Leading cells require LIMK for matrix degradation and invadopodia formation during collective cell migration.
Abstract
LIM kinases 1 and 2 (LIMK1/2) are centrally positioned regulators of actin cytoskeleton dynamics. Using siRNA-mediated knockdown or a novel small molecule inhibitor, we show LIMK is required for path generation by leading tumor cells and nontumor stromal cells during collective tumor cell invasion. LIMK inhibition lowers cofilin phosphorylation, F-actin levels, serum response factor transcriptional activity and collagen contraction, and reduces invasion in three-dimensional invasion assays. Although motility was unaffected, LIMK inhibition impairs matrix protein degradation and invadopodia formation associated with significantly faster recovery times in FRAP assays indicative of reduced F-actin stability. When LIMK is knocked down in MDA-MB-231 cells, they lose the ability to lead strands of collectively invading cells. Similarly, when LIMK activity is blocked in cancer-associated fibroblasts, they are unable to lead the collective invasion of squamous carcinoma cells in an organotypic skin model. These results show that LIMK is required for matrix remodeling activities for path generation by leading cells in collective invasion.
Introduction
Cancer cell metastasis is a multi-stage process that commences with local invasion of surrounding tissue. The ability of metastatic cells to invade requires reorganization of the actin-myosin cytoskeleton to facilitate the necessary morphological rearrangements and ECM remodeling activities that permit movement from the primary site (Olson and Sahai, 2009). A sophisticated network of components, acting in response to Rho GTPase signal transduction pathways, is responsible for the dynamic regulation of the actin-myosin cytoskeleton (Hall, 2009). As a consequence, there has been considerable interest in identifying critical elements in Rho GTPase signaling and actin-myosin regulation that are required for cancer cell invasion, with the long-term goal of developing pharmacological inhibitors that could provide clinical benefit by suppressing local dissemination and metastatic spread (Walker and Olson, 2005; Olson, 2008).
The ability of tumor cells to move through tissues is the end product of two processes: ECM remodeling to generate a path of reduced mechanical resistance, and cell motility to follow the path (Sahai, 2005). Although considerable recent effort has been focused on understanding how tumor cells invade 3D environments as individual cells, epithelial cancer cells commonly invade collectively in strands, sheets, and clusters (Friedl and Wolf, 2008). The cell collective may remain in contact with the tumor mass, or may detach and spread away from the primary site. In this form of invasion, leading cells generate paths of low physical resistance by focalizing protease activity and generating force to remodel the ECM for the remainder of the collective to follow. As a result, distinct cells in strands may have different roles; for example path generation, which would require ECM remodeling activities, versus path following, which would only need cell motility. One advantageous aspect of this invasion mode is that stromal cells may be selected for, or co-opted, to provide the path-generating activity, thereby contributing to tumor progression (Gaggioli, 2008).
The LIM kinases 1 and 2 (LIMK1 and LIMK2) are activated downstream of the Rho-ROCK, Rac-PAK, and Cdc42-MRCK pathways, thereby acting as network nodes in the signaling pathways that regulate the actin cytoskeleton (Scott and Olson, 2007). There are numerous ways that activation of these Rho GTPases has been associated with human cancers, particularly with the progression to invasive and metastatic stages (Sahai and Marshall, 2002; Benitah et al., 2004; Vega and Ridley, 2008). As a result, LIM kinases might present a vulnerable locus in actin regulation, which when targeted could result in inhibition of invasion driven by any combination of upstream Rho GTPase signaling. When active, LIM kinases phosphorylate and inactivate the F-actin severing functions of cofilin proteins, resulting in F-actin stabilization (Van Troys et al., 2008).
We now show that inhibition of LIMK function by siRNA-mediated knockdown or selective pharmacological inhibition with a first-in-class LIMK inhibitor blocks the collective invasion of MDA-MB-231 breast carcinoma cells in 3D extracellular matrices. In addition, LIMK activity was required for the collective invasion of squamous carcinoma cells (SCCs) in a 3D organotypic skin model. Although LIMK was not required for cell motility in two dimensions (2D), both ECM degrading and deforming activities were LIMK dependent in 3D. Consistent with this observation, LIMK activity was required in leading cells in MDA-MB-231 collective invasion as well as in leading fibroblasts in an organotypic SCC collective invasion model. However, LIMK activity was not required for path-following MDA-MB-231 or SCCs. These results reveal a requirement for LIMK activity in the path-generation function of leading cells in collective invasion.
Results
Collective invasion is blocked by LIMK inhibition or increased cofilin
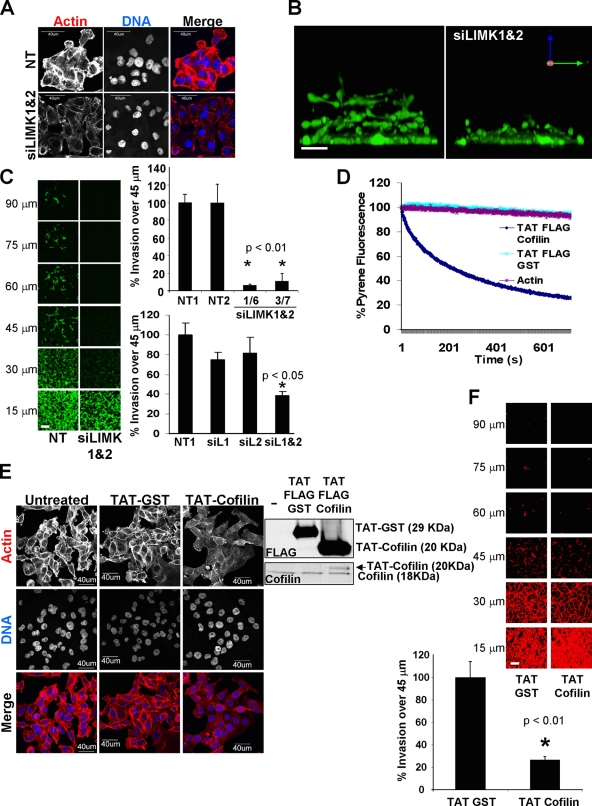
To examine the contribution of LIMK to cell invasion, several siRNA duplexes for LIMK1 and LIMK2 were characterized. Independent LIMK1 and LIMK2 siRNAs efficiently knocked down their target, either alone or in combinations, as determined by Western blotting and RT-PCR (Fig. S1 A). Individual knockdown of either LIMK1 or LIMK2 reduced phosphorylated cofilin levels; combined LIMK1/2 knockdown further decreased phospho-cofilin compared with nontargeting (NT) siRNA transfectants (Fig. S1 A) without affecting cell viability (Fig. S1 B). Consistent with a role in F-actin stabilization, combined LIMK1/2 knockdown reduced phalloidin staining without notably affecting cell morphology compared with control NT siRNA (Fig. 1 A). When the ability of MDA-MB-231 breast cancer cells to invade into a 3D matrix comprised of matrigel basement membrane proteins (Kleinman and Martin, 2005) was examined, cells invaded as strand-like collectives as previously observed (Wolf et al., 2007; Fig. 1 B and Video 1). After combined LIMK1/2 knockdown, collective invasion was dramatically decreased (Fig. 1 B). By examining the extent of matrigel invasion by confocal microscopy at 15-µm optical sections, it was observed that the combined LIMK1/2 knockdown using two independent siRNA pairs resulted in statistically significant reduction in invasion relative to NT control siRNAs when comparing the ratio of cells that had migrated beyond a 45-µm cutoff (Fig. 1 C and Video 1). When LIMK1 or LIMK2 were knocked down individually there were small but nonsignificant decreases in invasion, indicating that the combined output of both kinases contributes to invasion (Fig. 1 C).
Figure 1.
LIMK activity is required for collective invasion. (A) Double LIMK1/2 knockdown (siLIMK1&2) in MDA-MB-231 breast cancer cells cultured on glass coverslips dramatically reduced cytoskeletal F-actin staining. Microscope settings were kept constant between conditions to assess staining intensity differences. Bar, 40 µm. (B) X-Z stack view of MDA-MB-231 invasion into 3D matrix revealed inhibition by LIMK1/2 knockdown. Bar, 25 µm. (C) Confocal images taken at 15-µM intervals through the 3D matrix revealed significant inhibition of invasion for two independent siRNA duplex pairs against LIMK1/2 relative to NT control siRNA. Individual targeting of LIMK1 (siL1) or LIMK2 (siL2) did not significantly reduce invasion. Bar, 100 µm (average ± SEM, n = 3). (D) Recombinant TAT-Cofilin protein severed F-actin in vitro. (E) Recombinant TAT-Cofilin reduced phalloidin-stained F-actin structures in MDA-MB-231 breast cancer cells cultured on glass coverslips. Confocal microscope settings were kept constant between conditions. Bar, 40 µm. (F) Addition of cell-permeable TAT-Cofilin significantly inhibited 3D matrix invasion relative to equal mass of TAT-GST control. Bar, 100 µm (average ± SEM, n = 3).
Because LIMK knockdown reduced cofilin phosphorylation, the effect of introducing additional unphosphorylated cofilin by protein transduction on matrigel invasion was determined. An optimized protein transduction domain based on the HIV TAT protein (Ho et al., 2001) was fused to cofilin to promote cellular uptake. The F-actin severing activity (Hawkins et al., 1993) of TAT-cofilin was validated in vitro using a pyrene fluorescence assay (Cano et al., 1991; Fig. 1 D). Control TAT-GST protein was not different from actin alone in the rate of pyrene fluorescence decay (Fig. 1 D). After introduction into MDA-MB-231 cells of equivalent amounts of TAT-GST or unphosphorylated TAT-cofilin, comparable to endogenous cofilin levels, F-actin staining was reduced in the TAT-cofilin–treated cells compared with untreated or TAT-GST–treated cells (Fig. 1 E). In addition, a statistically significant effect of TAT-cofilin on inhibiting matrigel invasion compared with TAT-GST was observed (Fig. 1 F). These results indicate that inhibiting LIMK activity or perturbing the ratio of phosphorylated to nonphosphorylated cofilin had the same consequence of inhibiting 3D collective invasion.
