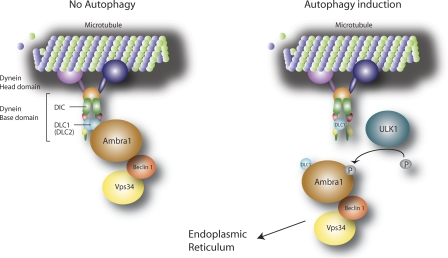
When autophagy is induced, ULK1 phosphorylates AMBRA1, releasing the autophagy core complex from the cytoskeleton and allowing its relocalization to the ER membrane to nucleate autophagosome formation.
Abstract
Autophagy is an evolutionary conserved catabolic process involved in several physiological and pathological processes such as cancer and neurodegeneration. Autophagy initiation signaling requires both the ULK1 kinase and the BECLIN 1–VPS34 core complex to generate autophagosomes, double-membraned vesicles that transfer cellular contents to lysosomes. In this study, we show that the BECLIN 1–VPS34 complex is tethered to the cytoskeleton through an interaction between the BECLIN 1–interacting protein AMBRA1 and dynein light chains 1/2. When autophagy is induced, ULK1 phosphorylates AMBRA1, releasing the autophagy core complex from dynein. Its subsequent relocalization to the endoplasmic reticulum enables autophagosome nucleation. Therefore, AMBRA1 constitutes a direct regulatory link between ULK1 and BECLIN 1–VPS34, which is required for core complex positioning and activity within the cell. Moreover, our results demonstrate that in addition to a function for microtubules in mediating autophagosome transport, there is a strict and regulatory relationship between cytoskeleton dynamics and autophagosome formation.
Introduction
Autophagy is a cellular process mediating degradation of bulk cytoplasm, long-lived proteins, and entire organelles (Klionsky, 2007; Mizushima, 2007). In this process, double-membraned vesicles, termed autophagosomes, wrap around portions of cytosol and transport them to the lysosome for degradation (Xie and Klionsky, 2007). In mammalian cells, mTOR is a nutrient sensor that represses autophagy and regulates protein synthesis (Hay and Sonenberg, 2004). The serine/threonine kinase ULK1 (a yeast ATG1 orthologue involved in mATG9 trafficking; Kuroyanagi et al., 1998; Young et al., 2006) plays an essential role downstream of mTOR on the regulation of autophagy. As described previously (Hosokawa et al., 2009), mTOR inhibits ULK1 activity by binding and phosphorylating it. After nutrient deprivation, mTOR dissociates from ULK1, allowing its activation. A few ULK1 targets have been identified so far, such as its interactors FIP200 and ATG13. However, a molecular link between ULK1 activity and autophagosome formation is still missing. Autophagosome formation requires phosphatidylinositol 3-phosphate (PI3P; Xie and Klionsky, 2007) and is believed to occur in specific subdomains of the ER, termed omegasomes (Axe et al., 2008; Hayashi-Nishino et al., 2009). Recent data suggest the mitochondria as additional sites of autophagosome formation (Hailey et al., 2010). BECLIN 1 and VPS34, which form a class III phosphatidylinositol 3-OH kinase (PI3K) complex, generate PI3P at the omegasome and are crucial for autophagosome nucleation (Suzuki and Ohsumi, 2007; Cecconi and Levine, 2008; Itakura et al., 2008; Levine and Kroemer, 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Recent findings provide strong biochemical evidence that mammalian BECLIN 1 exists in distinct class III PI3K complexes. Like in yeast, each complex seems to have a core consisting of BECLIN 1, VPS34, and VPS15 (Cecconi and Levine, 2008) and specific interactors, such as ATG14/BARKOR, UVRAG, or RUBICON, conferring them distinct functions in membrane trafficking (Itakura et al., 2008; Jahreiss et al., 2008; Kimura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). AMBRA1 has been identified as a crucial factor in regulating autophagy in vertebrates (Fimia et al., 2007). Its inactivation in vivo gives rise to defects in the developing nervous system and to embryonic death (Fimia et al., 2007; Cecconi et al., 2008). AMBRA1 promotes BECLIN 1 interaction with its target lipid kinase VPS34, thus mediating autophagosome nucleation (Fimia et al., 2007).
