Expression levels of SF2/ASF are controlled by Sam68 mediated activation of splicing-induced mRNA decay.
Abstract
Epithelial-to-mesenchymal transition (EMT) and its reversal (MET) are crucial cell plasticity programs that act during development and tumor metastasis. We have previously shown that the splicing factor and proto-oncogene SF2/ASF impacts EMT/MET through production of a constitutively active splice variant of the Ron proto-oncogene. Using an in vitro model, we now show that SF2/ASF is also regulated during EMT/MET by alternative splicing associated with the nonsense-mediated mRNA decay pathway (AS-NMD). Overexpression and small interfering RNA experiments implicate the splicing regulator Sam68 in AS-NMD of SF2/ASF transcripts and in the choice between EMT/MET programs. Moreover, Sam68 modulation of SF2/ASF splicing appears to be controlled by epithelial cell–derived soluble factors that act through the ERK1/2 signaling pathway to regulate Sam68 phosphorylation. Collectively, our results reveal a hierarchy of splicing factors that integrate splicing decisions into EMT/MET programs in response to extracellular stimuli.
Introduction
Alternative splicing (AS) is a key molecular mechanism in regulating gene expression and proteomic diversity. Recently, it has been shown that ∼95% of human genes undergo at least one AS event (Pan et al., 2008; Wang et al., 2008). Thus, AS is expected to play crucial roles in important biological events such as development, cell differentiation, organogenesis, and the response to environmental cues. Inclusion or exclusion of exons and alternative choices of 5′ or 3′ splice sites can give rise to different mRNAs from a single gene, which will be then translated into proteins with distinct functional activity or cellular localization. In addition, AS can affect the RNA stability by inclusion of premature translation termination codons (PTCs) that activate the nonsense-mediated mRNA decay (NMD) pathway (Isken and Maquat, 2008). In mammals, a termination codon is recognized as premature when located >50–55 nt upstream of the exon–exon junction marked by the exon junction complex (EJC). EJCs downstream of PTCs are no longer removed during the “pioneer” round of translation, and recruit essential NMD factors, including Upf1/Rent1, that in turn promote mRNA degradation (Isken and Maquat, 2008). Initially, NMD was considered only a mechanism for disposing of aberrant mRNAs arising from nonsense codon–containing alleles. It is now evident that NMD is involved in quantitative posttranscriptional regulation of gene expression through specific AS events (AS-activated NMD [AS-NMD], also termed regulated unproductive splicing and translation; Hillman et al., 2004). Recently, AS-NMD has been shown to regulate expression of specific gene families. Alternative “poison” exons containing premature in-frame stop codons, or introns in the 3′ untranslated region (UTR), are particularly frequent in mammalian genes for splicing regulators, such as SR factors and heterogeneous nuclear RNP (hnRNP) proteins (Lareau et al., 2007; Ni et al., 2007) and for many core spliceosomal proteins (Saltzman et al., 2008). Most of these proteins can regulate their own mRNA level by modulating AS-NMD in a feedback mechanism designed to maintain the protein homeostatic level (Saltzman et al., 2008). Interestingly, in several genes for SR factors and hnRNPs, AS-NMD cassettes overlap highly conserved or ultraconserved elements (UCEs; Lareau et al., 2007; Ni et al., 2007) longer than 200 bp, with 100% identity among rat, mouse, and human genomes. However, the role of UCEs in AS-NMD and their contribution to diseases, including cancer, is still hypothetical.
Epithelial-to-mesenchymal transition (EMT) is an important cell reorganization event that is crucially connected to developmental programs and occurs several times during embryogenesis in vertebrates. In adults, EMT is confined to a few physiological processes such as tissue regeneration and wound healing (Thiery and Sleeman, 2006). Notably, this program occurs during the progression of epithelial tumors and confers migratory and invasive properties to cancer cells (Thiery and Sleeman, 2006; Polyak and Weinberg, 2009). EMT describes a complex series of events during which epithelial cells lose many of their distinguishing features to acquire typical mesenchymal traits. Epithelial cells are rich in adherens junctions that are required for the formation of continuous cell layers. During EMT, epithelial cells acquire fibroblast-like morphology with a rearrangement of cytoskeletal architecture, reduced intercellular adhesion, and increased ability to invade nearby body districts (Thiery and Sleeman, 2006; Polyak and Weinberg, 2009). A variety of oncogenic pathways, activated by peptide growth factors, Src, Ras, ETS, integrin, Wnt/β-catenin, and Notch signaling, induce EMT (Thiery, 2003). A crucial event during EMT is the down-regulation of E-cadherin, a component of adherens junctions, which acts as a de facto tumor suppressor, inhibiting invasion and metastasis, and is frequently repressed or degraded during transformation. Down-regulation of E-cadherin is accompanied by increased expression of N-cadherin and vimentin by nuclear accumulation of β-catenin, and increased production of transcription factors, such as Snail and Slug, that inhibit E-cadherin production (Polyak and Weinberg, 2009). Phenotypic markers of EMT include an increased capacity of cell migration and three-dimensional invasion, as well as resistance to anoikis/apoptosis. Intriguingly, EMT is a transient process that occurs in a subset of cells at the invasive front of the metastasizing primary carcinomas and is reversed during the establishment of metastasis (Brabletz et al., 2001; Thiery and Sleeman, 2006). These cell morphology changes appear to be driven by several signals in the tumor microenvironment (Polyak and Weinberg, 2009). We have previously shown that EMT may be triggered by splicing factor and proto-oncogene SF2/ASF through the AS of the Ron proto-oncogene (Ghigna et al., 2005; Karni et al., 2007), which encodes the tyrosine kinase receptor for the macrophage-stimulating protein involved in control of cell scattering and motility (Trusolino and Comoglio, 2002). SF2/ASF overexpression promotes skipping of Ron exon 11 and results in the production of ΔRon, a constitutively active isoform that confers an invasive phenotype to the cells (Ghigna et al., 2005). In this paper, we have exploited an in vitro model to further investigate the impact of AS programs on EMT. EMT can be recapitulated in vitro by growing colon adenocarcinoma cells at different densities: at low density, sparse cells mimic the situation occurring at the invasive front of the tumor and display a mesenchymal-like phenotype, whereas at high density, the same cells switch to an epithelial-like phenotype (Brabletz et al., 2001). We have found that the level of splicing factor SF2/ASF is controlled during in vitro EMT through AS-NMD. This event is regulated by diffusible factors expressed by epithelial cells through the ERK1/2 pathway and the Sam68 splicing regulator.
Results
AS of the Ron gene and expression level of SF2/ASF are regulated during in vitro EMT
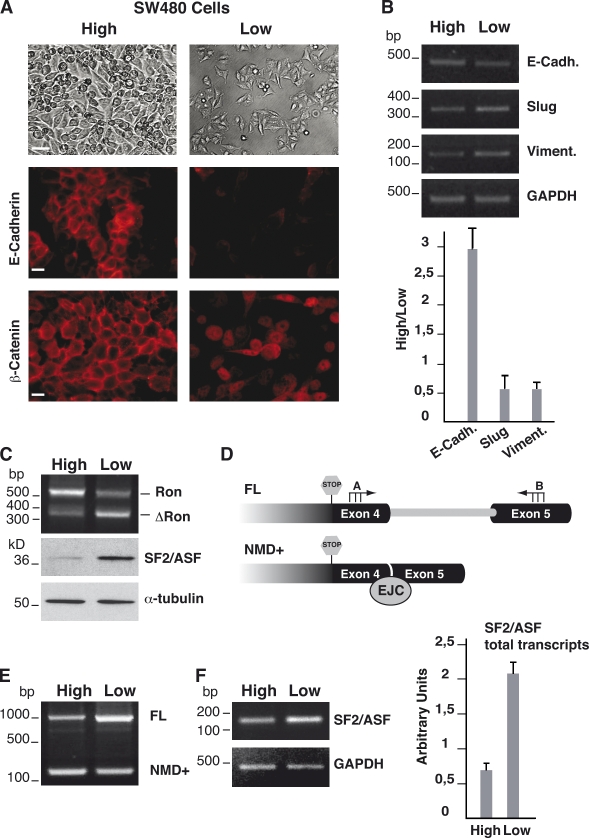
Morphological and molecular hallmarks of EMT can be recapitulated in vitro by growing colon adenocarcinoma SW480 cells at different densities: high-density (H) cells are similar to the central part of the tumor and display an epithelial-like phenotype, whereas low-density (L) cells resemble the invasive front of the tumor and have a mesenchymal-like phenotype (Brabletz et al., 2001). As shown in Fig. 1 A, L cells are poorly stained by an antibody against the epithelial marker E-cadherin and display nuclear accumulation of β-catenin. Accordingly, expression of the E-cadherin gene is drastically higher in H cells than in L cells (Fig. 1 B). On the contrary, expression of vimentin and Slug genes, two typical markers of mesenchymal cells, increases during EMT and is higher in L cells (Fig. 1 B). We decided to study the splicing profile of Ron transcripts in this in vitro model of EMT. As shown in Fig. 1 C, cell density affects the splicing profile of Ron exon 11, and the ΔRon mRNA is the prevailing splicing isoform in sparse mesenchymal-like L cells, which is consistent with the ability of this splicing isoform to induce EMT. We have previously shown that the production of the ΔRon isoform correlates with the level of the splicing factor SF2/ASF (Ghigna et al., 2005). In agreement with this, the switch in the splicing profile of the Ron gene transcripts driven by cell density is paralleled by a higher level (4.5-fold) of SF2/ASF in L cells compared with that in H cells (Fig. 1 C).
