed using DIC optics and a 63× Plan Apo 1.40 NA objective on a compound microscope (DM 6000; Leica). Images were acquired using a camera (DC350 FX; Leica) and the LAS AF software (Leica) at a rate of 15 s/frame for at least 40 min or 200 ms/frame for ∼5 min, as indicated. Tracking analysis of the nucleus was performed by designating the center of the nucleus and the two ends of the hyp7 precursor using the Manual Tracking plugin for ImageJ (National Institutes of Health). Hyp7 precursors were numbered according to the designations given in Sulston et al. (1983). Peak velocities were calculated by kymographs created in ImageJ. Lamin::GFP was imaged at a rate of 1 s/frame using the same microscope.
Time-lapse microscopy of β-tubulin::GFP, SP-12::GFP, and EB1::GFP–expressing embryos was performed using the 488-nm laser on a spinning-disc confocal microscope (Marianas Real Time Confocal SDC Workstation; Intelligent Imaging Innovations) using a scan head (CSU-X1; Yokogawa) and a modified electron multiplying charge-coupled device camera (Cascade QuantEM 512SC; Photometrics). Objective lenses used were either 63× NA 1.4 (Carl Zeiss, Inc.) or 100× NA 1.46 oil immersion (Carl Zeiss, Inc.). Images were acquired using SlideBook software (version 5.0; Intelligent Imaging Innovations) at a rate of ∼15 s/frame (β-tubulin::GFP and SP-12::GFP) or ∼100 ms/frame (EB1::GFP).
FRAP experiments were performed with a point-scanning laser confocal microscope (FV1000; Spectral Scan; Olympus) equipped with a 60× 1.42 NA oil objective and Fluoview software (version 1.7; Olympus). The embryos expressing β-tubulin::GFP were imaged using the 488-nm argon laser. A second scan head on the FV1000 equipped with a 405-nm diode laser was used to photobleach, which allowed simultaneous imaging and bleaching. The spindle was bleached in rectangular areas, and images were acquired every 0.7 s. The fluorescence intensities within the bleached region were measured using ImageJ. The fluorescence intensity of the bleached region over time was normalized with the prebleached fluorescence intensity corrected for bleaching and was plotted as a function of time. The recovery half-time was obtained by fitting a single exponential curve = F0 + (Finf − F0) (1 − e−kt) to the recovery time course curve (Salmon et al., 1984).
Antibodies and immunofluorescence
For immunofluorescence, late embryos were extruded from slightly starved hermaphrodites, permeabilized by the freeze-crack method, fixed for 10 min in −20°C methanol, and blocked in PBST (PBS and 0.1% Triton X-100) with 5% dry milk (Miller and Shakes, 1995). The fixed specimens were stained as described previously (Miller and Shakes, 1995). The mouse monoclonal antibody IFA diluted 1:50 in PBS was used to detect embryonic centrosomes (Leung et al., 1999). Adherens junctions were detected using rabbit polyclonal antibody NB600-308 against GFP at a 1:500 dilution (Novus Biologicals). Rabbit polyclonal antibody T1450 was used against γ-tubulin at a 1:200 dilution (Sigma-Aldrich; Ow et al., 2008). Cy3-conjugated donkey anti–rabbit or anti–mouse IgG and Cy2-conjugated donkey anti–mouse or anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.) diluted 1:200 in PBS were used as secondary antibodies. DNA was visualized by a 10-min stain in 1 µg/ml DAPI in PBS.