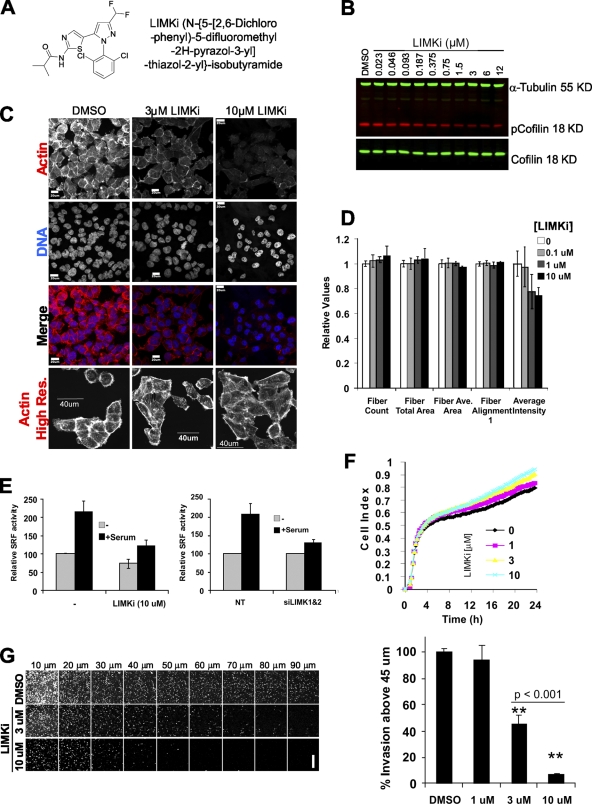
Stimulated by the potential of LIMK as a chemotherapeutic target, small molecule inhibitors have been developed (Ross-Macdonald et al., 2008). To determine whether LIMK inhibition would affect invasion, we tested the effects of N-{5-[2-(2,6-Dichloro-phenyl)-5-difluoromethyl-2H-pyrazol-3-yl]-thiazol-2-yl}-isobutyramide (Fig. 2 A; compound 3 in Ross-Macdonald et al., 2008; hereafter referred to as LIMKi), a highly selective, noncytotoxic and potent inhibitor of both LIMK1 and LIMK2. Treatment of MDA-MB-231 cells with LIMKi resulted in a dose-dependent inhibition of cofilin phosphorylation, with a cell-based IC50 of ∼1 µM (Fig. 2 B). Moreover, F-actin staining with phalloidin was similarly reduced by LIMK1/2 knockdown or by LIMKi treatment (Figs. 1 A and 2 C). When microscope settings were varied to acquire high resolution F-actin images of similar brightness, the only obvious effect was decreased intensity of cortical actin staining (Fig. 2 C). To more quantitatively examine F-actin intensity and organization, analysis of phalloidin-stained actin fibers using a Cellomics ArrayScan High Content Screening instrument followed by analysis using Morphology Explorer BioApplication software (which identifies intracellular fibers and measures morphology, alignment, and arrangement) indicated that there were no gross alterations in F-actin structures (Fig. 2 D). In agreement with observations made by microscopy (Fig. 2 C), LIMKi induced a dose-dependent reduction in F-actin signal intensity (Fig. 2 D). Additional morphometric analysis revealed no significant changes in size parameters or cell shape (Fig. S2, A and B). Consistent with published results (Ross-Macdonald et al., 2008), LIMKi had no effect on microtubule number or organization (Fig. S3). Cofilin phosphorylation by LIMK was previously reported to activate serum response factor (SRF) transcriptional activity (Geneste et al., 2002). Measurement of SRF activity using the 3D.Aluc reporter, which contains a luciferase reporter controlled by three SRF binding sites (Copeland and Treisman, 2002), showed that serum-stimulated SRF activity was reduced by LIMK1/2 knockdown or by LIMKi to a similar degree (Fig. 2 E). A kinetic adhesion/spreading assay, in which the electrical impedance of a set number of cells plated on a grid of electrodes was measured (cell index), revealed that after comparable patterns of initial adhesion there was an increase in cell spreading on fibronectin over 24 h proportional to LIMKi dose (Fig. 2 F). These results indicate that LIMKi potently inhibits LIMK activity and biological outputs comparable to LIMK1/2 knockdown. When tested in the 3D matrigel invasion assay, there was significant dose-dependent inhibition of matrigel invasion with a greater than 50% effect at 3 µM LIMKi (Fig. 2 G). These results confirm the dependence of MDA-MB-231 cell invasion on LIMK activity and demonstrate that pharmacological inhibition of LIMK activity reduces tumor cell invasion.
Figure 2.
Selective LIMK inhibitor affects actin dynamics and blocks collective invasion. (A) Structure of LIMK inhibitor (LIMKi). (B) LIMKi reduced cofilin phosphorylation in MDA-MB-231 breast cancer cells cultured on plastic in a dose-dependent manner as detected by Licor Odyssey two-color infrared scanning. (C) Cytoskeletal F-actin staining was reduced in MDA-MB-231 breast cancer cells cultured on glass coverslips by LIMK inhibitor. Confocal microscope settings were kept constant between the top three rows of images. Settings were varied in the actin high resolution (Res.) row to allow for comparable image acquisition of F-actin structures. Bar, 20 µm (top three rows) or 40 µm. (D) Actin fiber analysis by ArrayScan HCS Morphology Explorer BioApplication of MDA-MB-231 breast cancer cells grown on glass-bottomed dishes and treated with indicated LIMKi concentrations revealed no detectable changes in F-actin morphology but decreased overall staining intensity (averages ± SEM, n = 3). (E) Activation of the serum response factor transcriptional reporter by serum stimulation of MDA-MB-231 breast cancer cells cultured on plastic was comparably blocked by double LIMK1/2 knockdown or by LIMK inhibitor (averages ± SEM; left graph, n = 3; right graph, n = 5). (F) The cell index, measured by electrical impedance using the xCELLigence system, was measured over time for cells plated on fibronectin-coated wells in the presence of DMSO or increasing concentrations of LIMKi as indicated (averages of n = 3). (G) Confocal sectioning of 3D matrix invasion by MDA-MB-231 cells revealed a significant dose-dependent inhibition by LIMKi. Bar, 200 µm average ± SEM, n = 3).
LIMK inhibition does not affect 2D motility
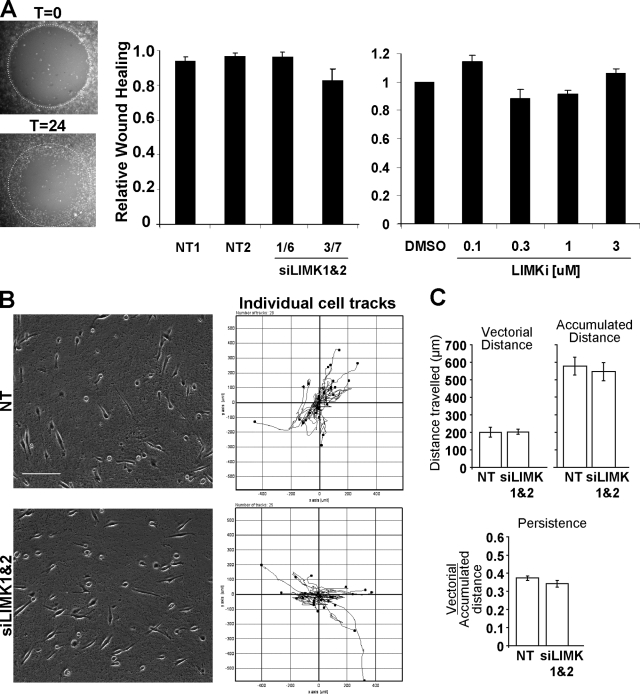
A circular wound-healing assay was used to determine the effects of LIMK inhibition on the migration of cells on a 2D surface. After the removal of a central stopper that blocks adherence to create a uniform cell-free zone, effects on cell migration were monitored (Fig. 3 A). Knockdown of LIMK1/2 by two independent siRNA pairs had no effect on cell migration after 24 h (Fig. 3 A). Consistent with these results, treatment with LIMKi concentrations from 0.1 to 3 µM also had no effect on wound healing (Fig. 3 A).
Figure 3.
LIMK activity is not required for 2D cell motility. (A) A modified scratch-wound assay in which stoppers were used to produce regular and reproducible cell-free zones was used to monitor cell motility. Diameter of circle = 2 mm. LIMK1/2 double knockdown with two independent siRNA duplex pairs did not affect MDA-MB-231 wound closing on uncoated glass-bottomed multi-well plates relative to NT control siRNA. Treatment with increasing LIMKi doses also did not affect wound closing (average ± SEM, n = 4). (B) The random migration of MDA-MB-231 cells transfected with NT or LIMK1/2 siRNA on an oriented 3D fibrillar ECM generated by primary cultured human dermal fibroblasts was determined by time-lapse microscopy. Tracks of individual cells revealed no obvious differences in path length, direction, or velocity. Bar, 100 µm. (C) Analysis of vectorial distance, accumulated distance, and persistence with ImageJ revealed no differences in 2D cell motility on fibroblast-derived matrix between NT or LIMK1/2 siRNA transfectants (average ± SEM, n = 4).
One possible explanation for the lack of effect of LIMK inhibition was that 2D migration across a rigid substratum is principally propelled by actin polymerization and forward protrusion (Olson and Sahai, 2009), and because cofilin activation could result in F-actin severing and increased density of barbed ends available to provide sites for monomer addition and filament extension, LIMK inhibition and consequent cofilin activation would not necessarily impair protrusion-driven motility. We next examined the effects of blocking LIMK activity on cell motility on an oriented 3D fibrillar ECM generated by primary cultured human dermal fibroblasts (Cukierman et al., 2001). Cell motility in this model does not require the generation of low-resistance paths by ECM remodeling, but movement is dependent upon engagement of α5β1 integrins (Cukierman et al., 2001) and is guided by a meshwork of long fibronectin fibrils that promote uniaxial polarity resulting in efficient directional protrusion (Doyle et al., 2009). Tracks of individual cell movement did not differ after LIMK1/2 knockdown from NT siRNA-transfected cells (Fig. 3 B, Video 2). Analysis of individual cell motility from multiple experiments revealed that the vectorial distances (from start to end points) covered were not different; nor was the accumulated distance (total track length) different for NT or LIMK1/2 siRNA-transfected cells (Fig. 3 C). Persistence of movement, which is a directional ratio of vectorial over accumulated distances, also did not differ between treatments (Fig. 3 C). These results indicate that the directionality and velocity of cell motility driven by cell-intrinsic factors on fibroblast-derived oriented matrices were as unaffected by LIMK inhibition as was wound-healing–induced motility on uncoated substrata.