Figure 1.
Splicing profile of Ron and SF2/ASF transcripts during in vitro EMT. (A) H (High) and L (Low) cells were analyzed in immunofluorescence for the expression of E-cadherin and the distribution of β-catenin (bottom two rows; magnification 60×; bar, 10 µm); cell morphologies were examined under a phase contrast microscope (top row; magnification 10×; bar, 30 µm). (B) RT-PCR analysis of indicated EMT markers in SW480 cells at different densities. The histogram shows the ratio between the mRNA levels quantified in H and L cells by qRT-PCR. Error bars indicate mean ± SD. (C) RT-PCR analysis of Ron splicing and Western blotting of SF2/ASF and α-tubulin proteins in SW480 cells at different densities. (D) Schematic representation of AS in the 3′ UTR of SF2/ASF transcripts. Intron retention generates the stable FL transcript of SF2/ASF, whereas splicing leads to the assembly of an EJC downstream of the natural stop codon (NMD+ transcript). The position of primers ASF-A and -B is indicated. (E) Splicing profile of SF2/ASF transcripts in H and L SW480 cells. Total RNAs were analyzed by RT-PCR with primers ASF-A and -B flanking the 3′ UTR-intron (see D). (F) RT-PCR analysis of RNAs with primers annealing to constitutive SF2/ASF exons (exons 2 and 3) and with primers for the GAPDH gene. The histogram shows the qRT-PCR analysis, with primers ASF-tot-for and -rev, of the total SF2/ASF transcripts in SW480 cells at different densities. Error bars indicate SD calculated from three independent experiments.
Expression of SF2/ASF during EMT is regulated through AS-NMD
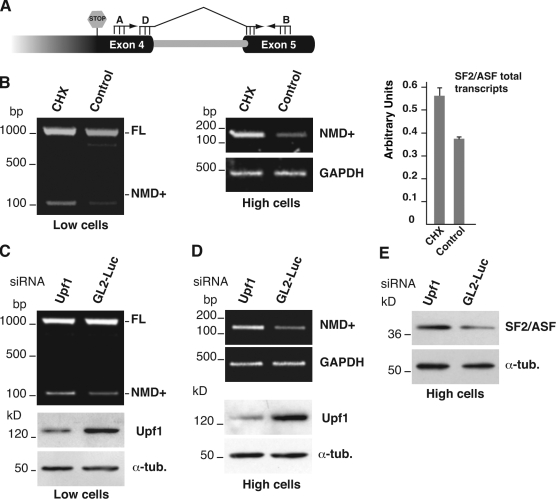
Recently, it has been found that AS-NMD regulates the expression of all SR splicing factors and several other RNA-binding proteins (Lareau et al., 2007; Ni et al., 2007; Saltzman et al., 2008). In particular, the 3′ UTR of the SF2/ASF gene contains an intron (3′ UTR-intron) that is normally retained in the mature transcript (full-length [FL] transcript). Splicing of this intron produces an RNA molecule (NMD+ transcript) in which the natural stop codon is recognized as a PTC and the mRNA is degraded by the NMD pathway (Fig. 1 D; Lareau et al., 2007). We asked whether this mechanism could be responsible for the lower level of SF2/ASF in H cells. RT-PCR analysis shows that splicing of the 3′ UTR-intron occurs in SW480 cells and is regulated during EMT: L cells express mainly the FL isoform, whereas the NMD+ transcript is predominant in H cells (Fig. 1 E). As predicted by the AS-NMD model, this splicing switch is accompanied by a lower level (about threefold as estimated by real-time quantitative RT-PCR [qRT-PCR] of SF2/ASF mRNAs in H cells (Fig. 1 F). NMD occurs during the pioneer round of translation, and NMD substrates are stabilized by the translation inhibitor cycloheximide (CHX; Isken and Maquat, 2008). To verify the involvement of the NMD pathway in the regulation of SF2/ASF mRNA levels, we treated SW480 cells with CHX. As shown in Fig. 2 B (left), CHX changes the ratio between the two splicing products in L cells and specifically increases the level of NMD+ transcripts. A similar effect of CHX is also detectable in H cells using the primer set D-B (Fig. 2 A), which specifically amplifies the NMD+ splicing product (Fig. 2 B, middle). Accordingly, qRT-PCR (Fig. 2 B, right) demonstrates that the total level of SF2/ASF transcripts in H cells increases when cells are treated with CHX.
Figure 2.
The NMD+ transcript of SF2/ASF is degraded by the NMD pathway. (A) Scheme of the SF2/ASF primers used in RT-PCR analysis. ASF-D is specific for the NMD+ transcript. (B) L cells (Low cells) were treated for 6 h with CHX, and RNAs were then analyzed by RT-PCR with primers ASF-A and -B (left). Control, untreated cells. H cells (High cells), treated with CHX for 6 h, were analyzed by RT-PCR with ASF-D and -B primers and with primers for the GAPDH gene (right). The histogram shows the relative quantification of the total SF2/ASF transcripts by qRT-PCR, performed as in Fig. 1 F, in H cells. Error bars indicate SD calculated from three independent experiments. (C) L cells were transfected with Upf1 siRNA or with GL2-Luciferase siRNA (GL2-Luc) and then analyzed in a Western blot with anti-Upf1 and anti–α-tubulin antibodies, and in RT-PCR with primers ASF-A and -B. (D) H cells, treated with the indicated siRNA oligos, were analyzed in RT-PCR with primers specific for the NMD+ transcript of SF2/ASF as in B or for GAPDH gene transcripts (E), and by Western blotting with anti–SF2/ASF and –α-tubulin antibodies.
To rule out that this result could be due to pleiotropic effects of CHX rather than stabilization of NMD+ transcripts, we specifically targeted the NMD pathway by siRNA-mediated knockdown of the key NMD player Upf1 (Isken and Maquat, 2008). Similarly to CHX, down-regulation of Upf1 changes the ratio between the two splicing products (Fig. 2 C) in L cells and augments the level of the NMD+ form in both L and H cells (Fig. 2 D). Consistent with this, down-regulation of Upf1 in H cells also increases the SF2/ASF protein level. Altogether, these results support the conclusion that AS-NMD plays a role in controlling the level of splicing factor SF2/ASF during in vitro EMT. Notably, this event is not limited to our in vitro cell system, and FL and NMD+ SF2/ASF splicing isoforms occur in normal human tissues as well (Fig. S2 A). The FL/NMD+ ratio varies in different tissues, which suggests that AS-NMD of SF2/ASF transcripts could play a physiological role in maintaining the homeostatic SF2/ASF level in different district of the organism. In this regard, it is worth noticing that the FL/NMD+ ratio is altered in a fraction of colon tumor specimens compared with normal control tissues (Fig. S2 B). An intriguing possibility, which deserves further investigation, is that this deregulation may have a role in the higher level of the oncogenic SF2/ASF protein described by Karni et al. (2007).
Soluble factors secreted by epithelial cells control AS-NMD of SF2/ASF transcripts and induce the mesenchymal-to-epithelial transition (MET)
Growth factors control the interaction and communication between epithelial and mesenchymal cells during development (Shannon and Hyatt, 2004). Recently, it has been reported that soluble factors secreted by tumor mammary mesenchymal cells regulate the AS profile of the fibronectin gene in normal mammary epithelial cells (Blaustein et al., 2004). Thus, we asked whether factors secreted by mesenchymal L cells or epithelial H cells could control splicing in the 3′ UTR of the SF2/ASF transcripts.
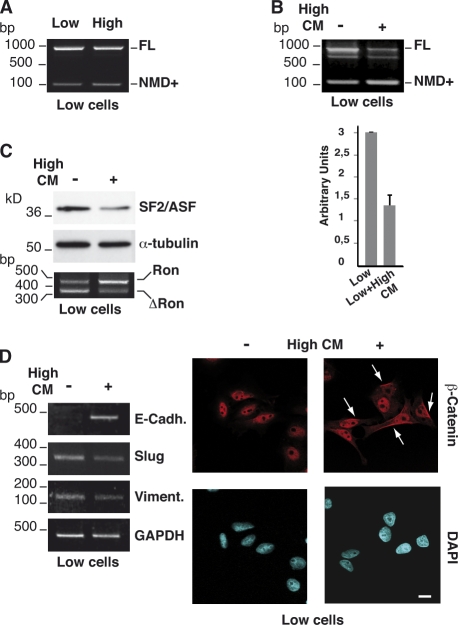
To reproduce the epithelial–mesenchymal interaction and assess its influence on AS, we performed co-culture experiments with tissue culture inserts that allow the interchange of soluble factors without cell–cell contacts. Interestingly, co-culture with H cells, plated on top of the insert, is sufficient to increase the level of the NMD+ transcripts in L cells in the lower chamber (Fig. 3 A), whereas co-culture with L cells had no effect on H cells (not depicted). It is plausible that this unidirectionality reflects the stoichiometry of factors expressed by L and H cells in co-culture experiments.