Online supplemental material
Fig. S1 shows that wild-type nuclei move bidirectionally but nud-2(ok949);bicd-1(RNAi) and unc-83(e1408);klc-2::KASH nuclei do not. Fig. S2 shows that centrosome position is random during nuclear migration. Video 1 shows hyp7 precursor intercalation and nuclear migration in a wild-type embryo imaged by DIC as in Fig. 1 A. Video 2 shows nuclear migration through the ER in a wild-type embryo imaged with SP-12::GFP as in Fig. 3. Video 3 shows nuclear rolling during hyp7 nuclear migration imaged by DIC as in Fig. 4 A. Video 4 shows nuclear rolling during hyp7 nuclear migration imaged with lamin::GFP as in Fig. 4 B. Video 5 shows hyp7 intercalation and nuclear migration in an unc-83(e1408) embryo imaged by DIC as in Fig. 1 B. Video 6 shows hyp7 intercalation and nuclear migration in a klc-2(km11) embryo imaged by DIC as in Fig. 5 A. Video 7 shows hyp7 intercalation and nuclear migration in a nud-2(ok949) embryo imaged by DIC. Video 8 shows hyp7 intercalation and nuclear migration in a nud-2(ok949);bicd-1(RNAi) embryo imaged by DIC as in Fig. 5 B. Video 9 shows hyp7 intercalation and nuclear migration in an unc-83(e1408);klc-2::KASH embryo imaged by DIC as in Fig. 6. Video 10 shows microtubule organization early in intercalation and migration imaged by GFP::β-tubulin as in Fig. 7. Video 11 shows direction of microtubule growth early in intercalation imaged by EBP-1::GFP as in Fig. 8 A. Video 12 shows direction of microtubule growth during nuclear migration imaged by EBP-1::GFP as in Fig. 8 B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004118/DC1.
Acknowledgments
We thank Michael Paddy and Daniel Elnatan (University of California, Davis, Davis, CA) for technical support with microscopy. We thank John Plenefisch for providing the lbp-1 promoter. We thank Erin Tapley and Lesilee Rose (University of California, Davis) for helpful comments on the manuscript.
This research was supported by the National Institutes of Health (grant R01GM073874). The spinning-disc confocal microscope was purchased with an equipment grant from the National Institutes of Health (grant S10RR024543), and H.N. Fridolfsson was supported by a National Institutes of Health training grant (5YT32GM007377).
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- KASH
- klarsicht/ANC-1/Syne homology
- SUN
- Sad1p/UNC-84
References
- Ally S., Larson A.G., Barlan K., Rice S.E., Gelfand V.I. 2009. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J. Cell Biol. 187:1071–1082 10.1083/jcb.200908075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Roux K.J. 2009. Nuclei take a position: managing nuclear location. Dev. Cell. 17:587–597 10.1016/j.devcel.2009.10.018 [DOI] [PubMed] [Google Scholar]
- Cheerambathur D.K., Civelekoglu-Scholey G., Brust-Mascher I., Sommi P., Mogilner A., Scholey J.M. 2007. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 177:995–1004 10.1083/jcb.200611113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 125:59–69 10.1016/j.cell.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Crisp M., Burke B. 2008. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 582:2023–2032 10.1016/j.febslet.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F., Wehman A.M., Link B.A., Baier H. 2008. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 134:1055–1065 10.1016/j.cell.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 12:863–872 10.1016/j.devcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Francis R., Waterston R.H. 1991. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114:465–479 10.1083/jcb.114.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson H.N., Ly N., Meyerzon M., Starr D.A. 2010. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol. 338:237–250 10.1016/j.ydbio.2009.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., Gundersen G.G. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., Hyman A.A. 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147:135–150 10.1083/jcb.147.1.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F., Dupré N., Dion P., Fox M.A., Laurent S., Verreault S., Sanes J.R., Bouchard J.P., Rouleau G.A. 2007. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat. Genet. 39:80–85 10.1038/ng1927 [DOI] [PubMed] [Google Scholar]
- Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. 2006. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26:3738–3751 10.1128/MCB.26.10.3738-3751.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H.R., Sulston J.E. 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 96:435–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko M.C., Williams B.D., Waterston R.H. 1994. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 124:491–506 10.1083/jcb.124.4.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi H.C., Palevitz B.A. 1996. gamma-Tubulin and microtubule organization in plants. Trends Cell Biol. 6:41–44 10.1016/0962-8924(96)81008-7 [DOI] [PubMed] [Google Scholar]