LIMK inhibition impairs matrix degradation
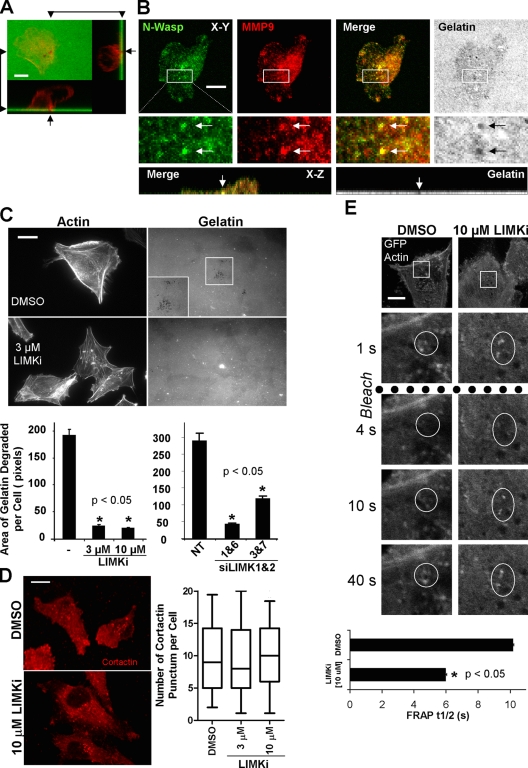
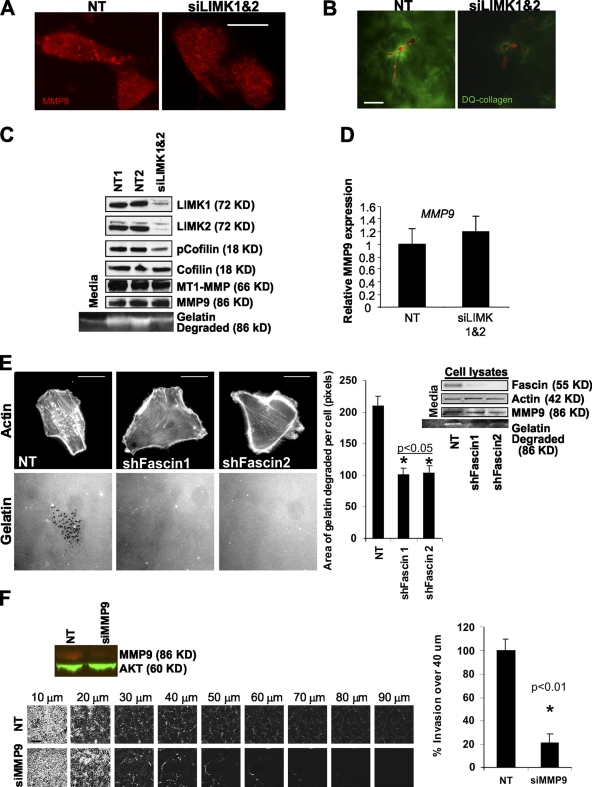
Because LIMK inhibition significantly reduced invasion into a 3D matrix without affecting cell motility, one possibility was that the processes required for path generation were affected. Collective invasion requires proteolytic remodeling of ECM for leading cells to initiate path generation. To quantitate the requirement for LIMK in focalized ECM proteolysis, MDA-MB-231 cells were plated on top of a thin layer of fluorescent gelatin to assess invadopodia formation (Buccione et al., 2009). Consistent with previous reports (Lizárraga et al., 2009), MDA-MB-231 cells had large numbers of actin-rich projections into the gelatin matrix, which were evident by discrete dark foci resulting from matrix metalloprotease (MMP)–mediated degradation of the protein matrix (Stylli et al., 2008; Fig. 4 A, arrows). Colocalization of matrix degradation sites with MMP9 and the actin polymerization regulator N-Wasp (Fig. 4 B) or with phalloidin-stained F-actin (Fig. S4 A) further validates these structures as invadopodia. In addition, sites of matrix degradation also showed colocalization of cortactin with F-actin (Fig. S4 B) or with N-Wasp (Fig. S4 C). LIMKi treatment at 3 or 10 µM or LIMK1/2 knockdown significantly reduced the area of gelatin degradation per cell (Fig. 4 C). Generation of active invadopodia requires the accumulation of cortactin to invadopodia precursors followed by F-actin stabilization during maturation (Artym et al., 2006; Oser et al., 2009). Treatment with 10 µM LIMKi, which significantly reduced gelatin degradation in invadopodia structures (Fig. 4 C), did not affect cortactin accumulation into distinct punctae (Fig. 4 D). One alternative possibility, therefore, was that LIMK inhibition did not allow for sufficient F-actin stabilization for invadopodia maturation into fully functional structures. Cells were transfected with GFP-actin and FRAP was used to determine the half times (t1/2) of recovery in punctate F-actin structures typical of invadopodia. Treatment with LIMKi significantly reduced the t1/2 of recovery (Fig. 4 E), indicating that LIMK activity was required for F-actin stabilization in these structures. These results indicate that LIMK inhibition significantly affected ECM degradation in 2D contexts, likely by inhibiting the formation of stable actin structures necessary for invadopodia maturation that focalizes MMP activity for matrix degradation.
Figure 4.
LIMK inhibition blocks invadopodia formation by decreasing F-actin stability. (A) X-Y, X-Z, and Z-Y views of a fixed MDA-MB-231 cell stained for F-actin (red) plated on FITC-labeled gelatin (green). Invadopodia are actin-rich projections within dark regions, indicative of gelatin degradation. Bar, 10 µM. (B) X-Y and X-Z views of a fixed MDA-MB-231 cell stained for N-Wasp (green) and MMP9 (red) plated on Oregon Green 488–labeled gelatin matrix (gray). Invadopodia are regions of gelatin degradation that colocalize with N-Wasp and MMP9. Bar, 10 µM. (C) LIMKi or LIMK1/2 knockdown with two independent duplex pairs significantly reduced invadopodia formation and consequent area of gelatin degradation relative to vehicle control and NT controls, respectively. Microscope settings were varied to acquire images of similar intensity for comparison purposes. Insets show higher magnification. Bar, 10 µM (averages ± SEM, n = 3). (D) Cortactin localization in cells plated on gelatin was unaffected by LIMKi treatment (box equals upper and lower quartiles with line indicating median, whiskers are 5 and 95% confidence intervals, n = 90 per treatment). Bar, 10 µM. (E) FRAP analysis of cells plated on gelatin and expressing GFP-actin revealed that LIMKi significantly decreased the t1/2 of recovery relative to vehicle DMSO-treated cells. Bar, 10 µM (average ± SD; untreated n = 16; LIMKi n = 26).
Consistent with MMP9 localization to invadopodia (Fig. 4 B) and the reduction in invadopodia formation by LIMK knockdown or inhibition (Fig. 4 C), LIMK1/2 knockdown resulted in loss of focalized MMP9 immunostaining (Fig. 5 A). Because MMP activity in invadopodia is required for direct proteolytic actions on the ECM (Artym et al., 2006; Poincloux et al., 2009), but also for the secretion and/or activation of soluble proteases such as MMP9 (Itoh and Seiki, 2004), we examined matrix degradation in a 3D context. Cells were plated at low density in a 3D matrigel environment mixed with dye-quenched (DQ) collagen, which is so heavily conjugated with fluorescein that the fluorescent signal is quenched until proteolysis separates collagen fragments, thereby lowering the local fluorescein concentration and permitting fluorescence. As shown in Fig. 5 B, RFP-labeled MDA-MB-231 cells transfected with NT siRNA were associated with strong fluorescence both adjacent to the cells and more distant. In marked contrast, cells transfected with both LIMK1/2 siRNA had much lower levels of fluorescence, both distal and proximal to the cells, indicating reduced proteolytic activity (Fig. 5 B). Conditioned media from NT or LIMK1/2 siRNA-transfected cells was collected and concentrated before electrophoresis in gelatin-containing polyacrylamide zymography gels; in parallel whole-cell lysates were prepared for Western blotting. LIMK1/2 knockdown reduced both proteins and cofilin phosphorylation without affecting MT1-MMP or MMP9 expression (Fig. 5, C and D). However, LIMK1/2 knockdown reduced the 86-KD gelatin-degradation activity (the size of activated MMP9; Lindenmeyer et al., 1997) almost to the background levels observed in concentrated serum-containing media (Fig. 5 C). To determine whether the effect on secreted MMP9 activity was specific to LIMK inhibition or due to reduced invadopodia formation, cells in which Fascin had been knocked down with two independent shRNA hairpins were examined for invadopodia formation and secreted MMP9 activity. Consistent with previous results (Li et al., 2010), Fascin knockdown significantly reduced invadopodia formation (Fig. 5 E). In addition, although MMP9 levels in whole-cell lysates were not affected, Fascin knockdown did reduce the level of secreted MMP9 activity in concentrated media (Fig. 5 E). Consistent with previous results (Kunigal et al., 2007), MMP9 knockdown significantly reduced MDA-MB-231 invasion (Fig. 5 F). These results indicate that 3D ECM degradation, secretion/activation of MMP9, and 3D invasion are dependent on the formation of the LIMK-dependent stable actin structures that are also required for invadopodia formation. The inability to remodel ECM by proteolysis, without affecting 2D motility, again suggests that LIMK inhibition likely affects path generation in collective invasion rather than path following.
Figure 5.