Figure 3.
Factors secreted by H cells control the AS of SF2/ASF transcripts and EMT markers in L cells. (A) Analysis of AS in SF2/ASF 3′ UTR in a co-culture experiment. L (Low) or H (High) cells (as indicated above the image of the gel) were grown on a porous filter placed inside a tissue culture well containing L cells. After 24 h, L cells in the bottom chamber were analyzed by RT-PCR as in Fig. 1 E. (B) L cells were plated in 3% serum into 35-mm tissue culture wells, and after 24 h, they were left untreated (−) or treated with conditioned medium from H cells (High-CM) for an additional 24 h. Total RNAs were analyzed by RT-PCR as in Fig. 1 E. The histogram shows the analysis of the total SF2/ASF transcripts by qRT-PCR in L cells treated or not treated with High-CM. Error bars indicate SD calculated from three independent experiments. (C) Total RNAs from L cells treated or not treated with High-CM were analyzed in a Western blot with SF2/ASF and α-tubulin antibodies and in RT-PCR for Ron splicing. (D) Total RNAs were also analyzed by RT-PCR with primers for the indicated EMT markers and GAPDH gene transcripts. The same cells were stained with an antibody specific to β-catenin and counterstained with DAPI (magnification 63×). Arrows indicate the cytoplasmic accumulation of β-catenin. Bar, 10 µm.
To further investigate this aspect, we grew H cells in the presence of conditioned medium from L cells and vice versa. In accord with the co-culture experiment, conditioned medium from L cells did not change the AS profile in the 3′ UTR of SF2/ASF mRNA (unpublished data), whereas conditioned medium from H cells (High-CM) shifts the splicing profile of SF2/ASF transcripts in L cells toward the production of the NMD+ form (Fig. 3 B), with a concomitant reduction of SF2/ASF mRNA (twofold) and protein (2.3-fold) levels and, as a consequence, of ΔRon transcripts (Fig. 3 C). More importantly, High-CM affected molecular markers that distinguish mesenchymal from epithelial cells, leading to the increased expression of E-cadherin and the concomitant reduction in the Slug and vimentin mRNA levels (Fig. 3 D). Moreover, a partial cytoplasmic accumulation of β-catenin was visible (Fig. 3 D, right). Collectively, these observations suggest that soluble factors released in the medium by high-density epithelial-like SW480 cells act dominantly in activating MET in L cells.
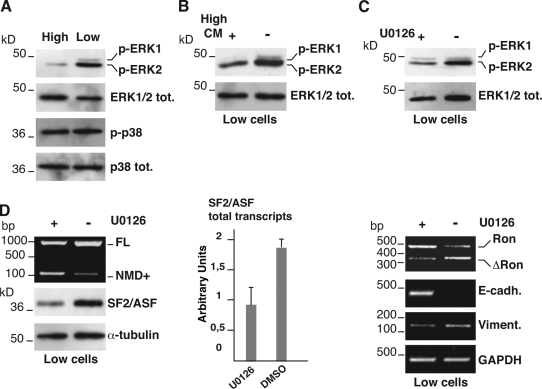
Soluble factors act through the ERK1/2 pathway to regulate AS-NMD of SF2/ASF transcripts and production of the ΔRon mRNA
Several secreted growth factors exert their action by activating the ERK1/2 pathway (Dhillon et al., 2007), which stimulates most forms of epithelial invasive motility that occur during cell scattering, wound healing, EMT, malignant invasion, and metastasis (Thiery and Sleeman, 2006). However, the mechanisms by which ERK1/2 controls motile and invasive capacities of epithelial cells are not well understood. We decided to investigate the involvement of the ERK1/2 pathway in the regulation of SF2/ASF splicing during EMT. As shown in Fig. 4 A, phosphorylation of ERK1/2 kinase is higher in L cells compared with H cells, whereas the phosphorylation status of the stress kinase p38, which identifies another pathway activated by growth factors, is not affected by cell density. Moreover, treatment of L cells with conditioned medium from H cells, as in Fig. 3, is accompanied by a decreased ERK1/2 phosphorylation (Fig. 4 B). Collectively these observations suggest an involvement of ERK1/2 in the regulation of AS-NMD of SF2/ASF transcripts. To directly verify this possibility, we specifically inhibited the ERK1/2 activity in L cells with the small molecule U0126 (Fig. 4 C). As shown in Fig. 4 D, U0126 changes the splicing profile of SF2/ASF mRNA in L cells and favors the production of the NMD+ isoform typical of epithelial H cells. As a result, the SF2/ASF protein level decreases and the splicing profile of Ron transcripts switches to the epithelial program with reduction of the ΔRon splicing product (Fig. 4 D). Importantly, inhibition of ERK1/2 in L cells activates the epithelial expression program characterized by increased expression of E-cadherin and down-regulation of vimentin mRNAs (Fig. 4 D). Altogether, our results implicate the ERK1/2 signaling cascade in the choice between the epithelial and mesenchymal status of SW480 cells. Inhibition of this pathway by soluble factors expressed by epithelial H cells or with the ERK1/2 inhibitor U0126 uncouples the occurrence of MET from cell density, and regulates AS-NMD of SF2/ASF transcripts and ΔRon mRNA production.
Figure 4.
Factors secreted by H cells act through the ERK1/2 pathway. (A) Western blot with phospho-specific (p-) and pan-antibodies (tot) against ERK1/2 and p38 kinases in SW480 cells grown at different densities. (B and C) Western blot analyses of the ERK1/2 phosphorylation status in L cells (Low) treated or untreated with High-CM (B), or treated with the specific ERK1/2 inhibitor U0126 (+) or with DMSO (−) (C) . (D) L cells treated with U0126 were also analyzed for the splicing profile of the 3′ UTR-intron (RT-PCR with primers ASF-A and -B), the total level of SF2/ASF transcripts by qRT-PCR (histogram), the SF2/ASF and α-tubulin protein levels, Ron splicing, and the mRNA expression levels of the indicated EMT markers and GAPDH gene. Error bars indicate SD calculated from three independent experiments.
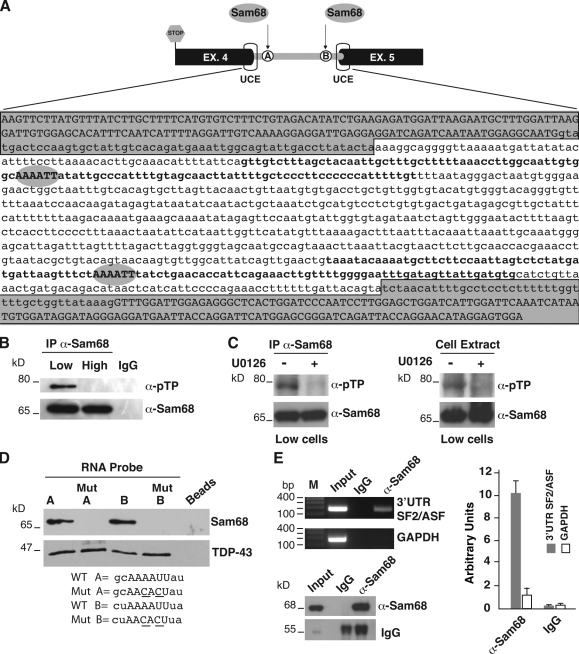
Sam68 interacts with the 3′ UTR of the SF2/ASF transcript
To understand the molecular mechanism regulating AS-NMD of SF2/ASF transcripts, we analyzed the SF2/ASF 3′ UTR sequence. This region contains two UCEs that overlap the exon junctions (Fig. 5 A). Two putative binding sites (AAAAUU) for the splicing regulator Sam68 (Matter et al., 2002) are located near the UCEs (A and B in Fig. 5 A). Sam68, the 68-kD Src-associated protein in mitosis, is a member of the STAR (signal transduction activator of RNA) family of RNA-binding proteins proposed to link signaling cascades to RNA metabolism (Lukong and Richard, 2003). Notably, Sam68 is a substrate of ERK1/2 kinase and controls splicing of exon v5 of the CD44 gene after Ras activation (Matter et al., 2002). In agreement with the activation of the ERK1/2 pathway, we found that Sam68 is hyperphosphorylated in L cells (Fig. 5 B), and its phosphorylation level is reduced upon treatment with the extracellular signal–regulated kinase (ERK) inhibitor U0126 (Fig. 5 C).
Figure 5.