Once formed, the autophagosome moves toward the lysosome along the microtubules by means of the dynein motor complex (Ravikumar et al., 2005; Köchl et al., 2006; Jahreiss et al., 2008; Kimura et al., 2008). Besides its role as a cytoskeletal motor (King, 2000; Höök and Vallee, 2006), the dynein complex is also a docking system for regulatory factors involved in several signaling pathways (Jaffrey and Snyder, 1996; Crépieux et al., 1997; Campbell et al., 1998; Puthalakath et al., 1999, 2001; Herzig et al., 2000; Kaiser et al., 2003; Machado et al., 2003; Vadlamudi et al., 2004; Varadi et al., 2004). In particular, dynein light chain 1 (DLC1) and DLC2 are involved in cell death regulation by sequestering proapoptotic proteins (Puthalakath et al., 1999, 2001). In this study, we report that autophagy induction is regulated by a dynamic interaction between the BECLIN 1 core complex and the dynein motor complex, mediated by a direct binding between AMBRA1 and DLC1/2. Autophagy induction leads to the release of the BECLIN 1 core complex from dynein through an ULK1-dependent AMBRA1 phosphorylation. This event allows the translocation of the core complex to the ER, where it can prime autophagosome formation.
Results
AMBRA1 interacts with DLC1
To dissect the molecular mechanisms of AMBRA1 function, we performed a yeast two-hybrid assay using a cDNA encoding the C-terminal 533–1,269 amino acids of the human AMBRA1 protein. By screening a human brain cDNA library, we isolated, as an AMBRA1 interactor, the DLC 1 protein (also known as DYNLL1, LC8, and PIN).
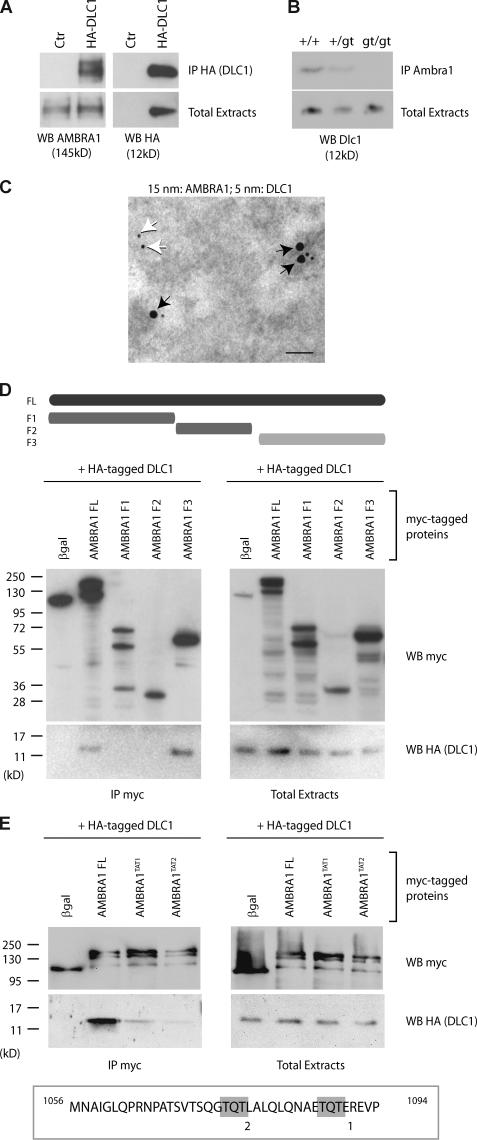
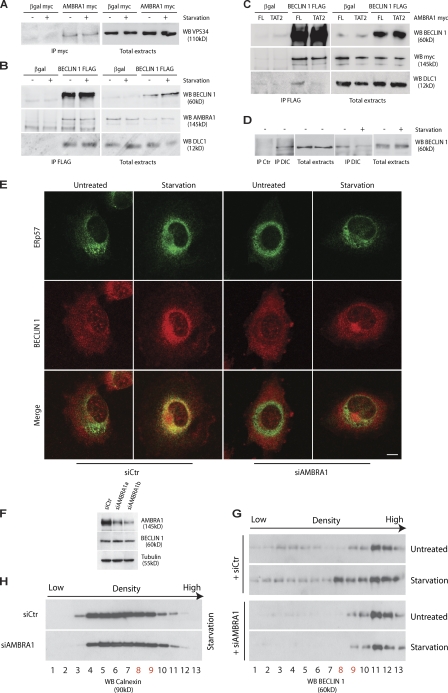
First, we decided to confirm the AMBRA1–DLC1 interaction in vivo by a coimmunoprecipitation (co-IP) assay. DLC1 and AMBRA1 associated effectively with each other when coexpressed in human 2FTGH (2F) fibroblasts (Fig. 1 A). Their binding was also tested in embryonic tissues, using protein extracts prepared from wild-type and Ambra1 mutant (Ambra1gt) embryos (Fimia et al., 2007). Dlc1 copurified with AMBRA1 in wild-type and Ambra1+/gt samples but not in samples where both Ambra1 alleles are mutated (Ambra1gt/gt; Fig. 1 B). Consistent with their interaction in vivo, DLC1 and AMBRA1 showed a significant colocalization by immunogold assay (Fig. 1 C and Fig. S1 A).