Inhibition of invadopodia formation reduces levels of active secreted MMP9 and matrix degradation. (A) Focalized MMP9 immunostaining in MDA-MB-231 cells plated on gelatin was reduced after LIMK1/2 knockdown. Bar, 10 µM. (B) RFP-labeled MDA-MB-231 cells plated in matrix-containing dye-quenched (DQ) collagen reveal considerable proteolytic activity that was reduced by LIMK1/2 knockdown. Bar, 20 µm. (C) LIMK1/2 knockdown in MDA-MB-231 cells plated on plastic, which decreased cofilin phosphorylation, did not alter MT1-MMP or MMP9 protein levels in cell lysates but did reduce levels of MMP9 activity in conditioned media. (D) Although LIMK1/2 knockdown in MDA-MB-231 cells plated on plastic reduced soluble MMP9-like activity, there was no effect on MMP9 expression as determined by RT-PCR. (E) Inhibition of invadopodia formation on gelatin by stable Fascin knockdown was associated with reduced levels of MMP9 activity in conditioned media but not in lysates from cells plated on plastic. Microscopy settings were varied to acquire images of similar intensities. Bar, 10 µm (averages ± SD, n = 180). (F) Knockdown of MMP9 significantly inhibited invasion into 3D matrix relative to control NT siRNA. Western blot shows MMP9 knockdown compared with AKT loading controls. Bar, 200 µm (averages ± SEM, n = 3).
LIMK is required in leading path-generating cells for collective invasion
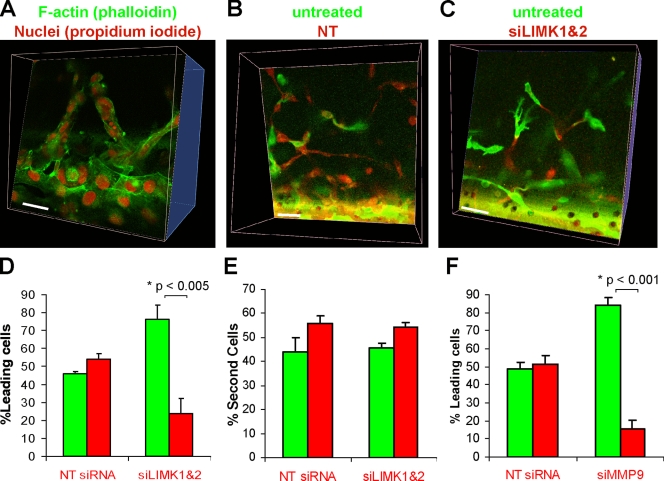
The invasion of MDA-MB-231 cells into 3D matrigel was via collective strands (Fig. 1 B and Video 1; Wolf et al., 2007). Confirming that there were multiple cells within strands, fixation and staining with FITC-phalloidin and propidium iodide (PI) revealed numerous PI-stained nuclei within each strand (Fig. 6 A). Because MDA-MB-231 cells are capable of providing the path-generating activity required in leading cells during collective invasion, we labeled pools of cells with either membrane-tethered GFP or RFP. While GFP-expressing cells were left untreated, RFP-expressing cells were transfected either with NT or LIMK1/2 siRNA. Consistent with the results in Fig. 1, B and D, NT siRNA did not affect matrigel invasion (Fig. 6 B) and the occurrence of RFP expressers as leading cells in collective strands was ∼50% (Fig. 6 D). However, when RFP-expressing cells were transfected with LIMK1/2 siRNA there were fewer RFP-positive cells invading into matrigel, and the RFP-expressing cells observed were largely restricted to following GFP-positive cells (Fig. 6 C), with the occurrence of RFP expressers as leading cells significantly dropping to less than 20% (Fig. 6 D). When the identities of the “second cell” following immediately after the leading cell were assessed, there was no difference in the incidence of RFP-expressing cells in this position between NT or LIMK1/2 siRNA transfectants (Fig. 6 E). Consistent with the critical role of MMP9 in MDA-MB-231 invasion (Fig. 5 F), MMP9 knockdown in RFP expressers significantly impaired their ability to be leading cells (Fig. 6 F). These results indicate that LIMK and MMP9 activity are required for path generation by leading cells in collective invasion. However, similar to the lack of effect on 2D motility (Fig. 3), LIMK activity was dispensable for path following (Fig. 6 E).
Figure 6.
LIMK activity is required for path generation in collective invasion. (A) To confirm that MDA-MB-231 strands invading 3D matrix were comprised of cell collectives, fixation and staining of F-actin (green) and DNA (red) revealed multiple cells within strands. Bar, 20 µM. (B) Separate pools of MDA-MB-231 cells were labeled either with RFP or GFP, then RFP-expressing cells were transfected either with NT or LIMK1/2 siRNA targeting duplexes as indicated. Untreated GFP-expressing cells were combined with siRNA-transfected RFP-expressing cells and the mixture assayed for 3D matrix invasion. Strands were equally led by GFP-expressing or NT-transfected RFP-expressing cells. Bar, 20 µM. (C) LIMK1/2 knockdown blocked the occurrence of leading RFP-expressing cells, although RFP-labeled cells were still observed after GFP-expressing cells. Bar, 20 µM. (D) 3D reconstructions of confocal image stacks were assembled with Volocity software, and used to assess the incidence of GFP or RFP cells leading collective invasion strands. Although there was no difference when cells were transfected with NT siRNA, LIMK1/2 knockdown significantly reduced the ability of RFP-expressing cells to lead collective invasion (average ± SEM, n = 4). (E) When the identity of the cell immediately after the leading cell was determined, there were no differences in the NT or LIMK1/2 siRNA-transfected RFP-expressing cells to follow in the second position (average ± SEM, n = 5). (F) Although there was no difference when cells were transfected with NT siRNA, MMP9 knockdown significantly reduced the ability of RFP-expressing cells to lead collective invasion (average ± SEM, n = 3).
Fibroblast-led collective invasion requires LIMK for path generation
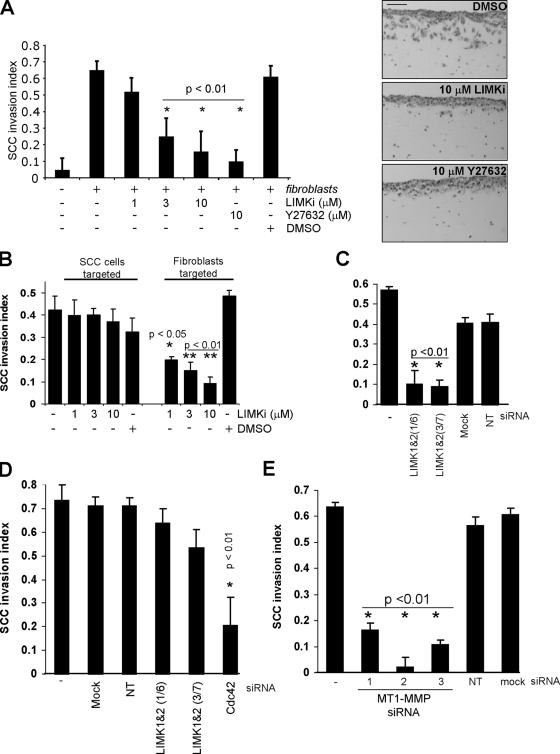
The full repertoire of activities necessary for collective invasion is intrinsic to MDA-MB-231 cells (Wolf et al., 2007). It is also possible for noninvasive cells to become invasive by following tracks laid down by other path-generating cells; in fact path generation may occur temporally in advance of invasion (Gaggioli et al., 2007). We used an organotypic culture model in which the invasiveness of SCCs, which retain epithelial markers and do not invade on their own, is dependent upon stromal fibroblasts to remodel the ECM and generate paths (Gaggioli et al., 2007; Gaggioli, 2008). SCCs were grown, supported by a metal bridge to allow the upper surface to be exposed to air and the lower surface to be in contact with media, on a dense matrix composed primarily of fibrillar collagen I. Although unable to invade alone, SCCs were able to efficiently invade when cancer-associated fibroblasts (CAF) were present in the matrix (Fig. 7 A). Although DMSO vehicle had no effect on invasion, there was a dose-dependent decrease in SCC invasion in response to increasing LIMKi concentrations attaining comparable inhibition to ROCK inhibitor Y-27632 at 10 µM (Fig. 7 A). To determine the LIMKi target cells, CAF cells were embedded in matrix either in the absence or presence of increasing doses of LIMKi, allowed to generate paths for five days, then killed with puromycin. After extensive washes, SCCs were seeded on top of the matrix and allowed to invade. Alternatively, CAF cells were allowed to generate tracks for five days, killed with puromycin, and then SCCs were seeded in the absence or presence of increasing LIMKi concentrations and allowed to invade. When CAFs were incubated with LIMKi, there was a dose-dependent inhibition in SCC invasion (Fig. 7 B). In contrast, treatment of SCCs with LIMKi at the same doses that had inhibited invasion in Fig. 7 A had no effect on SCC invasion (Fig. 7 B), indicating again that path following was not affected by LIMK inhibition. To confirm that LIMK was the critical target, knockdown of LIMK1/2 in CAFs with two independent siRNA pairs significantly inhibited SCC invasion, whereas neither mock transfection or NT siRNA affected invasion (Fig. 7 C). In contrast, knockdown of LIMK1/2 in SCCs did not significantly affect invasion, although Cdc42 knockdown significantly reduced invasion (Fig. 7 D), again consistent with LIMK activity not being required for path following (Gaggioli et al., 2007). Unlike MDA-MB-231 cells (Fig. 5 F), CAFs did not express detectable levels of MMP9; instead, MT1-MMP was highly expressed (Fig. S5). Given that LIMK was required for collective invasion (Fig. 1) and ECM degradation (Figs. 4 C and 5 B), we examined whether direct reduction of MMP activity in path-generating CAFs would reduce SCC invasion as was the case for leading MDA-MB-231 cells (Fig. 6 F). Knockdown of MT1-MMP expression with three individual siRNA duplexes in CAFs significantly reduced SCC invasion (Fig. 7 E). These results indicate that the path-generating activity of ECM degradation provided by CAFs was targeted by LIMK inhibition, and that the ability of SCCs to move through preformed paths through ECM was unaffected, reinforcing the conclusion that LIMK activity is critical for invasive path generation but not path following.
Figure 7.