Sam68 binds to the 3′ UTR-intron of SF2/ASF transcripts. (A) Schematic representation of the 3′ UTR region of the SF2/ASF gene. The relative positions of UCEs and Sam68 binding sites A and B are indicated. Below is the reported the sequence of the 3′ UTR region between the two UCEs (gray boxes). The sequence of the exons is indicated in uppercase, and the 3′ UTR-intron is in lowercase. The two putative Sam68 binding sites (AAAAUU, called A and B) are shown with gray ovals. The 100-mer riboprobes spanning the two binding sites are in bold. (B and C) Analysis of the phosphorylation status of Sam68 in H (High) and L (Low) cells (B) or in L cells untreated or treated with the specific ERK inhibitor U0126 (C). Sam68 was immunoprecipitated with anti-Sam68 or IgG antibodies and then analyzed in a Western blot with anti–phospho-threonine-proline (α-pTP) and Sam68 antibodies. The panel on the right shows the Western blot analysis of L cell extracts, untreated or treated with U0126, with anti-Sam68 and anti-pTP antibodies. (D) An in vitro pull-down assay with HeLa nuclear extracts and with 100-mer RNA probes (see A) containing the WT (A or B) Sam68 binding sites or the mutated sequences Mut A or Mut B. All probes contain at their 3′ end the binding sequence for the splicing factor TDP-43 as an internal control of pull-down efficiency. (E) Immunoprecipitation of the SF2/ASF 3′ UTR from in vivo cross-linked L cells using anti-Sam68 or IgG antibodies. The immunoprecipitated RNA was analyzed by RT-PCR with primers annealing to the 3′ UTR-intron (ASFbsB-for and ASFbsB-rev primers) and with primers for the GAPDH gene (left, top). Western blot analysis of the immunoprecipitated materials with anti-Sam68 and IgG antibodies (left, bottom) is shown. The immunoprecipitated RNAs were also analyzed by q-RT-PCR using ASFbsB-for and ASFbsB-rev primers and primers for GAPDH (histogram on the right). Error bars indicate SD calculated from three independent experiments.
As a first step to understanding whether Sam68 may control AS in the 3′ UTR of SF2/ASF transcripts, we performed in vitro pull-down assays using riboprobes spanning the Sam68 putative binding sites (Fig. 5 A). As shown in Fig. 5 D, Sam68 binds to both riboprobes, and binding is abrogated by mutating the Sam68 recognition sequence to AACACU. To determine if Sam68 binds to the SF2/ASF 3′ UTR in vivo, we performed RNA immunoprecipitation with an antibody that recognizes both the phosphorylated and unphosphorylated forms of Sam68. As shown in Fig. 5 E, Sam68 binds in vivo to the endogenous SF2/ASF transcript but not to GAPDH mRNA.
Sam68 controls AS-NMD of SF2/ASF transcripts and mesenchymal-to-epithelial cell transition
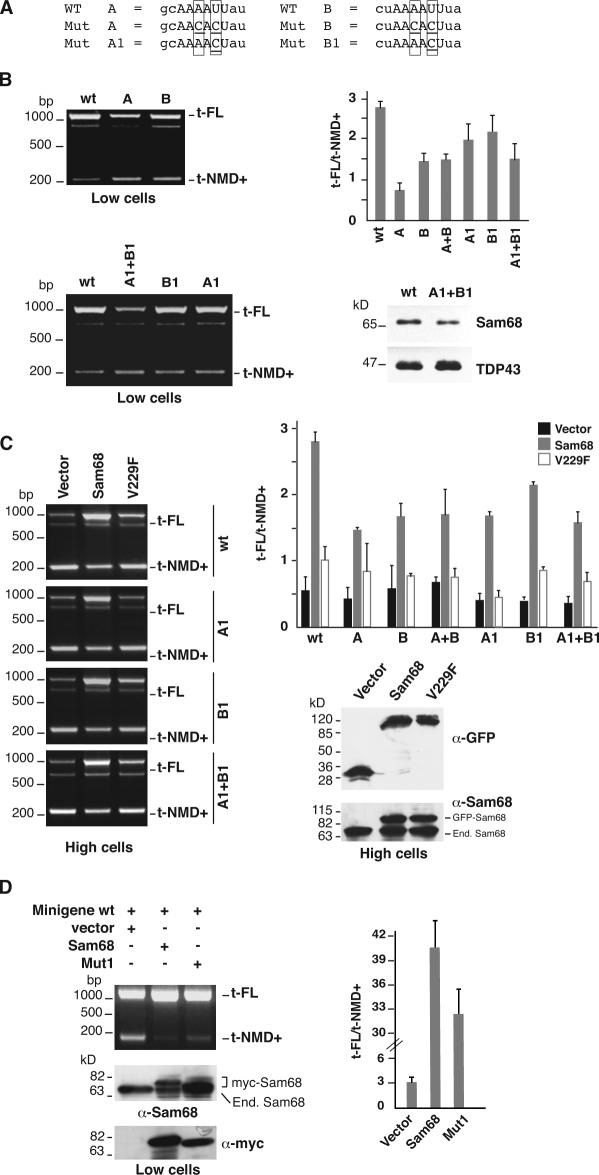
Based on the result in the previous paragraph, we hypothesized a role of Sam68 in controlling AS-NMD of SF2/ASF transcripts. To verify this possibility, we analyzed the in vivo splicing of minigene transcripts containing either the wild-type (WT) SF2/ASF 3′ UTR or mutated versions in which one or both Sam68 binding sites were substituted with AACACU (Mut A, Mut B, and Mut A+B). Similarly to endogenous SF2/ASF transcripts, RNAs encoded by the WT minigene mainly show 3′ UTR-intron retention in L-cells (t-FL in Fig. 6 B). Mutation of one Sam68 binding site reduces intron retention (t-NMD+); this is more evident in the case of Mut A, which points to a role of these sequence elements in the splicing decision. Unexpectedly, however, when both sites are contemporarily mutated (Mut A+B), the effect is lower than that observed with Mut A (Fig. S3 and the histogram in Fig. 6 B), as if Mut A+B is responding to some other splicing regulatory mechanism. In an attempt to circumvent this problem, we generated another set of mutants (Mut A1, Mut B1, and Mut A1+B1 in Fig. 6 A) in which a single nucleotide was substituted in the Sam68 consensus sequence (AAAACU). Although less marked than previous mutants, Mut A1 and Mut B1 also show a shift toward the production of t-NMD+ RNAs in L cells. Remarkably, the mutation of both sites (Mut A1+B1) gives the maximal effect on splicing (Fig. 6 B) and reduces the interaction with Sam68 in a pull-down assay (Fig. 6 B). We then tested all these minigenes in H cells. Similarly to the endogenous SF2/ASF RNA, WT minigene transcripts mainly undergo 3′ UTR-intron removal with the production of t-NMD+ molecules (Fig. 6 C). As expected from the fact that Sam68 sites are affected, the same preferential expression of t-NMD+ RNAs is observed with all mutants, even though some differences are visible (Fig. S4 and the histogram in Fig. 6 C).
Figure 6.
Sam68 triggers retention of the 3′ UTR-intron. (A) Comparison between WT and mutated A and B sites. Boxes highlight residues that are changed in the different mutants; underlined residues indicate a mutated nucleotide in a specific mutant. (B) L cells (Low cells) were transfected with a minigene containing the WT SF2/ASF 3′ UTR or with derivatives with one (Mut A, Mut B, Mut A1, and Mut B1) or both Sam68 binding sites (Mut A+B and Mut A1+B1) mutated. t-FL and t-NMD+, minigene transcripts generated by retention and splicing of the 3′ UTR-intron, respectively. The histogram on the right shows the ratio between t-FL and t-NMD+ splicing products calculated from three independent experiments. (B, bottom right) Western blot analysis with anti-Sam68 and anti-TDP43 antibodies of a pull-down experiment, as in Fig. 5 D, performed with a 965-mer riboprobe spanning the 3′ UTR-intron from 50 nt upstream of the Sam68-binding site A to 50 nt downstream of the Sam68 binding site B, in either WT or mutated. (C) WT, Mut A1, Mut B1, and Mut A1+B1 minigenes were cotransfected into H cells (High cells) along with a plasmid encoding the GFP-tagged WT Sam68, the GFP-tagged V229F mutant, or the empty vector (left). The same experiment was performed with Mut A, Mut B, and Mut A+B minigenes (Fig. S4). The histogram on the right shows the t-FL/t-NMD+ ratio calculated in three independent experiments. Expression of tagged Sam68 proteins was verified by Western blotting with anti-GFP and anti-Sam68 antibodies (left). (D) A minigene containing the WT sequence of the SF2/ASF 3′ UTR region was cotransfected into L cells along with a plasmid encoding the myc-tagged WT Sam68, the myc-tag Mut 1 (Matter et al., 2002), or the empty vector. The splicing profile of minigene transcripts was analyzed in RT-PCR, and the ratio between the two splicing products is shown in the histogram on the right. The expression of the tagged proteins was verified by Western blotting with antibodies against Sam68 and against the myc tag. Error bars indicate SD calculated from three independent experiments.