Figure 1.
AMBRA1 interacts with DLC1. (A) AMBRA1–DLC1 interaction in mammalian cells. 2F cells were infected with a retroviral vector encoding HA-DLC1 or an empty vector (control [Ctr]). Protein extracts were subjected to IP using an anti-HA antibody (IP HA; DLC1). Purified complexes and corresponding total extracts were analyzed by WB using anti-AMBRA1 (WB AMBRA1; left) or anti-HA antibodies (WB HA; right). (B) AMBRA1–Dlc1 interaction in mouse embryos. Protein extracts from embryos at developmental day 14.5 wild-type (+/+), heterozygous (+/gt), and homozygous (gt/gt) for the Ambra1 gene trap mutation were subjected to IP using an anti-AMBRA1 antibody (IP AMBRA1). Purified complexes and corresponding total extracts were analyzed by WB using an anti-DLC1 antibody (WB Dlc1). (C) AMBRA1–DLC1 colocalization in mammalian cells. Immunogold analysis of 2F cells coexpressing HA-DLC1 and AMBRA1. 15-nm and 5-nm gold particles label the AMBRA1 and DLC1 proteins, respectively. Black arrows point to AMBRA1–DLC1-colocalizing proteins, whereas white arrows indicate a single DLC1 molecule. A larger, low-magnification area of the same section is shown in Fig. S1 A. Bar, 45 nm. (D and E) Characterization of the DLC1-interacting domain of AMBRA1. (D) 2F cells were coinfected with retroviral vectors encoding HA-tagged DLC1 and the indicated Myc-tagged AMBRA1 proteins or Myc-tagged βgal as a negative control. FL, full length; F1–3, fragments 1–3 (a scheme of the AMBRA1 mutants with the corresponding amino acid sequence boundary is reported; the light gray bar indicates the DLC1-interacting fragment). Protein extracts were subjected to IP using an anti-Myc antibody (IP Myc). Purified complexes and corresponding total extracts were analyzed by WB using anti-Myc (WB Myc; top) and anti-HA antibody (WB HA [DLC1]; bottom). (E) 2F cells were coinfected with retroviral vectors encoding HA-tagged DLC1 and the Myc-tagged AMBRA1 FL or AMBRA1 mutants TAT1 and TAT2. The positions within the AMBRA1 sequence of the two TQT domains (1 and 2), which were mutated in the TAT mutants, are shown (gray boxes). Protein extracts were subjected to IP using an anti-Myc antibody (IP Myc). Purified complexes and corresponding total extracts were analyzed by WB using an anti-Myc (WB Myc; top) and anti-HA antibody (WB HA [DLC1]; bottom).