Collective invasion of squamous cell carcinoma requires LIMK activity in leading stromal fibroblasts. (A) The ability of squamous carcinoma cells (SCC) that retain epithelial markers to invade 3D matrix is dependent on stromal fibroblasts. LIMKi significantly inhibited SCC invasion in a dose-dependent manner. Photomicrographs indicate effects of LIMKi or Y27632 on SCC invasion. Bar, 100 µm. (B) Paths were allowed to be generated by fibroblasts, which were then removed by puromycin extraction before the addition of the SCC layer. Increasing LIMKi doses were either included only after path generation by fibroblasts was allowed to occur (SCCs targeted) or during fibroblast-mediated path generation (Fibroblasts targeted), as indicated. Significant inhibition of SCC invasion occurred specifically when fibroblasts were targeted, indicating that path generation but not path following required LIMK activity (averages ± SEM, n = 5). (C) To determine whether LIMK activity was required in fibroblasts for SCC invasion, two independent siRNA duplex pairs targeting LIMK1/2 were used to knock down expression. Relative to mock or NT siRNA-transfected cells, LIMK1/2 knockdown significantly reduced SCC invasion (averages ± SEM, n = 4). (D) To corroborate the dispensability of LIMK in SCC for invasion, two independent siRNA duplex pairs targeting LIMK1/2 were used to knockdown expression. Relative to mock or NT siRNA-transfected cells, LIMK1/2 knockdown did not significantly affect invasion, although Cdc42 knockdown did reduce invasion (averages ± SEM, n = 4). (E) Knockdown of MT1-MMP with three independent siRNA duplexes significantly reduced SCC invasion (averages ± SEM, n = 15).
LIMK activity is required for ECM deformation
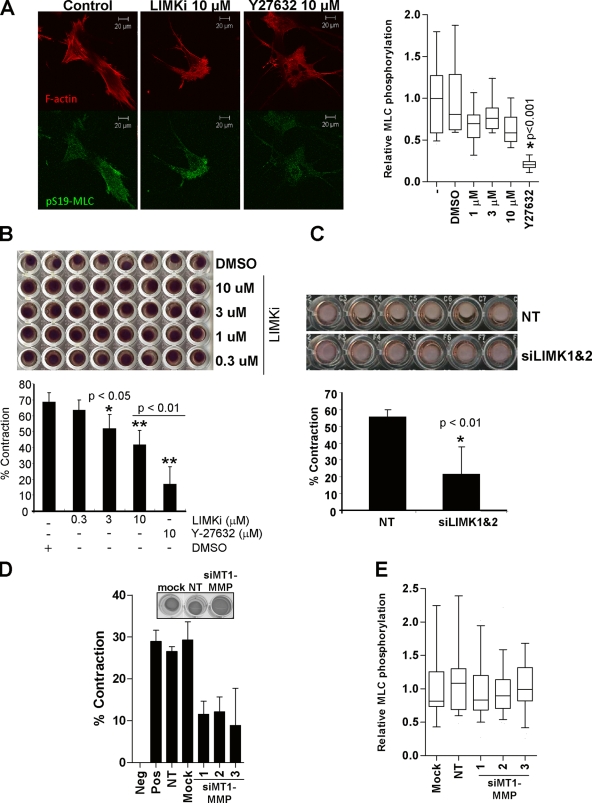
In addition to ECM proteolysis, force-mediated remodeling contributes to the path generation that permits collective invasion (Gaggioli et al., 2007). Key drivers of actin-myosin contractile force and matrix deformation enabling tumor cell invasion are the ROCK kinases (Wyckoff et al., 2006), which phosphorylate a number of substrates that lead to increased myosin light chain phosphorylation (pMLC) (Riento and Ridley, 2003). Treatment of MDA-MB-231 cells plated on a pliable collagen matrix with LIMKi did not affect pMLC levels, although ROCK inhibitor Y27632 significantly lowered pMLC (Fig. 8 A), indicating that LIMK does not directly trigger force generation by this mechanism. When CAFs were mixed with ECM and plated in the wells of 96-well tissue culture plates, there was a significant dose-dependent decrease in ECM contraction in response to increasing LIMKi concentrations (Fig. 8 B). In addition, there was a significant decrease in ECM contraction when LIMK1/2 were knocked down relative to NT controls (Fig. 8 C). Because LIMK inhibition blocked both ECM degradation (Figs. 4 C and 5 B) and ECM contraction (Fig. 8, B and C), we directly examined whether there was a requirement for MMP activity in ECM contraction. Consistent with MT1-MMP promoting collagen contraction (Deryugina et al., 1998), knockdown of MT1-MMP with three independent siRNA duplexes reduced contraction of collagen by CAFs (Fig. 8 D). Similar to the lack of effect of LIMKi on pMLC, MT1-MMP knockdown did not affect pMLC levels (Fig. 8 E). These results indicate that both LIMK and MT1-MMP are required for matrix deformation without affecting MLC phosphorylation, suggesting MMPs facilitate contraction by breaking down the cross-linking that makes the ECM rigid to allow for subsequent contraction, which additionally contributes to the ECM remodeling required for invasive path generation.
Figure 8.
Matrix contraction by cancer-associated fibroblasts requires LIMK activity. (A) Phosphorylation of myosin light chain on Ser19 (pS19-MLC) was significantly reduced by the ROCK inhibitor Y27632 but not by LIMKi (box equals upper and lower quartiles with line indicating median; whiskers are 5 and 95% confidence intervals; n = 24–30). (B) Force-mediated remodeling by fibroblasts contracts matrix protein. Scoring the percentage of contraction area per well revealed significant inhibition of contraction by increasing LIMKi doses (average ± SEM, n = 3). (C) Cells transfected with LIMK1/2 siRNA relative to NT siRNA transfectants had reduced matrix contraction (average ± SEM, n = 3). (D) Knockdown of MT1-MMP reduced matrix contraction. (E) MT1-MMP knockdown did not affect MLC phosphorylation (box equals upper and lower quartiles with line indicating median; whiskers are 5 and 95% confidence intervals; n = 24–30 per treatment).
Discussion
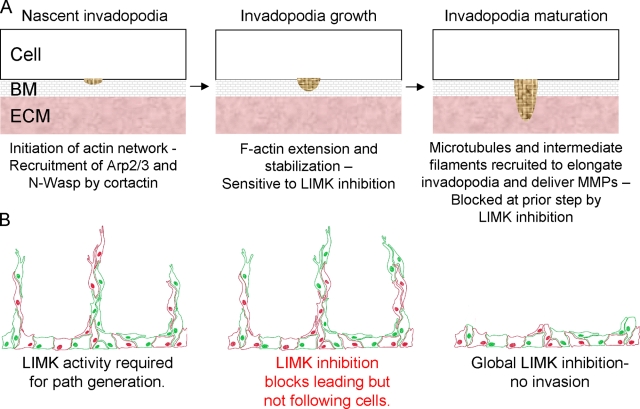
The ability of tumor cells to spread from a primary mass is dependent on ECM remodeling to generate paths that permit movement, and on motility to move through the paths. Many cancer cells, when plated on substrates composed of matrix proteins, form ventral actin-rich structures called invadopodia that are associated with ECM proteolytic activity (Yamaguchi et al., 2006; Linder, 2007; Buccione et al., 2009). The ability of cancer cells to make invadopodia often correlates with their ability to enter the vasculature (Yamaguchi et al., 2006), and as such likely are a representation of the ECM degrading activities that facilitate invasive path generation. The formation of invadopodia requires the activity of the actin-nucleating Arp2/3 complex, which is regulated by cortactin (Yamaguchi et al., 2005; Oser and Condeelis, 2009; Oser et al., 2009). The birth, growth, and maturation of invadopodia takes place over a number of stages (Artym et al., 2006; Oser et al., 2009; Schoumacher et al., 2010; Fig. 9 A): (1) Accumulation of cortactin and recruitment of actin polymerization machinery including Arp2/3 and N-Wasp at sites of membrane adherence to ECM; (2) F-actin extension and stabilization; and (3) recruitment of microtubules and intermediate filaments for invadopodia elongation and delivery of MMPs. Although cortactin foci were still formed (Fig. 4 D), the stability of F-actin structures was significantly reduced when LIMK was inhibited (Fig. 4 E). In addition, the accumulation of discrete subcellular structures containing MMP9 and ECM proteolytic activity were blocked by LIMK inhibitor (Figs. 4 C and 5 A). The results from this study indicate that the latter two steps involving invadopodia growth and maturation were sensitive to LIMK inhibition. Although the association of cortactin with cofilin was reported to be sufficient to F-actin stabilization and invadopodia maturation (Oser et al., 2009), LIMK activity apparently also is required for adequate cofilin inactivation to complete maturation and formation of functional invadopodia.
Figure 9.
Model of LIMK requirement in path-generating leading cells in collective invasion. (A) Leading cells degrade basement membrane (BM) and extracellular matrix (ECM) to generate a path of low mechanical resistance. Invadopodia undergo a growth and maturation process, developing at their site of birth. LIMK activity is required in the second step of invadopodia growth for stabilization of F-actin to allow for extension of the cytoskeleton. If the ability of the actin cytoskeleton to extend is impaired by LIMK inhibition, maturation cannot occur, which also impacts upon MMP delivery and results in reduced ECM proteolysis. (B) The path-generating activity may be provided by tumor or nontumor stromal cells. Following cells move through paths created by leading cells. LIMK activity allows for cytoskeletal structures required for ECM degradation and deformation to be formed. If LIMK activity is blocked in a cell subpopulation, either tumor or nontumor, they no longer have the ability to generate paths, but will follow paths generated by unaffected cells. Global LIMK inhibition blocks path generation leading to decreased invasion. By targeting the path-generating activity the contributions of tumor and nontumor stromal cells are inhibited.
The results from this study reveal that LIMK activity is crucial for the cellular processes that contribute to path generation, namely ECM degradation and force-mediated deformation (Fig. 9 B). The path-generating function may be provided by leading tumor cells or by stromal cells; in either case LIMK inhibition blocked their ability to make paths, but not to follow preformed paths or the paths made by noninhibited cells in collective invasion (Fig. 9 B). Global LIMK inhibition blocked invasion because the ECM remodeling processes necessary for path generation were no longer active (Fig. 9 B).