To directly assess the effect of Sam68 on the splicing of the 3′ UTR-intron, the various Mut constructs were cotransfected in H cells with plasmids directing the expression of Sam68 or of the Sam68 V229F mutant, which is impaired in RNA binding (Paronetto et al., 2007). As shown in Fig. 6 C, overexpression of Sam68 inhibits splicing of the 3′ UTR-intron, with the production of t-FL molecules, and gives rise to a profile similar to that observed in L-cells. This result clearly involves Sam68 in the regulatory circuit that controls splicing decision in the 3′ UTR region of SF2/ASF transcripts. However, Sam68 also promotes intron retention, although to a lower extent, in transcripts encoded by mutated minigenes. It is plausible that this behavior actually reflects the interaction of Sam68 with sequence elements in the 3′ UTR-intron related to the consensus binding site. Consistent with this hypothesis, the mutation of both sites decreases, but does not abrogate, the interaction with Sam68 in the pull-down assay (Fig. 6 B). The necessity of a direct protein–RNA interaction is also supported by the behavior of the V229F mutant (Fig. 6 C), which displays a strongly reduced ability to stimulate 3′ UTR-intron retention. The residual activity may reflect the interaction with some other, still unidentified, proteins that bind to the minigene transcript. This possibility has been recently advanced to explain the ability of the V229F mutant to stimulate splicing of the RTL4-T transcripts (Liu et al., 2009).
Because the level of Sam68 does not appreciably change during EMT (unpublished data), we asked whether phosphorylation of this factor by ERK1/2 may be critically linked to the switch in the splicing profile of SF2/ASF transcripts. To verify this hypothesis, we cotransfected L cells with the WT SF2/ASF 3′ UTR minigene and with plasmids directing the expression of either Sam68 WT or Sam68 in which the eight ERK phosphorylation sites were mutated to alanine (Mut 1; Matter et al., 2002). As shown in Fig. 6 D, mutation of the ERK1/2 phosphorylation sites reduced the ability of Sam68 to modulate the splicing profile of WT minigene transcripts.
Altogether, these results suggest that differential splicing of minigene transcripts during EMT is controlled by the interaction of Sam68 with the two binding sites in the 3′ UTR-intron and is modulated by protein phosphorylation.
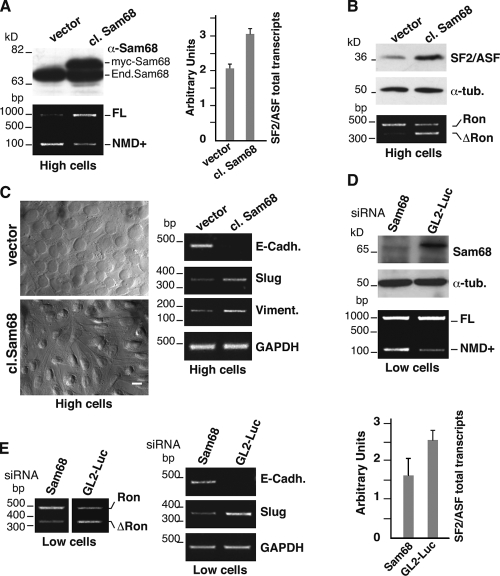
To investigate the effect of Sam68 on the splicing profile of the endogenous SF2/ASF transcripts, we generated a clone of SW480 cells stably overexpressing Myc-tagged Sam68 (cl. Sam68). As shown in Fig. 7 A, a twofold increase in Sam68 expression is sufficient to augment the ratio of FL to NMD+ mRNAs, leading to a higher level of SF2/ASF total mRNA. This is accompanied by an increased level of SF2/ASF protein and by a switch in the splicing profile of Ron gene transcripts (Fig. 7 B). Strikingly, cl. Sam68 cells are unable to undergo MET. Indeed they do not acquire the typical epithelial morphology of SW480 cells and display elongated and spindle-shaped morphology typical of mesenchymal cells even at high density. Moreover, they fail to up-regulate E-cadherin and show constitutive expression of Slug and vimentin (Fig. 7 C). To confirm the involvement of Sam68 in AS-NMD of SF2/ASF transcripts, we used siRNA to down-regulate endogenous Sam68 expression in L cells, which display high levels of FL mRNA. As shown in Fig. 7 D, down-regulation of Sam68 results in a shift of the splicing profile toward the NMD+ isoform and in an overall reduction of SF2/ASF transcription levels. Accordingly, we observed a switch in the splicing pattern of Ron transcripts, increased expression of E-cadherin, and reduced transcription of Slug (Fig. 7 E). On the basis of these observations, we suggest that the level and/or activity of the splicing regulator Sam68 contributes to determine the choice between the epithelial and mesenchymal status. Notably, a higher level of Sam68 mRNA is observed in a fraction of colon tumors (Fig. S2).
Figure 7.
Sam68 controls AS in the SF2/ASF 3′ UTR and the activation of the MET program. (A) SW480 cells were stably transfected with myc-tagged Sam68 (cl. Sam68) or with the empty vector (vector). Western blot analysis of extracts from cl. Sam68 and “vector” H cells (High cells) with anti-Sam68 antibody. H cells were also analyzed to determine the splicing profile of the 3′ UTR-intron (RT-PCR with primers ASF-A and B) and the level of the total SF2/ASF transcripts by qRT-PCR (histogram). (B) The protein levels of SF2/ASF and α-tubulin and the splicing profile of the Ron gene transcripts. (C) The morphology of cl. Sam68 and “vector” cells grown at high density was examined under a phase-contrast microscope (magnification 60×), and the mRNA expression levels of the indicated EMT markers and GAPDH gene were determined by RT-PCR. Bar, 10 µm. (D) L cells (Low cells) were transfected with Sam68 or with GL2-Luciferase siRNAs. Cells were analyzed by Western blotting with anti-Sam68 and α-tubulin antibodies and by RT-PCR to reveal the splicing profile of the 3′ UTR region of the endogenous SF2/ASF transcripts (primers ASF-A and -B). The histogram shows the relative quantification of the total SF2/ASF transcripts by qRT-PCR. (E) L cells knocked down for Sam68 were analyzed in RT-PCR to determine the Ron splicing. Note that images of Ron splicing derive from different part of the same agarose gel. The same cells were also analyzed for the expression level of the indicated EMT markers. Error bars indicate SD calculated from three independent experiments.
Discussion
In this manuscript, we have investigated the relevance of AS to the EMT or its reversal MET. Both EMT and MET are reversible processes that result from the activation of gene expression programs crucial for normal development. However, they are pathologically exploited during the progression of well-differentiated epithelial tumors (Polyak and Weinberg, 2009). EMT-like cell dedifferentiation takes place at the invasive front of carcinomas, favoring detachment, migration, and dissemination of cancer cells. Redifferentiation with regain of epithelial features in a MET-like process occurs at the final metastatic site (Thiery and Sleeman, 2006). Our analysis indicates that cell growth conditions and external cues act through the ERK1/2 signaling pathway to modulate the activity and expression levels of two splicing regulators, Sam68 and SF2/ASF, critically involved in EMT/MET programs in SW480 cells.
AS, NMD pathway, and expression of the SF2/ASF proto-oncogene
A recent work on splicing factor SF2/ASF has shown that multiple posttranscriptional and translational mechanisms operate to fine-tune the level of this protein (Sun et al., 2010). AS-NMD is one of the mechanisms that control the expression of a large number of pre-mRNA processing factors, many of which directly modulate AS-NMD of their own transcript, thus maintaining, through a sort of feedback mechanism, their homeostatic level (Isken and Maquat, 2008). Splicing regulators may also modulate AS-NMD in transcripts of other RNA-binding proteins. This is the case of two pairs of closely related hnRNP proteins—polypyrimidine tract-binding protein (PTB)/neural PTB (nPTB) and hnRNP L/hnRNP LL—that control their relative abundance during cell differentiation through AS-NMD (Spellman et al., 2007; Rossbach et al., 2009). We have identified another example of regulated AS-NMD in which the level and/or phosphorylation status of Sam68 affects the level of SF2/ASF by regulating unproductive splicing in the 3′ UTR of SF2/ASF transcripts. As observed for PTB/nPTB and hnRNPL/hnRNPLL pairs, regulation of the SF2/ASF protein level by Sam68 is involved in cell differentiation programs, namely the EMT–MET transitions. The identification of a regulative circuit in which a splicing regulator, Sam68, controls the level of an unrelated one, SF2/ASF, opens a new layer of complexity in splicing regulation. In particular, it unveils a hierarchy of actions between splicing factors in which Sam68 links the signaling pathway to downstream effectors such as SF2/ASF. Thus far, we know only one target of this circuit, i.e., the Ron proto-oncogene that, through AS, produces the ΔRon isoform able to trigger EMT. However, it is likely that other transcripts for proteins involved in the EMT program are also controlled.