To map the AMBRA1 region responsible for DLC1 binding, different AMBRA1 cDNA deletion constructs were transduced in 2F cells and tested for their capacity to co-IP with DLC1. As shown in Fig. 1 D, the AMBRA1 F3 fragment (C terminal) is sufficient to bind DLC1, whereas the AMBRA1 N-terminal (F1) and central regions (F2) show no interaction. By in silico analysis, we identified two putative DLC1-binding motifs (TQT; Lo et al., 2001) on the AMBRA1 C-terminal sequence. To verify whether these motifs were indeed required for interaction, we generated two mutated cDNA constructs, namely AMBRA1TAT1 and AMBRA1TAT2, carrying a Q1088→A and a Q1076→A point mutation within the putative binding sites 1 and 2, respectively. By co-IP assays, we observed that both AMBRA1TAT1 and AMBRA1TAT2 interact with DLC1 to a very low extent compared with the AMBRA1 wild-type (full length [FL]) protein (Fig. 1 E).
AMBRA1 is released from the dynein motor complex upon autophagy induction
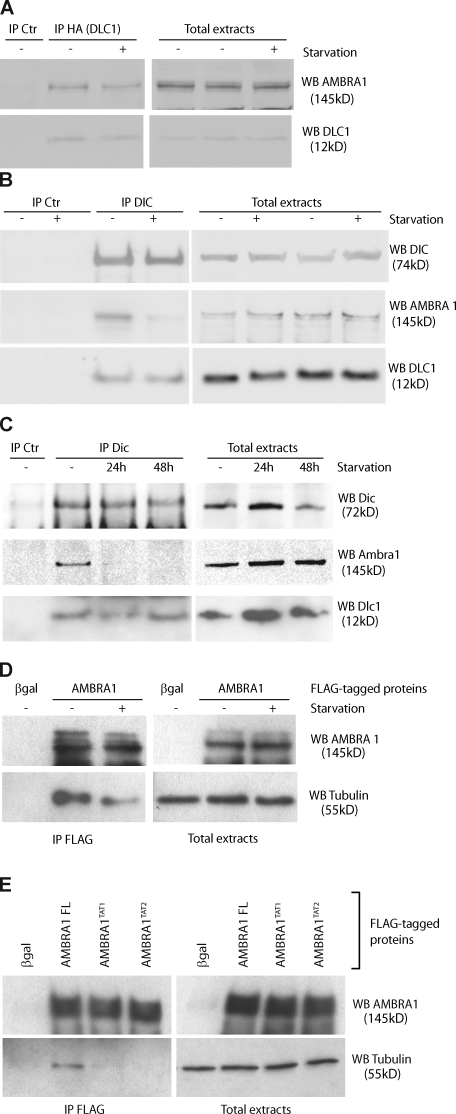
Because of the important role played by AMBRA1 in regulating autophagy (Cecconi and Levine, 2008; Cecconi et al., 2008), we decided to analyze its interaction with DLC1 upon autophagy induction. 2F cells coexpressing AMBRA1 and DLC1 were incubated in a nutrient-free medium for 4 h and analyzed by reciprocal co-IP assays. As shown in Fig. 2 A, AMBRA1–DLC1 interaction is not altered by autophagy stimulation. Accordingly, AMBRA1 shows a significant colocalization with DLC1 in 2F cells independent of nutrient supply (Fig. S1 B).
Figure 2.