The ability of tumor cells to invade and spread is critically dependent upon dynamic and regulated rearrangements to the actin-myosin cytoskeleton (Olson and Sahai, 2009). F-actin filaments are in a constant state of flux with new monomers being added at the plus end, depolymerization at the minus end, and F-actin severing occurring within the filament. Due to the complexity of these dynamic processes and their importance to cellular homeostasis, a vast number of proteins contribute in numerous ways to their regulation. Although it might seem that the effect of altering the input from a single protein would be unlikely to produce a significant change in output, data from clinical and basic studies indicate that LIM kinases are important factors in cancer cell invasion and metastasis (Davila et al., 2003; Yoshioka et al., 2003; Wang et al., 2004; Okamoto et al., 2005; Bagheri-Yarmand et al., 2006). In support of this conclusion, results from in vivo studies indicated that interfering with LIMK function inhibited metastasis (Yoshioka et al., 2003; Vlecken and Bagowski, 2009). These reports are consistent with the hypothesis that LIM kinases are important regulators of actin cytoskeleton dynamics and that LIMK inhibition would effectively reduce cancer cell invasion and metastasis. The results from this study also support the possibility that pharmacological LIMK inhibition will be efficacious in blocking the spread of metastatic cancers.
Although numerous lines of basic and clinical research support LIMK as a positive factor in cell motility and/or invasion (Yoshioka et al., 2003; Nishita et al., 2005; Kobayashi et al., 2006; Ding et al., 2008; Horita et al., 2008; Mishima et al., 2010), the field has not been without controversy as it has also been reported that LIMK expression may have an inhibitory effect on cell motility and invasion. Indeed, we found that expression of full-length LIMK1 inhibited MDA-MB-231 invasion (unpublished data) and decreased the motility of Ras-transformed Swiss 3T3 fibroblasts (Sahai et al., 2001). In addition, overexpression of LIMK1 in MTLn3 rat mammary carcinoma cells inhibited lamellipod extensions (Zebda et al., 2000), EGF-stimulated motility, and in vivo invasion (Wang et al., 2006). These seemingly contradictory observations may be explained if the relationship between LIMK and cofilin is not considered as a simple linear pathway in which more LIMK activity equals more cofilin phosphorylation equals more invasion. Instead, the dynamic cycling of cofilin between inactive and active states (the “cofilin activity cycle”) influenced significantly by phosphorylation/dephosphorylation must be regarded as a key determinant of invasive and metastatic potential (Oser and Condeelis, 2009; van Rheenen et al., 2009). Depending on the initial starting point, small changes in LIMK activity and consequent cofilin phosphorylation could induce increased or decreased cell invasiveness depending on whether the cofilin activity cycle was moving toward or away from an optimal state. If the hypothesis that cancer cells acquire the ability to invade and metastasize as one of the “hallmarks of cancer” (Hanahan and Weinberg, 2000), then it would be predicted that the cofilin activity cycle should approach the most favorable operational state for increased invasive ability as cells progress from normal to cancerous, in which case tipping the levels of cofilin phosphorylation in either direction and restricting the ability of cofilin to dynamically cycle between active and inactivate states should decrease invasion. Consequently, robust LIMK inhibition should consistently decrease the invasiveness of highly metastatic cells by skewing the cofilin activity cycle away from optimal by driving cofilin toward the dephosphorylated state and by limiting cycling. The results of this study indicate that the abilities of MDA-MB-231 breast cancer cells and CAFs to generate paths required for invasiveness were indeed inhibited by knockdown or pharmacological inhibition of LIMK.
In conclusion, we have shown that inhibition of LIMK activity by siRNA-mediated knockdown or pharmacological inhibition blocked the collective invasion of tumor cells by blocking activities required for path generation. A major challenge for the future will be to design clinical trials to test the effectiveness of anti-invasive therapeutic agents within a practical and cost-effective time frame. One possibility is that LIMK inhibitors would reduce the ability of tumor cells to move away from the primary tumor to the point where they would be able to intravasate and enter the circulation. In a tumor such as prostate cancer, where the presence of a localized tumor may not be life threatening but mortality results from metastatic spread, the effect of LIMK inhibition on the levels of circulating tumor cells might be an intermediate endpoint in clinical trials to monitor disease status and progression that ultimately could be correlated with survival to evaluate efficacy (Scher et al., 2009).
Materials and methods
Cell culture
MDA-MB-231 breast cancer cells were grown in DME with 10% fetal bovine serum (FBS) and 2 mM l-glutamine plus 10 U/ml penicillin and 10 µg/ml streptomycin at 37°C in 5% CO2 in a humidified incubator. Cancer-associated fibroblasts were cultured in DME supplemented with 10% FBS, 1% ITS (insulin, transferrin, selenium), 2 mM l-glutamine, 10 U/ml penicillin, and 10 µg/ml streptomycin. Organotypic skin model was performed as described in Gaggioli et al. (2007), with the change that puromycin was used to kill cancer-associated fibroblasts rather than detergent extraction. Matrix contraction was quantified by scanning the plates and the diameter of the gel to well was measured using ImageJ software (http://rsbweb.nih.gov/ij). The extent of contraction was assessed by subtracting the gel area from the well area and percentage changes were calculated as described previously (Hooper et al., 2010).
siRNA transfection
All siRNA reagents were obtained from Thermo Fisher Scientific. Oligofectamine was used for transfection of siRNA on subconfluent cells, according to the manufacturer’s instructions. siRNA reference numbers and sense sequences are as follows: LIMK1-1, D-007730-01, GAACGCAGACCCUGACUACUU; LIMK1-3, D-007730-03, UCAAGGAGGUGAAGGUCAUUU; LIMK2-6, D-003311-06, GAUCUGAACUCGCACAACUUU; LIMK2-7, D-003311-07, UGACUGAGGUGAAAGUGAUUU.
Retrovirus production and transduction
Phoenix ecotropic packaging cells were transfected with pBABE puro GFP or RFP using Effectene (QIAGEN) according to the manufacturer’s instructions. 24 h later cells were washed twice and 1.5 ml of DMEM plus 10% FBS was added per well. Virus was collected after 48 h and centrifuged to remove any cells, and supernatant was stored at −80°C. Ecotropic receptor–expressing MDA-MB-231 cells were infected with 1 ml of virus and 4 µl polybrene (4 mg/ml). The next day the media was replaced and cells were subsequently selected with puromycin. Uniform GFP or RFP expression was selected by flow cytometric sorting.
Recombinant protein preparation
Overnight cultures of pGEX-KG TAT-FLAG-COFILIN or pGEX-KG TAT-FLAG in BL21 (DE3) pLyss were each diluted 1:10 and grown to OD600 0.6–1.0 at 37°C before induction with 100 µM IPTG for 3 h at 37°C. Cells were pelleted, resuspended in 5 ml of PBS with 1x Complete protease inhibitor cocktail, and then disrupted by three 1-min rounds of sonication at 20% intensity using a digital sonifier (model 250; Branson Ultrasonics). Debris was removed by centrifugation and clarified supernatants were incubated with PBS-washed glutathione–Sepharose (GE) bead slurry overnight at 4°C. Beads were washed with PBS and cofilin released from the GST moiety by incubation with 100 units of bovine thrombin (Sigma-Aldrich) overnight at 4°C. Alternatively, recombinant GST was eluted with 5 mM reduced glutathione overnight at 4°C. Supernatant was removed and incubated with 30 µl of washed p-aminobenzamidine beads (Sigma-Aldrich) for 1 h at room temperature to remove thrombin, before snap-freezing aliquots and storing at −80°C.
Cell migration assay
Cell migration assay was performed in Oris 96-well plates with rubber plugs in each well occluding a central portion. Once cells had settled, plugs were removed to create an identical central cell-free zone. After 24 h, cells were imaged on an upright tissue culture microscope using QcapturePro. To quantify the level of migration into the center of the well, cells were stained Calcein AM and scanned with a plate reader (Safire2; Tecan Group Ltd.).
Inverse invasion assay
Cell invasion assays were a modification of a method described previously (Hennigan et al., 1994). Complete matrigel was thawed on ice, diluted 1:1 with PBS and then 100 µl of diluted matrigel pipetted into Transwell inserts in a 24-well tissue culture plate and left to incubate for 30 min at 37°C to set. During this time, cell suspensions of 3 × 105 cells per ml from each condition were prepared in their normal growth medium. When the matrigel had set, the Transwell inserts were inverted and 100 µl of cell suspension pipetted onto the filter. The Transwell inserts were then carefully covered with the base of the 24-well tissue culture plate, making contact with each cell suspension droplet, and the plate then incubated in the inverted state for 4 h to allow cell attachment. After this time, plates were turned right-side-up and each Transwell insert washed with 3 × 1 ml serum-free DMEM and finally placed in a well containing any additional inhibitors as indicated. DMEM plus 10% FBS was gently pipetted on top of the set matrigel/PBS mixture and incubated for 5 d at 37°C. Transwell inserts were placed in fresh 24-well dishes and 1 ml of 4 µM Calcein AM stain solution pipetted on top. After 1 h of incubation at 37°C, cells were imaged by confocal microscopy with a 20x objective with optical Z-sections scanned at 15-µm intervals moving up from the underside of the filter into the matrigel. ImageJ software was used for quantification. Cells stably expressing RFP or GFP were paraformaldehyde fixed before imaging. Alternatively, propidium iodide (PI) was used to visualize nuclei after fixation with paraformaldehyde; F-actin staining was optionally stained with FITC-conjugated phalloidin.
Gelatin degradation assay
Sterilized acid-washed 13-mm glass coverslips were placed on top of 100 µl of poly-l-lysine for 15 min at room temperature. Excess liquid was removed and the coated side placed on top of 100 µl of 0.5% gluteraldehyde (in PBS) for 10 min. The uncoated surfaces were then placed on 60 µl of 488-nm fluorescent gelatin for 15 min. The coverslips were washed three times with PB and then with 5 mg/ml sodium borohydrate solution in PBS for 3 min, and six further PBS washes before sterilizing the coverslips in 70% EtOH for 5 min. Finally, the EtOH was aspirated and the coverslips quenched in DMEM with 10% FBS for 1 h at 37°C. 3 × 104 cells were plated on fluorescent gelatin–coated coverslips in media containing 10 µM GM6001 MMP inhibitor and allowed to adhere at 37°C overnight. Cells were washed three times with PBS to remove the MMP inhibitor before the addition of complete media containing inhibitors as indicated. After 4 h, the coverslips were washed before 4% paraformaldehyde fixation and permeabilization with 0.1% Triton X-100 in PBS for 15 min. After blocking for 30 min in 5% BSA, F-actin was stained with Texas red–conjugated phalloidin. Coverslips were mounted using ProLong Gold Antifade reagent and viewed on an upright fluorescent microscope.