- Kandert S., Lüke Y., Kleinhenz T., Neumann S., Lu W., Jaeger V.M., Munck M., Wehnert M., Müller C.R., Zhou Z., et al. 2007. Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum. Mol. Genet. 16:2944–2959 10.1093/hmg/ddm255 [DOI] [PubMed] [Google Scholar]
- Kardon J.R., Vale R.D. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 133:1188–1201 10.1016/j.cell.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. 2007. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J. Cell Biol. 176:147–154 10.1083/jcb.200609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B., Hermann G.J., Priess J.R. 1999. Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216:114–134 10.1006/dbio.1999.9471 [DOI] [PubMed] [Google Scholar]
- Levy J.R., Holzbaur E.L. 2008. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121:3187–3195 10.1242/jcs.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Fixsen W.D., Horvitz H.R., Han M. 1999. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 126:3171–3181 [DOI] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- Marmé A., Zimmermann H.P., Moldenhauer G., Schorpp-Kistner M., Müller C., Keberlein O., Giersch A., Kretschmer J., Seib B., Spiess E., et al. 2008. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int. J. Cancer. 123:2048–2056 10.1002/ijc.23763 [DOI] [PubMed] [Google Scholar]
- Martin M., Iyadurai S.J., Gassman A., Gindhart J.G., Jr., Hays T.S., Saxton W.M. 1999. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell. 10:3717–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee M.D., Rillo R., Anderson A.S., Starr D.A. 2006. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell. 17:1790–1801 10.1091/mbc.E05-09-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W., Mushika Y., Ichii T., Takeichi M. 2008. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 135:948–959 10.1016/j.cell.2008.09.040 [DOI] [PubMed] [Google Scholar]
- Meyerzon M., Fridolfsson H.N., Ly N., McNally F.J., Starr D.A. 2009. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 136:2725–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M., Shakes D.C. 1995. Immunofluorescence microscopy. Methods Cell Biol. 48:365–394 10.1016/S0091-679X(08)61396-5 [DOI] [PubMed] [Google Scholar]
- Morrison E.E., Wardleworth B.N., Askham J.M., Markham A.F., Meredith D.M. 1998. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 17:3471–3477 10.1038/sj.onc.1202247 [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop K.L., Li Q., Patterson L., Fischer J.A. 1999. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9:1211–1220 10.1016/S0960-9822(99)80501-6 [DOI] [PubMed] [Google Scholar]
- Motegi F., Velarde N.V., Piano F., Sugimoto A. 2006. Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Dev. Cell. 10:509–520 10.1016/j.devcel.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Murphy S.M., Stearns T. 1996. Cytoskeleton: microtubule nucleation takes shape. Curr. Biol. 6:642–644 10.1016/S0960-9822(09)00437-0 [DOI] [PubMed] [Google Scholar]
- Ostlund C., Folker E.S., Choi J.C., Gomes E.R., Gundersen G.G., Worman H.J. 2009. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 122:4099–4108 10.1242/jcs.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J.A., Rossing M.A., Cushing-Haugen K.L., Chen C., Van Den Berg D.J., Wu A.H., Pike M.C., Ness R.B., Moysich K., Chenevix-Trench G., et al. 2010. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiol. Biomarkers Prev. 19:245–250 10.1158/1055-9965.EPI-09-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Martinez N.J., Olsen P.H., Silverman H.S., Barrasa M.I., Conradt B., Walhout A.J., Ambros V. 2008. The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev. 22:2520–2534 10.1101/gad.1678808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock S.W., Albrecht-Buehler G. 1986a. The degree of coupling of nuclear rotation in binucleate 3T3 cells. Exp. Cell Res. 166:113–126 10.1016/0014-4827(86)90512-4 [DOI] [PubMed] [Google Scholar]
- Paddock S.W., Albrecht-Buehler G. 1986b. Distribution of microfilament bundles during rotation of the nucleus in 3T3 cells treated with monensin. Exp. Cell Res. 163:525–538 10.1016/0014-4827(86)90083-2 [DOI] [PubMed] [Google Scholar]
- Padmakumar V.C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A.A., Gotzmann J., Foisner R., Karakesisoglou I. 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118:3419–3430 10.1242/jcs.02471 [DOI] [PubMed] [Google Scholar]
- Plenefisch J., Xiao H., Mei B., Geng J., Komuniecki P.R., Komuniecki R. 2000. Secretion of a novel class of iFABPs in nematodes: coordinate use of the Ascaris/Caenorhabditis model systems. Mol. Biochem. Parasitol. 105:223–236 10.1016/S0166-6851(99)00179-6 [DOI] [PubMed] [Google Scholar]