We found that Sam68, an ERK1/2 target, binds to A/U-rich motifs near the UCEs that flank the 3′ UTR-intron. Interestingly, the association between UCEs and Sam68 binding sites in the SF2/ASF gene is evolutionarily conserved (Fig. S1). In particular, the distance between binding site B (Fig. 5) and the downstream UCE is identical (117 bp) in the rat and human genes, and the B site is the only one conserved in mice (Fig. S1). This suggests a relevant function of this element in the AS of SF2/ASF transcripts that, as indicated by EST analysis, occurs in mice and rats as well.
Connection between the ERK1/2 signaling pathway and splicing control
The mechanisms used by oncogenes or tumor suppressors to regulate malignant transformation are becoming progressively better understood. Recently, the emphasis on crucial events during tumor progression is changing, as many molecular and epigenetic changes seem to cooperate in altering tumor cell behavior. Despite of its prevalence and importance (Ghigna et al., 2008), very little is known about the role of AS in tumorigenesis.
A remarkable finding of our analysis is that morphological and molecular markers of mesenchymal cells and AS-NMD of SF2/ASF transcripts are controlled by soluble factors present in the epithelial cell–conditioned media. We have shown that these factors repress the activity of the extracellular signal–regulated kinase ERK1/2 and Sam68 phosphorylation, thus leading to a switch in the splicing profile of the SF2/ASF transcript. Interestingly, the role of the ERK1/2 pathway seems to be specific, as we did not detect alteration of the signaling pathway identified by p38 kinase, another cascade regulated by extracellular cues.
The ERK1/2 pathway mediates the response to growth signals and controls cell proliferation and survival, cell adhesion, angiogenesis, and cancer metastasis (Fang and Richardson, 2005). This signaling cascade includes several proto-oncogenes and is deregulated in approximately one third of all human cancers (Dhillon et al., 2007). Interestingly, ERK1/2 activation is associated to induction of EMT (Thiery and Sleeman, 2006). Among the hundreds of ERK1/2 substrates, very few direct effectors have been identified with respect to induction of EMT. Even though the molecular details remain poorly understood, it has been suggested that ERK1/2 operates through regulation of transcription (Fang and Richardson, 2005; Thiery and Sleeman, 2006). Notably, during epidermal differentiation, ERK1/2 activation induces higher levels of transcripts for RNA splicing proteins (Gazel et al., 2008). Our data suggest that, at least for SF2/ASF, the increment in mRNA levels may reflect inhibition of unproductive splicing, raising the possibility that posttranscriptional regulation of RNA stability by AS-NMD might account for part of these changes in mRNA levels. Altogether, our results therefore suggest that ERK1/2 may contribute to tumor metastasis by modulating AS events critically linked to EMT.
It is commonly accepted that tumor cells accumulate widespread chromosomal changes and somatic mutations that predispose to EMT and metastasis. Although the highly aberrant genomes of tumor cells are consistent with this idea, our data support an alternative model in which a subset of the tumor metastasizes as a consequence of its unique responsiveness to diverse microenvironmental cues. An unresolved issue is whether diffusible factors in the tumor microenvironment are also involved in MET induction. It has been suggested that, in the absence of signals that actively promote EMT, mesenchymal tumor cells could revert to the epithelial state once they reached the final metastatic location as a consequence of a transcriptional default mechanism (Polyak and Weinberg, 2009). Interestingly, our data indicate that, at least in the case of SW480 cells, factors secreted by epithelial cells act dominantly to activate MET. It is, therefore, plausible that cancer cells may undergo MET in response to signals coming from surrounding epithelial cells.
Although MET is induced in mesenchymal-like SW480 cells by factors present in the conditioned medium of epithelial-like H-cells, we failed to observe the opposite phenomenon, i.e., induction of mesenchymal markers in epithelial-like H cells in response to factors expressed by mesenchymal-like L cells (unpublished data). This could be simply due to the fact that relevant growth factors expressed by L cells are too diluted to trigger EMT in H cells. An alternative explanation takes into account the fact that SW480 cells express a constitutively active Ras (K-RasV12) mutant that activates ERK1/2 (Dhillon et al., 2007) and expression of mesenchymal markers. Indeed, pharmacological inhibition of ERK1/2 activity in L cells induces MET. Interestingly, the transition of mesenchymal SW480 cells to an epithelial-like state (MET) is accompanied by inactivation of the ERK1/2 pathway (Fig. 4). Thus, in line with recent findings (Gibbons et al., 2009) and despite having numerous somatic genetic mutations, SW480 cells retain a marked plasticity and may undergo reversible EMT. Different possible molecular mechanisms by which H cells bypass constitutive K-RasV12 can be proposed, and their relevance will be investigated in the future. Interestingly, some of them involve the expression of specific microRNAs or AS events in kinases acting downstream of Ras.
The splicing regulator Sam68 is a master controller of AS in response to ERK1/2 activation
Sam68 is a member of the STAR family of RNA-binding proteins that links extracellular signals to RNA metabolism (Lukong and Richard, 2003). The molecular mechanism through which Sam68 regulates AS decisions in response to signaling cascades is poorly understood. Previously, it has been shown that phosphorylation of Sam68 by ERK1/2 stimulates inclusion of the CD44 exon v5 (Matter et al., 2002), an exon frequently included during tumor progression, and the production of the oncogenic cyclin D1b variant in the human CCND1 gene (Paronetto et al., 2010). Moreover, Sam68 modulates AS of human BCL-X (Paronetto et al., 2007) and murine Sgce (Chawla et al., 2009) gene transcripts. Here, we have added the SF2/ASF proto-oncogene to the list of Sam68 substrates. Our data indicate that, in addition to being a direct target of signaling pathways, Sam68 may transduce the signal to a downstream splicing regulator, SF2/ASF, by controlling its mRNA and protein levels through AS-NMD. Moreover, similarly to ERK1/2, Sam68 impact on EMT/MET programs and its overexpression prevents cell density–driven MET in SW480. Because of the relevance of EMT/MET in the formation of metastases, it is tempting to propose a role for Sam68 in cancer progression. In accord with this hypothesis, tyrosine phosphorylation of Sam68 is elevated in human breast and prostate tumor tissues and cell lines (Paronetto et al., 2004; Babic et al.,2004; Lukong et al., 2005). Moreover, the expression levels of Sam68 are higher in human prostate cancer tissues (Busà et al., 2007; Rajan et al., 2008), and Sam68 supports prostate cancer cell proliferation and resistance to chemotherapeutic agents (Busà et al., 2007). Further indications of a role in cell transformation come from studies in mutant mice in which Sam68 haplo-insufficiency delays the onset of induced breast tumors and decreases the number of metastasis in vivo (Richard et al., 2008). Collectively, these data suggest that the Sam68 expression level and phosphorylation status play a role in tumorigenesis and point to Sam68 as a proto-oncogene. As indicated by our analysis, Sam68 could contribute to the malignant transformation of epithelial cancers by up-regulating the expression of the proto-oncogene SF2/ASF and inducing a posttranscriptionally regulated EMT.
Materials and methods
Cell culture, transfection, treatments, and immunofluorescence
Human colon carcinoma-derived SW480 cells (American Type Culture Collection, No. CCL-228) were grown in RPMI media supplemented with 10% FBS, 2 mM l-glutamine, and 25 mM Hepes. To obtain high-density (H) and low-density (L) conditions, SW480 cells were plated at 8,000 and 500 cells/mm2, respectively. For the transient transfection, we used Lipofectamine 2000 (Invitrogen) as recommended by the provider. After 24 h from transfection, H or L cells were plated into 35-mm tissue culture wells and analyzed after an additional 24 h. SW480 cells, stably transfected with the cDNA of mouse Myc-tagged Sam68 or with the empty vector pcDNA3.1 (Invitrogen), were selected in 800 µg/ml G418.
Immunofluorescence was performed on cells fixed in 3% paraformaldehyde and stained with antibodies to β-catenin (Millipore) or E-cadherin (BD) followed by indirect immunofluorescence using TRITC-conjugated anti–mouse antibody (Jackson ImmunoResearch Laboratories, Inc.). Nuclei were stained with 0.1 µg/ml DAPI (Sigma-Aldrich). Slides were then mounted with MOWIOL reagent (EMD) and analyzed by using an optical microscope (IX71; Olympus) equipped with a 60× NA1.25 oil immersion objective lens (Olympus; Fig. 1 A). Immunofluorescence images were acquired at room temperature with a digital camera (CoolSNAP; Photometrics) and processed using the MetaMorph software (version 6.0/6.1; MDS Analytical Technologies). Analysis in Fig. 3 D was performed with a digital scanning confocal microscope apparatus (TCS-SP2; Leica) equipped with a 63× NA 1.32 oil immersion objective. We used the 543-nm laser line for the TRITC fluorescence (detected at λ > 570 nm). Images were exported to Photoshop (Adobe). No manipulations were performed other than adjustments in brightness and contrast.
The cell co-culture experiment was performed with tissue culture inserts bearing a 0.4-µm pore size polyethylene terephthalate membrane (Corning). H or L cells were plated into 12-mm tissue culture wells. After 24 h, cell monolayers were rinsed, and an insert, containing L or H cells cultured on top of the porous filter, was placed inside each well. Cells in the bottom well were analyzed after 24 h of co-culture.