Motility on cell-derived matrix
Sterilized coverslips were incubated with 0.2% gelatin for 1 h at 37°C, washed twice with PBS, then cross-linked with 1% glutaraldehyde for 30 min at room temperature. After two washes, cross-linker was quenched with 1 M glycine in PBS (pH 7) for 20 min at room temperature, washed twice with PBS, and then incubated with growth media for 30 min at 37°C before seeding with human dermal fibroblasts at 5 × 104 cells per well of a 24-well plate. Cells were cultured at 37°C in 5% CO2 overnight before changing the media to complete media supplemented with 50 µg/ml ascorbic acid. The ascorbic acid media was changed every second day to increase collagen production and stabilize the matrix. Cells were removed with prewarmed extraction buffer (20 mM NH4OH and 0.5% Triton X-100 in PBS). Residual DNA was digested with 10 µg/ml DNase I in PBS containing calcium and magnesium and incubated for 30 min at 37°C. Matrix-coated coverslips were washed twice then stored at 4°C in PBS containing penicillin, streptomycin, and fungizone. Before use, coverslips were washed with PBS and incubated for 1 h at 37°C with DMEM. After cells were plated subconfluently, tissue culture dishes were transferred to a time-lapse microscope with a 37°C, 5% CO2 incubation chamber for 1 h to allow cells to settle and for the plasticware to equilibrate. Images were taken using an inverted microscope system (Carl Zeiss, Inc.) with a 10x objective and bright-field channels at 15-min intervals over up to 24 h. Analysis of experimental data was performed using ImageJ, or AndorIQ software.
Protein and mRNA preparation
RNA was isolated from MDA-MB-231 cells using the RNeasy kit according to the manufacturer’s instructions (QIAGEN). RNA was eluted with RNase-free water and stored at −20°C. Protein lysates were made and Western blotted as described previously (Coleman et al., 2001). In some instances Western blots were visualized with infrared fluorophore-conjugated secondary antibodies and detected with an Odyssey system (LI-COR Biosciences).
cDNA synthesis and qPCR
RNA was reverse transcribed into cDNA with the Dynamo SYBR Green 2-step qRT-PCR kit according to the manufacturer’s instructions (Finnzymes). The qPCR reactions were performed as per the manufacturer’s instructions on a DNA Engine thermal cycler (Bio-Rad Laboratories) with a Chromo4 real-time PCR detector and Opticon Monitor 3 using the appropriate optimized protocol.
Antibodies
Antibodies were routinely used at 1:1,000 for Western blotting and 1:500 for immunofluorescence. Antibodies used were: cofilin (Cell Signaling Technology); cofilin (Cytoskeleton, Inc.); LIMK1 (Cell Signaling Technology); LIMK2 (Santa Cruz Biotechnology, Inc.); α-tubulin (Sigma-Aldrich); β-tubulin (Sigma-Aldrich); FLAG (Sigma-Aldrich); phospho-cofilin (Cell Signaling Technology); MT1-MMP (MMP14; Neomarkers); MMP9 (Neomarkers); cortactin (Millipore).
F-actin severing
G-actin was purified from rabbit muscle as described previously (Spudich and Watt, 1971) and maintained in G-buffer (2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 0.5 mM DTT, and 0.2 mM CaCl2). 1 mg/ml G-actin was labeled with 5 mg/ml pyrene in dimethylformamide for 15 h and dialyzed in G-buffer. Actin polymerization was initiated by the addition of 50 mM KCl, 2 mM MgCl2, and 0.1 mM EGTA, and then ultracentrifuged to pellet the F-actin. The F-actin pellet resuspended in G-buffer and concentration determined. For the severing assay, G-actin (95% unlabeled, 5% pyrene labeled) was polymerized at 2.5 µM (G-actin final concentration) overnight. Labeled F-actin (50 µl at 2.5 µM) was added to a PTI glass cuvette (with or without the addition of 10 µM TAT-FLAG-Cofilin or TAT-GST) and steady-state fluorescence (excitation = 339 nm; emission = 384 nm) was measured on a spectrofluorimeter for 1,000 s. Data were normalized to allow comparison between datasets, and reported as percent fluorescence over time.
Cell adhesion/spreading assay
Kinetics of cell adhesion and spreading were assayed using the xCELLigence Real-Time Cell Analyzer (RTCA DP; Roche). E-plate 16 wells were incubated with 100 µl of 25 µg/ml fibronectin for 1 h at 37°C, then 5,000 cells were plated in each well with DMSO vehicle or indicated concentration of LIMKi. Cells were allowed to settle for 30 min at room temperature before being placed in the RTCA DP in a humidified incubator at 37°C with 5% CO2. Readings were taken every 15 min for 24 h and plotted curves represent the averages from four independent wells.
Zymography
Concentrated conditioned media samples were prepared in nonreducing SDS buffer and electrophoresed on a 10% Tris-Glycine polyacrylamide gel with 0.1% gelatin incorporated as a substrate, using Tris-Glycine SDS running buffer. Gelatinase activity was renatured by incubation in Zymogram renaturing buffer containing a nonionic detergent, and then equilibration in Zymogram developing buffer to add divalent cation for enzymatic activity. To visualize protein, the gel was stained using Safestain (Invitrogen).
Immunofluorescence microscopy
Cells were grown on glass coverslips and fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 and blocked in 1% BSA. Primary antibodies were used at a 1:1,000 dilution in blocking buffer. After 1–24 h of incubation at room temperature (depending on the antibody) cells were washed in blocking buffer, then secondary antibody added at 1:500 dilution in blocking buffer (plus fluorescently labeled phalloidin if required) for 1 h. Coverslips were washed and then mounted on a glass slide with Vectashield (with or without DAPI) or ProLong Gold antifade and sealed with clear nail varnish. The slides were imaged using a confocal microscope and LCS software (Leica). Alternatively, image acquisition and analysis was performed with a Cellomics Arrayscan using the Morphology Explorer BioApplication according to the manufacturer’s instructions. For the descriptions of actin fiber count, area, and alignment, the Morphology Explorer BioApplication detects the patterns of fluorescent staining within each cell and classifies structures over a certain minimum cut-off size as either spots or fibers, depending on criteria such as object length and length/width ratio. Having identified fibers, the software calculates the number of fibers per cell, average area of fibers per cell, average area per fiber, and alignment relative to other fibers in the cell. In each case, the measurements were normalized to the DMSO vehicle–treated group (value = 1 ± SEM; n = 3). Quantification of pMLC levels was performed with ImageJ.
FRAP imaging and analysis
All FRAP experiments were performed on a confocal microscope (FV1000; Olympus) equipped with a UplanS Apo 60x 1.35 oil objective. The 488 laser line was used for imaging and existing standard dye settings for EGFP excitation and emission were chosen. A separate SIM scanner controlling an independent 405 laser enabled the bleaching to occur simultaneously with the imaging scan. The cells were kept at 37°C in a heating chamber placed on the stage. Acquisition was done using Fluoview software (Olympus). Scan parameters for imaging included an image size of 512 × 512 pixels with a scan rate of 1.107 s per frame. The region of interest for the bleach was created with the circular region tool and bleached for around 20 ms.
Image analysis was done in ImageJ, where a circular region was fitted into the bleached area and a recovery curve was created using the “intensity versus time” plugin. The data of each curve were exported and fitted in SigmaPlot using the following equation: I = Io + a(1 − exp(−bx)) + c(1 − exp(−dx). The resulting parameters b and d were extracted. b represents the fast, cytoplasmic recovery and d represents the recovery of the actin in structures, and the latter was therefore used to obtain the presented lifetimes. The average of d under each condition was taken and the correlating lifetime was calculated using the equation: t1/2 = ln2/d. A two-tailed, homoscedastic Student’s t test was performed in Excel to compare the resulting t1/2.
Online supplemental material
Fig. S1 shows the validation of the LIMK1 and LIMK2 siRNA duplexes used, and the lack of effect on cell viability. Fig. S2 shows the lack of effect of the LIMK inhibitor on cell size and shape parameters. Fig. S3 shows the lack of effect of LIMK inhibitor on microtubule numbers or organization. Fig. S4 shows the colocalization of MMP9, cortactin, N-Wasp, and F-actin with invadopodia. Fig. S5 shows MMP9 and MT1-MMP expression in SCC12 cells and CAFs. Video 1 shows rotating views of MDA-MB-231 invasion into 3D matrix. Stacks of confocal images were rendered into a rotating view of NT siRNA or LIMK1/2-transfected MDA-MB-231 cells using Volocity software. Strands of associated cells collectively invaded upward into matrix protein. Video 2 shows a time-lapse video of NT siRNA and LIMK1/2 siRNA transfected MDA-MB-231 cell motility on fibroblast-derived matrix. Images were acquired at 10-min intervals for 16 h. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002041/DC1.
Acknowledgments
Thanks to Martin Stockley for LIMKi synthesis. Diane Crighton and Elisabeth Trivier are employed by Cancer Research Technology Ltd., a private company wholly owned by the nonprofit medical charity Cancer Research UK.
This research was supported by Cancer Research UK and a grant to M.F. Olson from the National Institutes of Health (CA030721).