Modulation of AMBRA1–dynein interaction during autophagy. (A) AMBRA1–DLC1 interaction upon autophagy induction. 2F cells coinfected with retroviral vectors encoding HA-tagged DLC1 and AMBRA1 proteins were nutrient starved for 4 h (+) or left untreated (−). Protein extracts were subjected to IP with an anti-HA–tagged antibody (IP HA; DLC1) or, as a negative control, with an unrelated antibody (IP control [Ctr]). Purified complexes were analyzed together with the corresponding total extracts by WB using anti-AMBRA1 (WB AMBRA1; top) and anti-DLC1 antibodies (WB DLC1; bottom). (B) AMBRA1 dissociates from the dynein motor complex during autophagy. 2F cells were nutrient starved for 4 h or left untreated. Protein extracts were subjected to IP using an anti-DIC (IP DIC) or an unrelated antibody (IP Ctr) as a negative control. The purified complexes were analyzed by WB together with the corresponding total extracts by using anti-DIC (WB DIC; top), anti-AMBRA1 (WB AMBRA1; middle), and anti-DLC1 antibodies (WB DLC1; bottom). (C) AMBRA1 dissociates from the dynein motor complex in mouse tissues after autophagy induction. Mice from the same litter were kept without food for 24 and 48 h or fed ad libitum (−) before sacrifice and necropsy. Kidneys from these mice were homogenized and subjected to IP analysis by using an anti-DIC antibody (IP DIC) or with an unrelated antibody (IP Ctr). Protein immunocomplexes were probed together with the corresponding total extracts using anti-DIC (WB DIC, top), anti-AMBRA1 (WB AMBRA1; middle), and anti-DLC1 (WB DLC1; bottom) antibodies. (D) AMBRA1 dissociates from tubulin upon autophagy induction. 2F cells infected with retroviral vectors encoding Flag-tagged βgal or AMBRA1 proteins were nutrient starved for 4 h or left untreated. Protein extracts were subjected to IP with an anti-Flag antibody (IP Flag). Purified proteins were eluted using the Flag peptide and analyzed by WB together with the corresponding total extracts using anti-AMBRA1 (WB AMBRA1; top) and anti–α-tubulin antibodies (WB tubulin; bottom). (E) AMBRA1 interacts with tubulin via DLC1. 2F cells were coinfected with retroviral vectors encoding Flag-tagged AMBRA1 FL, AMBRA1TAT1, or AMBRA1TAT2. Protein extracts were subjected to IP using an anti-Flag antibody (IP Flag). Purified complexes and corresponding total extracts were analyzed by WB using an anti-AMBRA1 (WB AMBRA1; Top) or anti–α-tubulin antibody (WB tubulin; bottom).
Because other factors, such as the proapoptotic BH3-only protein BIM, have been shown to be regulated by a dynamic interaction with the dynein motor complex via DLC1 (Puthalakath et al., 1999), we set out to investigate (a) whether AMBRA1 is also part of this complex and, this being the case, (b) whether AMBRA1 association to this complex is modified during autophagy. To this end, co-IP experiments were performed in 2F cells using an antibody specific for the dynein intermediate chain (DIC; Lo and Pfister, 2007). We observed that under control conditions, AMBRA1 is associated with DIC and is therefore part of the wider dynein machinery (Fig. 2 B, third lane). Moreover, upon autophagy induction obtained by nutrient starvation, AMBRA1 partially dissociated from the dynein complex (Fig. 2 B, fourth lane). However, only a fraction of DLC1 interacts with AMBRA1 because most DLC1 is still present in the complex, where it plays alternative functions (Fig. 2 B; King, 2000). Importantly, the same interaction dynamics were also observed in kidney tissues from mice kept in the absence of food for 24–48 h, thus emphasizing the physiological occurrence of this regulation (Fig. 2 C).
Consistently with its dynamic interaction with the dynein motor complex, AMBRA1 is associated to the microtubule component tubulin, from which it is released upon nutrient starvation (Fig. 2 D). AMBRA1–tubulin interaction is disrupted by mutating the DLC1-binding sites on the AMBRA1 moiety, confirming that AMBRA1’s relationship with the cytoskeleton strictly depends on this accessory protein (Fig. 2 E).