Conditioned medium (CM) was obtained by plating H or L SW480 cells into 35-mm tissue culture dishes in RPMI with 3% FBS. CM was collected 24 h later and centrifuged to discard cell debris.
H cells were treated with 10 µg/ml cycloheximide (Sigma-Aldrich) for 6 h before analysis. L cells were treated with 40 µM U0126 (Sigma-Aldrich) and analyzed after 24 h. As a control, L cells were treated with DMSO.
Normal human tissues and cancer specimens
RNAs prepared from pathologically confirmed primary nonfamilial colon adenocarcinomas and neighboring normal colon tissues have been described previously (Ghigna et al., 2005). 3 µg of total RNA from normal human tissues (Clontech) and from colon tumors was analyzed with RT-PCR.
Plasmids
To generate the plasmid containing the WT 3′ UTR region of SF2/ASF transcripts, this sequence was PCR amplified with primers ASF-A and -B (Table S1) by using cDNA from L cells. The amplification band was then cloned into the EcoRV site of pcDNA3.1(+) (Invitrogen) and then sequenced. The mutated minigenes Mut A, Mut B, Mut A+B, Mut A1, Mut B1, and Mut A1+B1, all in pcDNA3.1(+), were generated by PCR-mediated mutagenesis and sequenced. The GFP-tagged Sam68 expression plasmids were provided by D. Elliott (Institute of Human Genetics, Newcastle University, Centre for Life, Newcastle upon Tyne, England, UK), whereas Myc-tagged Sam68-mut1 was provided by H. Köning (Forschungszentrum Karlsruhe GmbH, Institut für Toxikologie und Genetik, Karlsruhe, Germany).
RT-PCR
3 µg of DNase-treated total RNA was retro-transcribed with d(T)18 and Superscript II RT (Invitrogen). An aliquot (1/20th) was then PCR amplified. For analysis of AS-NMD of SF2/ASF RNA, we used primers ASF-A or ASF-D in combination with ASF-B, whereas to detect total SF2/ASF transcripts, we used ASF_cost_for and ASF_cost_rev (Table S1). Total RNA from SW480 cells, transfected with SF2/ASF 3′ UTR minigenes, were RT-PCR analyzed with the primers ASF-A and BGHrev (Ghigna et al., 2005), the latter of which anneals to the vector sequence. Primers used to amplify Ron, E-cadherin, and GAPDH transcripts have been described previously (Ghigna et al., 2005). For other EMT markers, we used VimEX6 with VimEX6-7 and SlugEx2 with SlugEx2-3. All primers are listed in Table S1.
qRT-PCR
An aliquot (1/20th) of the RT reaction was used in a quantitative PCR with the QuantiTec SYBR Green PCR kit (QIAGEN). PCR was performed on LightCycler 480 (Roche). The level of GAPDH or HP0 transcripts was used to normalize the mRNA levels of SF2/ASF and Sam68, and the EMT markers E-cadherin, Slug, and vimentin (primers listed in Table S2). The expression of each gene was measured in triplicate in at least three independent experiments.
Western blot analysis
Cells were lysed in Laemmli buffer and analyzed in a Western blot by standard procedures (Rossi et al., 1999). The following primary antibodies were used: anti-SF2/ASF mAb96 (Invitrogen), anti-ERK1/2 (EMD), anti–phospho-ERK1/2 (EMD), anti–phospho-p38 (EMD), anti-p38 (EMD), anti–α-tubulin (Sigma-Aldrich), anti-Upf1/RENT1 (Bethyl Laboratories, Inc.), anti-Myc (Roche), and anti-GFP (Roche). Immunostained bands were detected by the chemiluminescent method (Thermo Fisher Scientific).
Immunoprecipitation
After being washed once with PBS, 2.3 × 107 H cells (corresponding to 3 wells of a 6-well tissue culture plate) and L cells (corresponding to 6 × 100-mm tissue culture dishes) were lysed in 1.5 ml of radio-immunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors (complete tablet; Roche) and phosphatase inhibitors (PhosSTOP tablet; Roche). The lysate was sonicated twice at 50 W for 20 s and centrifuged at 12,000 rpm for 10 min at 4°C to remove cell debris. Cell lysates were precleared by incubating for 2 h at 4°C with 40 µl of A/G plus–agarose beads (Santa Cruz Biotechnology, Inc.). 10 µl of anti-Sam68 (C-20; Santa Cruz Biotechnology, Inc.) or anti-IgG (Santa Cruz Biotechnology, Inc.) antibodies were conjugated with 40 µl of A/G plus–agarose beads (Santa Cruz Biotechnology, Inc.) in RIPA buffer for 1 h at 4°C. Precleared lysates were divided in half and incubated overnight at 4°C with anti-Sam68 or with anti-IgG beads. After centrifugation, the beads were washed three times with RIPA buffer and incubated for 10 min at 95°C with 24 µl of SDS-PAGE protein sample buffer. All the immunoprecipitated materials were then analyzed by Western blotting with anti-Sam68 antibody C1-7 (Santa Cruz Biotechnology, Inc.) in parallel with a fraction corresponding to 1/10 of the total lysate (input). Western blots were analyzed with an antibody directed against Phospho-Thr-Pro (Cell Signaling Technology).
In vitro RNA transcription and pull-down assay
100-mer RNA probes containing the first or the second Sam68 binding site in the SF2/ASF 3′ UTR region or the entire 3′ UTR-intron (695 mer) were transcribed according to established protocols (Buratti et al., 2004), using as templates the minigenes SF2/ASF 3′ UTR WT, Mut A+B, and Mut A1+B1 and the following set of oligos: T7bsA with bsA, T7bsB with bsB, and T7bsA with bsB (Table S1). Sense and antisense oligos carried a T7 polymerase promoter sequence and a consensus binding motif for TDP-43, respectively. For in vitro pull-down assays, 500 pmol of the target RNA was placed in a 400-µl reaction mixture containing 100 mM NaOAC, pH 5.0, and 5 mM sodium m-periodate (Sigma-Aldrich); incubated for 1 h in the dark at RT; ethanol precipitated; and resuspended in 100 µl of 0.1 M NaOAC, pH 5.0. 300 µl of adipic acid dihydrazide agarose bead 50% slurry (Sigma) was added, and the mixture was equilibrated in 100 mM NaOAC, pH 5.0, then incubated for 12 h at 4°C on a rotator. The RNA beads were then pelleted, washed three times with 1 ml of 2 M NaCl, and equilibrated in washing buffer (5 mM Hepes, pH 7.9, 1 mM MgCl2, and 0.8 mM magnesium acetate). They were then incubated on a rotator with a protein mixture containing approximately 1 mg of HeLa cell nuclear extract for 30 min at RT in 1 ml final volume. The beads were then pelleted by centrifugation at 3,000 rpm for 3 min and washed four times with 1.5 ml of washing buffer before the addition of SDS sample buffer and loading on a 10% SDS-PAGE gel. The materials were analyzed by Western blotting with anti-Sam68 antibody C1-7 (Santa Cruz Biotechnology, Inc.) and with a rabbit polyclonal antibody directed against TDP-43 (made in the F.E. Baralle laboratory according to standard immunization procedures).
Ribonucleoprotein immunoprecipitation (RIP)
RNA immunoprecipitation was performed as described in Niranjanakumari et al. (2002). In brief, 1.8 × 107 L cells (corresponding to 2 × 150-mm tissue culture dishes) were fixed in 1% formaldehyde for 10 min at RT, and then the cross-link reaction was quenched by adding glycine for 5 min (0.25 M final). Cells were washed with PBS, scraped, collected by centrifugation, and resuspended in 10 ml of PBS. 1 ml was saved as input. The remaining cells were pelleted and then lysed in 1.5 ml of RIPA buffer, sonicated and then centrifuged to remove cell debris. Cell lysates were precleared by incubating for 2 h at 4°C with 40 µl of A/G plus–agarose beads (Santa Cruz Biotechnology, Inc.). 4 µg of anti-Sam 68 (C-20; Santa Cruz Biotechnology, Inc.) or anti-IgG (Santa Cruz Biotechnology, Inc.) was conjugated with 40 µl of A/G plus–agarose beads (Santa Cruz Biotechnology, Inc.) for 1 h at 4°C. Precleared lysates were divided in half and incubated overnight at 4°C with anti-Sam68 beads or with anti-IgG beads. The beads were washed three times with RIPA buffer and the cross-linking was reversed by incubating the beads in 40 µl of PBS at 70°C for 45 min. RNA was extracted with TRIzol (Invitrogen) according to manufacturer’s instructions, and DNase-treated RNA was used in reverse transcriptase with random hexamers and Superscript II RT (Invitrogen). Quantitative PCR was performed with ASFbsB-for and ASFbsB-rev primers (Table S2).