Footnotes
Abbreviations used in this paper:
- CAF
- cancer-associated fibroblast
- LIMK
- LIM kinase
- MMP
- matrix metalloprotease
- NT
- nontargeting
- pMLC
- myosin light chain phosphorylation
- SCC
- squamous carcinoma cell
References
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., Mueller S.C. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Bagheri-Yarmand R., Mazumdar A., Sahin A.A., Kumar R. 2006. LIM kinase 1 increases tumor metastasis of human breast cancer cells via regulation of the urokinase-type plasminogen activator system. Int. J. Cancer. 118:2703–2710 10.1002/ijc.21650 [DOI] [PubMed] [Google Scholar]
- Benitah S.A., Valerón P.F., van Aelst L., Marshall C.J., Lacal J.C. 2004. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim. Biophys. Acta. 1705:121–132 [DOI] [PubMed] [Google Scholar]
- Buccione R., Caldieri G., Ayala I. 2009. Invadopodia: specialized tumor cell structures for the focal degradation of the extracellular matrix. Cancer Metastasis Rev. 28:137–149 10.1007/s10555-008-9176-1 [DOI] [PubMed] [Google Scholar]
- Cano M.L., Lauffenburger D.A., Zigmond S.H. 1991. Kinetic analysis of F-actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulation increases actin filament number without altering the filament length distribution. J. Cell Biol. 115:677–687 10.1083/jcb.115.3.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M.L., Sahai E.A., Yeo M., Bosch M., Dewar A., Olson M.F. 2001. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3:339–345 10.1038/35070009 [DOI] [PubMed] [Google Scholar]
- Copeland J.W., Treisman R. 2002. The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol. Biol. Cell. 13:4088–4099 10.1091/mbc.02-06-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., Yamada K.M. 2001. Taking cell-matrix adhesions to the third dimension. Science. 294:1708–1712 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Davila M., Frost A.R., Grizzle W.E., Chakrabarti R. 2003. LIM kinase 1 is essential for the invasive growth of prostate epithelial cells: implications in prostate cancer. J. Biol. Chem. 278:36868–36875 10.1074/jbc.M306196200 [DOI] [PubMed] [Google Scholar]
- Deryugina E.I., Bourdon M.A., Reisfeld R.A., Strongin A. 1998. Remodeling of collagen matrix by human tumor cells requires activation and cell surface association of matrix metalloproteinase-2. Cancer Res. 58:3743–3750 [PubMed] [Google Scholar]
- Ding Y., Milosavljevic T., Alahari S.K. 2008. Nischarin inhibits LIM kinase to regulate cofilin phosphorylation and cell invasion. Mol. Cell. Biol. 28:3742–3756 10.1128/MCB.01832-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A.D., Wang F.W., Matsumoto K., Yamada K.M. 2009. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 184:481–490 10.1083/jcb.200810041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Wolf K. 2008. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 68:7247–7249 10.1158/0008-5472.CAN-08-0784 [DOI] [PubMed] [Google Scholar]
- Gaggioli C. 2008. Collective invasion of carcinoma cells: when the fibroblasts take the lead. Cell Adh Migr. 2:45–47 10.4161/cam.2.1.5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., Sahai E. 2007. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9:1392–1400 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- Geneste O., Copeland J.W., Treisman R. 2002. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell Biol. 157:831–838 10.1083/jcb.200203126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. 2009. The cytoskeleton and cancer. Cancer Metastasis Rev. 28:5–14 10.1007/s10555-008-9166-3 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2000. The hallmarks of cancer. Cell. 100:57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hawkins M., Pope B., Maciver S.K., Weeds A.G. 1993. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 32:9985–9993 10.1021/bi00089a014 [DOI] [PubMed] [Google Scholar]
- Hennigan R.F., Hawker K.L., Ozanne B.W. 1994. Fos-transformation activates genes associated with invasion. Oncogene. 9:3591–3600 [PubMed] [Google Scholar]
- Ho A., Schwarze S.R., Mermelstein S.J., Waksman G., Dowdy S.F. 2001. Synthetic protein transduction domains: enhanced transduction potential in vitro and in vivo. Cancer Res. 61:474–477 [PubMed] [Google Scholar]
- Hooper S., Gaggioli C., Sahai E. 2010. A chemical biology screen reveals a role for Rab21-mediated control of actomyosin contractility in fibroblast-driven cancer invasion. Br. J. Cancer. 102:392–402 10.1038/sj.bjc.6605469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita Y., Ohashi K., Mukai M., Inoue M., Mizuno K. 2008. Suppression of the invasive capacity of rat ascites hepatoma cells by knockdown of Slingshot or LIM kinase. J. Biol. Chem. 283:6013–6021 10.1074/jbc.M706538200 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Seiki M. 2004. MT1-MMP: an enzyme with multidimensional regulation. Trends Biochem. Sci. 29:285–289 10.1016/j.tibs.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Kleinman H.K., Martin G.R. 2005. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15:378–386 10.1016/j.semcancer.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Nishita M., Mishima T., Ohashi K., Mizuno K. 2006. MAPKAPK-2-mediated LIM-kinase activation is critical for VEGF-induced actin remodeling and cell migration. EMBO J. 25:713–726 10.1038/sj.emboj.7600973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunigal S., Lakka S.S., Gondi C.S., Estes N., Rao J.S. 2007. RNAi-mediated downregulation of urokinase plasminogen activator receptor and matrix metalloprotease-9 in human breast cancer cells results in decreased tumor invasion, angiogenesis and growth. Int. J. Cancer. 121:2307–2316 10.1002/ijc.22962 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li A., Dawson J.C., Forero-Vargas M., Spence H.J., Yu X., König I., Anderson K., Machesky L.M. 2010. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20:339–345 10.1016/j.cub.2009.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmeyer F., Legrand Y., Menashi S. 1997. Upregulation of MMP-9 expression in MDA-MB231 tumor cells by platelet granular membrane. FEBS Lett. 418:19–22 10.1016/S0014-5793(97)01336-7 [DOI] [PubMed] [Google Scholar]
- Linder S. 2007. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 17:107–117 10.1016/j.tcb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Lizárraga F., Poincloux R., Romao M., Montagnac G., Le Dez G., Bonne I., Rigaill G., Raposo G., Chavrier P. 2009. Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 69:2792–2800 10.1158/0008-5472.CAN-08-3709 [DOI] [PubMed] [Google Scholar]
- Mishima T., Naotsuka M., Horita Y., Sato M., Ohashi K., Mizuno K. 2010. LIM-kinase is critical for the mesenchymal-to-amoeboid cell morphological transition in 3D matrices. Biochem. Biophys. Res. Commun. 392:577–581 10.1016/j.bbrc.2010.01.075 [DOI] [PubMed] [Google Scholar]
- Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., Mizuno K. 2005. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 171:349–359 10.1083/jcb.200504029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I., Pirker C., Bilban M., Berger W., Losert D., Marosi C., Haas O.A., Wolff K., Pehamberger H. 2005. Seven novel and stable translocations associated with oncogenic gene expression in malignant melanoma. Neoplasia. 7:303–311 10.1593/neo.04514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.F. 2008. Applications for ROCK kinase inhibition. Curr. Opin. Cell Biol. 20:242–248 10.1016/j.ceb.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.F., Sahai E. 2009. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis. 26:273–287 10.1007/s10585-008-9174-2 [DOI] [PubMed] [Google Scholar]
- Oser M., Condeelis J. 2009. The cofilin activity cycle in lamellipodia and invadopodia. J. Cell. Biochem. 108:1252–1262 10.1002/jcb.22372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C.C., Bravo-Cordero J.J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A.J., Condeelis J. 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186:571–587 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Lizárraga F., Chavrier P. 2009. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122:3015–3024 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- Riento K., Ridley A.J. 2003. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4:446–456 10.1038/nrm1128 [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P., de Silva H., Guo Q., Xiao H., Hung C.Y., Penhallow B., Markwalder J., He L., Attar R.M., Lin T.A., et al. 2008. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol. Cancer Ther. 7:3490–3498 10.1158/1535-7163.MCT-08-0826 [DOI] [PubMed] [Google Scholar]
- Sahai E. 2005. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 15:87–96 10.1016/j.gde.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. 2002. RHO-GTPases and cancer. Nat. Rev. Cancer. 2:133–142 10.1038/nrc725 [DOI] [PubMed] [Google Scholar]
- Sahai E., Olson M.F., Marshall C.J. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755–766 10.1093/emboj/20.4.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher H.I., Jia X., de Bono J.S., Fleisher M., Pienta K.J., Raghavan D., Heller G. 2009. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 10:233–239 10.1016/S1470-2045(08)70340-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., Vignjevic D.M. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.W., Olson M.F. 2007. LIM kinases: function, regulation and association with human disease. J. Mol. Med. 85:555–568 10.1007/s00109-007-0165-6 [DOI] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871 [PubMed] [Google Scholar]
- Stylli S.S., Kaye A.H., Lock P. 2008. Invadopodia: at the cutting edge of tumour invasion. J. Clin. Neurosci. 15:725–737 10.1016/j.jocn.2008.03.003 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Condeelis J., Glogauer M. 2009. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 122:305–311 10.1242/jcs.031146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. 2008. Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87:649–667 10.1016/j.ejcb.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Vega F.M., Ridley A.J. 2008. Rho GTPases in cancer cell biology. FEBS Lett. 582:2093–2101 10.1016/j.febslet.2008.04.039 [DOI] [PubMed] [Google Scholar]
- Vlecken D.H., Bagowski C.P. 2009. LIMK1 and LIMK2 are important for metastatic behavior and tumor cell-induced angiogenesis of pancreatic cancer cells. Zebrafish. 6:433–439 10.1089/zeb.2009.0602 [DOI] [PubMed] [Google Scholar]
- Walker K., Olson M.F. 2005. Targeting Ras and Rho GTPases as opportunities for cancer therapeutics. Curr. Opin. Genet. Dev. 15:62–68 10.1016/j.gde.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Wang W., Goswami S., Lapidus K., Wells A.L., Wyckoff J.B., Sahai E., Singer R.H., Segall J.E., Condeelis J.S. 2004. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 64:8585–8594 10.1158/0008-5472.CAN-04-1136 [DOI] [PubMed] [Google Scholar]
- Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A.R., Condeelis J.S. 2006. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 173:395–404 10.1083/jcb.200510115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Wu Y.I., Liu Y., Geiger J., Tam E., Overall C., Stack M.S., Friedl P. 2007. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9:893–904 10.1038/ncb1616 [DOI] [PubMed] [Google Scholar]
- Wyckoff J.B., Pinner S.E., Gschmeissner S., Condeelis J.S., Sahai E. 2006. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 16:1515–1523 10.1016/j.cub.2006.05.065 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Pixley F., Condeelis J. 2006. Invadopodia and podosomes in tumor invasion. Eur. J. Cell Biol. 85:213–218 10.1016/j.ejcb.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Foletta V., Bernard O., Itoh K. 2003. A role for LIM kinase in cancer invasion. Proc. Natl. Acad. Sci. USA. 100:7247–7252 10.1073/pnas.1232344100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebda N., Bernard O., Bailly M., Welti S., Lawrence D.S., Condeelis J.S. 2000. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151:1119–1128 10.1083/jcb.151.5.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]