- Puckelwartz M.J., Kessler E., Zhang Y., Hodzic D., Randles K.N., Morris G., Earley J.U., Hadhazy M., Holaska J.M., Mewborn S.K., et al. 2009. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet. 18:607–620 10.1093/hmg/ddn386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich W.B., Agbunag C., Hardin J. 1999. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr. Biol. 9:1139–1146 10.1016/S0960-9822(00)80015-9 [DOI] [PubMed] [Google Scholar]
- Reed N.A., Cai D., Blasius T.L., Jih G.T., Meyhofer E., Gaertig J., Verhey K.J. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16:2166–2172 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Rolls M.M., Hall D.H., Victor M., Stelzer E.H., Rapoport T.A. 2002. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell. 13:1778–1791 10.1091/mbc.01-10-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J.L., Ali M.Y., Warshaw D.M. 2008. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 20:41–47 10.1016/j.ceb.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., Burke B. 2009. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 106:2194–2199 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A., Hisamoto N., Hyodo J., Tanaka-Hino M., Matsumoto K., Bargmann C.I. 2001. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 105:221–232 10.1016/S0092-8674(01)00313-0 [DOI] [PubMed] [Google Scholar]
- Sakamoto R., Byrd D.T., Brown H.M., Hisamoto N., Matsumoto K., Jin Y. 2005. The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization. Mol. Biol. Cell. 16:483–496 10.1091/mbc.E04-07-0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E.D., Leslie R.J., Saxton W.M., Karow M.L., McIntosh J.R. 1984. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 99:2165–2174 10.1083/jcb.99.6.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., Dernburg A.F. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubeita G.T., Tran S.L., Xu J., Vershinin M., Cermelli S., Cotton S.L., Welte M.A., Gross S.P. 2008. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 135:1098–1107 10.1016/j.cell.2008.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A.G., Orita S., Malone C.J., Han M. 2001. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 98:13132–13137 10.1073/pnas.241504098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A. 2009. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122:577–586 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Han M. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 298:406–409 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Han M. 2005. A genetic approach to study the role of nuclear envelope components in nuclear positioning. Novartis Found. Symp. 264:208–219, discussion :219–230 10.1002/0470093765.ch14 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Hermann G.J., Malone C.J., Fixsen W., Priess J.R., Horvitz H.R., Han M. 2001. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 128:5039–5050 [DOI] [PubMed] [Google Scholar]
- Stearns T., Kirschner M. 1994. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 76:623–637 10.1016/0092-8674(94)90503-7 [DOI] [PubMed] [Google Scholar]
- Strome S., Powers J., Dunn M., Reese K., Malone C.J., White J., Seydoux G., Saxton W. 2001. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell. 12:1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. 1981. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82:41–55 10.1016/0012-1606(81)90427-9 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Bremner K.H., Vallee R.B. 2007. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10:970–979 10.1038/nn1934 [DOI] [PubMed] [Google Scholar]
- Vallee R.B., Tsai J.W. 2006. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 20:1384–1393 10.1101/gad.1417206 [DOI] [PubMed] [Google Scholar]
- Welte M.A. 2004. Bidirectional transport along microtubules. Curr. Biol. 14:R525–R537 10.1016/j.cub.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Masson E.M., Heid P.J., Lavin C.A., Hardin J. 1998. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev. Biol. 204:263–276 10.1006/dbio.1998.9048 [DOI] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M. 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 149:1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bethmann C., Worth N.F., Davies J.D., Wasner C., Feuer A., Ragnauth C.D., Yi Q., Mellad J.A., Warren D.T., et al. 2007. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 16:2816–2833 10.1093/hmg/ddm238 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 64:173–187 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Rolls M.M., Hall D.H., Malone C.J., Hanna-Rose W. 2009. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 186:229–241 10.1083/jcb.200902101 [DOI] [PMC free article] [PubMed] [Google Scholar]