RNA interference
RNA interference was performed as described previously (Elbashir et al., 2001). For Upf1, SW480 cells were transfected at 50% confluence with Upf1 oligo (5′-TTCCACTGCTAAAACTGAA-3′) using Oligofectamine (Invitrogen). After 24 h, cells were seeded at high density into 35-mm tissue culture wells and assayed after an additional 24 h. For Sam68, 5 × 105 SW480 cells were transfected three times at 24 h intervals with Sam68-1 oligo (Cheng and Sharp, 2006) and assayed 24 h after the third transfection. As a control in all experiments, cells were transfected with GL2-luciferase oligo (Elbashir et al., 2001).
Online supplemental material
Fig. S1 shows the association between UCEs and Sam68 binding sites in mouse, rat, and human SF2/ASF genes. Fig. S2 shows the analysis of SF2/ASF AS-NMD and Sam68 expression in normal human tissues and colon cancer. Fig. S3 shows the effect of mutations in Sam68 binding sites on SF2/ASF AS-NMD. Fig. S4 shows the effect of Sam68 overexpression in H cells on the splicing profile of transcripts encoded by SF2/ASF minigenes. Tables S1 and S2 show the sequences of the oligonucleotides used for the PCR and qPCR reactions, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001073/DC1.
Acknowledgments
This paper is dedicated to the memory of Prof. Arturo Falaschi, who devoted his life to the development of molecular biology in Italy. We thank D. Elliott for providing GFP-tagged Sam68 expression plasmids, H. Köning for Myc-tagged Sam68-mut1, and Cristiana Stuani for technical assistance.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC), from the European Network of Excellence on Alternative Splicing (EURASNET), and from the Fondazione Cariplo to G. Biamonti. E. Buratti and F.E. Baralle are supported by Telethon and EURASNET.
Footnotes
Abbreviations used in this paper:
- AS
- alternative splicing
- AS-NMD
- AS-activated NMD
- CHX
- cycloheximide
- EJC
- exon junction complex
- EMT
- epithelial-to-mesenchymal transition
- ERK
- extracellular signal–regulated kinase
- FL
- full length
- hnRNP
- heterogeneous nuclear RNP
- MET
- mesenchymal-to-epithelial transition
- NMD
- nonsense-mediated mRNA decay
- PTB
- polypyrimidine tract-binding protein
- PTC
- premature translation termination codon
- qRT-PCR
- quantitative RT-PCR
- RIPA
- radio-immunoprecipitation assay
- UCE
- ultraconserved element
- UTR
- untranslated region
- WT
- wild type
References
- Babic I., Jakymiw A., Fujita D.J. 2004. The RNA binding protein Sam68 is acetylated in tumor cell lines, and its acetylation correlates with enhanced RNA binding activity. Oncogene. 23:3781–3789 10.1038/sj.onc.1207484 [DOI] [PubMed] [Google Scholar]
- Blaustein M., Pelisch F., Coso O.A., Bissell M.J., Kornblihtt A.R., Srebrow A. 2004. Mammary epithelial-mesenchymal interaction regulates fibronectin alternative splicing via phosphatidylinositol 3-kinase. J. Biol. Chem. 279:21029–21037 10.1074/jbc.M314260200 [DOI] [PubMed] [Google Scholar]
- Brabletz T., Jung A., Reu S., Porzner M., Hlubek F., Kunz-Schughart L.A., Knuechel R., Kirchner T. 2001. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA. 98:10356–10361 10.1073/pnas.171610498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. 2004. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 32:4224–4236 10.1093/nar/gkh752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busà R., Paronetto M.P., Farini D., Pierantozzi E., Botti F., Angelini D.F., Attisani F., Vespasiani G., Sette C. 2007. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 26:4372–4382 10.1038/sj.onc.1210224 [DOI] [PubMed] [Google Scholar]
- Chawla G., Lin C.H., Han A., Shiue L., Ares M., Jr., Black D.L. 2009. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol. Cell. Biol. 29:201–213 10.1128/MCB.01349-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Sharp P.A. 2006. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 26:362–370 10.1128/MCB.26.1.362-370.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon A.S., Hagan S., Rath O., Kolch W. 2007. MAP kinase signalling pathways in cancer. Oncogene. 26:3279–3290 10.1038/sj.onc.1210421 [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- Fang J.Y., Richardson B.C. 2005. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327 10.1016/S1470-2045(05)70168-6 [DOI] [PubMed] [Google Scholar]
- Gazel A., Nijhawan R.I., Walsh R., Blumenberg M. 2008. Transcriptional profiling defines the roles of ERK and p38 kinases in epidermal keratinocytes. J. Cell. Physiol. 215:292–308 10.1002/jcp.21394 [DOI] [PubMed] [Google Scholar]
- Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P.M., Green M.R., Riva S., Biamonti G. 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell. 20:881–890 10.1016/j.molcel.2005.10.026 [DOI] [PubMed] [Google Scholar]
- Ghigna C., Valacca C., Biamonti G. 2008. Alternative splicing and tumor progression. Curr. Genomics. 9:556–570 10.2174/138920208786847971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons D.L., Lin W., Creighton C.J., Rizvi Z.H., Gregory P.A., Goodall G.J., Thilaganathan N., Du L., Zhang Y., Pertsemlidis A., Kurie J.M. 2009. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 23:2140–2151 10.1101/gad.1820209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman R.T., Green R.E., Brenner S.E. 2004. An unappreciated role for RNA surveillance. Genome Biol. 5:R8 10.1186/gb-2004-5-2-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9:699–712 10.1038/nrg2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185–193 10.1038/nsmb1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 446:926–929 10.1038/nature05676 [DOI] [PubMed] [Google Scholar]
- Liu Y., Bourgeois C.F., Pang S., Kudla M., Dreumont N., Kister L., Sun Y.H., Stevenin J., Elliott D.J. 2009. The germ cell nuclear proteins hnRNP G-T and RBMY activate a testis-specific exon. PLoS Genet. 5:e1000707 10.1371/journal.pgen.1000707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong K.E., Richard S. 2003. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta. 1653:73–86 [DOI] [PubMed] [Google Scholar]
- Lukong K.E., Larocque D., Tyner A.L., Richard S. 2005. Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J. Biol. Chem. 280:38639–38647 10.1074/jbc.M505802200 [DOI] [PubMed] [Google Scholar]
- Matter N., Herrlich P., König H. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 420:691–695 10.1038/nature01153 [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21:708–718 10.1101/gad.1525507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 26:182–190 10.1016/S1046-2023(02)00021-X [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40:1413–1415 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Paronetto M.P., Farini D., Sammarco I., Maturo G., Vespasiani G., Geremia R., Rossi P., Sette C. 2004. Expression of a truncated form of the c-Kit tyrosine kinase receptor and activation of Src kinase in human prostatic cancer. Am. J. Pathol. 164:1243–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M.P., Achsel T., Massiello A., Chalfant C.E., Sette C. 2007. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 176:929–939 10.1083/jcb.200701005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M.P., Cappellari M., Busà R., Pedrotti S., Vitali R., Comstock C., Hyslop T., Knudsen K.E., Sette C. 2010. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 70:229–239 10.1158/0008-5472.CAN-09-2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Weinberg R.A. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9:265–273 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Rajan P., Gaughan L., Dalgliesh C., El-Sherif A., Robson C.N., Leung H.Y., Elliott D.J. 2008. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J. Pathol. 215:67–77 10.1002/path.2324 [DOI] [PubMed] [Google Scholar]
- Richard S., Vogel G., Huot M.E., Guo T., Muller W.J., Lukong K.E. 2008. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene. 27:548–556 10.1038/sj.onc.1210652 [DOI] [PubMed] [Google Scholar]
- Rossbach O., Hung L.H., Schreiner S., Grishina I., Heiner M., Hui J., Bindereif A. 2009. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell. Biol. 29:1442–1451 10.1128/MCB.01689-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi R., Villa A., Negri C., Scovassi I., Ciarrocchi G., Biamonti G., Montecucco A. 1999. The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J. 18:5745–5754 10.1093/emboj/18.20.5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A.L., Kim Y.K., Pan Q., Fagnani M.M., Maquat L.E., Blencowe B.J. 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28:4320–4330 10.1128/MCB.00361-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon J.M., Hyatt B.A. 2004. Epithelial-mesenchymal interactions in the developing lung. Annu. Rev. Physiol. 66:625–645 10.1146/annurev.physiol.66.032102.135749 [DOI] [PubMed] [Google Scholar]
- Spellman R., Llorian M., Smith C.W. 2007. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 27:420–434 10.1016/j.molcel.2007.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Zhang Z., Sinha R., Karni R., Krainer A.R. 2010. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 17:306–312 10.1038/nsmb.1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15:740–746 10.1016/j.ceb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Thiery J.P., Sleeman J.P. 2006. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7:131–142 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- Trusolino L., Comoglio P.M. 2002. Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nat. Rev. Cancer. 2:289–300 10.1038/nrc779 [DOI] [PubMed] [Google Scholar]
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature. 456:470–476 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]