Dissociation of AMBRA1 from the dynein motor complex requires ULK1 kinase activity
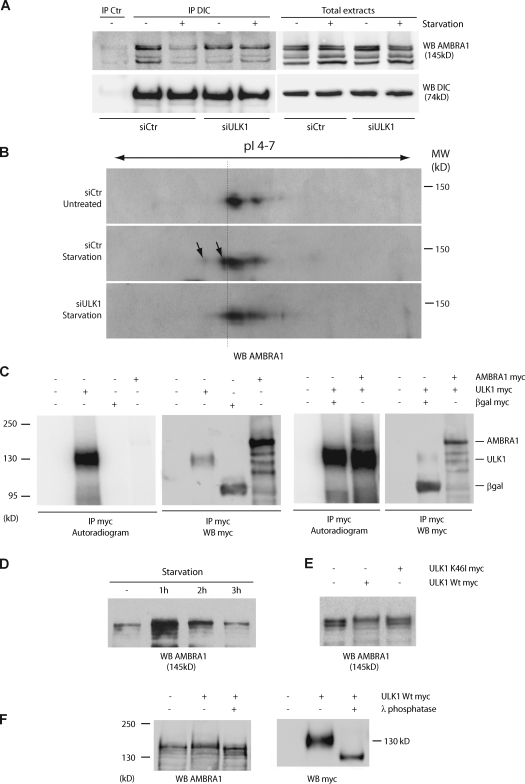
To investigate the upstream regulation of AMBRA1–DLC1 release from DIC, we checked whether the serine/threonine kinase ULK1 (Kuroyanagi et al., 1998), whose yeast orthologue Atg1 is a key regulator of the preautophagosomal structure formation (Cheong et al., 2008; Kawamata et al., 2008), was essential for this dissociation upon autophagy induction. When ULK1 is down-regulated by siRNA oligonucleotides (Fig. S2 A), we observed a remarkable reduction of the dissociation of AMBRA1 from DIC upon starvation conditions (Fig. 3 A) accompanied by autophagy impairment (Fig. S2 B). Moreover, as revealed by Western blot (WB) analysis of bidimensional gel electrophoresis, AMBRA1 is modified upon autophagy induction, and this process is inhibited by ULK1 down-regulation (Fig. 3 B; and Fig. S2, C and D). In line with this observation, we found that AMBRA1 is phosphorylated by an ULK1-immunopurified complex (but not by its kinase-mutated form, K46I; Chan et al., 2009), as revealed by (a) an in vitro kinase assay, (b) AMBRA1 mobility shift assays, and (c) phosphatase treatment experiments (Fig. 3, C–F; and Fig. S2 E). In contrast, no modifications were detected by bidimensional gel electrophoresis for both DIC and DLC1 (Fig. S2 F). Consistent with this finding, inhibition of JNK, a crucial regulator of DLC1 phosphorylation during apoptosis induction (Lei and Davis, 2003), does not interfere with the dissociation process (Fig. S2 G). Also, as expected, inhibition of BECLIN 1–VPS34 activity, a downstream event in autophagosome formation, does not interfere with the AMBRA1–DLC1 dissociation from DIC (Fig. S2 H). Altogether, these data showed that AMBRA1–DLC1 dissociates from the dynein complex upon ULK1-dependent AMBRA1 phosphorylation.
Figure 3.
AMBRA1 is phosphorylated during autophagy in an ULK1-dependent manner. (A) Down-regulation of ULK1 expression prevents AMBRA1 dissociation from the dynein motor complex during autophagy. ULK1 was down-regulated in 2F cells using specific siRNA oligonucleotides (siULK1). ULK1 expression level was analyzed by quantitative PCR and WB analyses (Fig. S2 A). siCtr, unrelated oligonucleotides. 48 h after transfection, 2F cells were starved for 4 h or left untreated. Protein extracts were prepared and subjected to IP using an anti-DIC (IP DIC) or an unrelated antibody (IP control [Ctr]) as a negative control. The purified complexes were analyzed by WB together with the corresponding total extracts using anti-DIC (WB DIC; bottom) and anti-AMBRA1 antibodies (WB AMBRA1; top). (B) Analysis of AMBRA1 modification after autophagy induction in control and ULK1 down-regulated cells. Protein extracts from samples as described in A were resolved by bidimensional gel electrophoresis (2DE) and analyzed by WB using an anti-AMBRA1 antibody. pI 4–7, isoelectric pH gradient 4–7; MW, molecular weight. AMBRA1 protein spots induced by starvation are indicated (arrows). Broken line highlights the isoelectric shift of the AMBRA1 isoforms produced by ULK1 activity. (C) Immunopurified ULK1 phosphorylates AMBRA1 in vitro. HEK293 cells were transfected with expression vectors encoding either AMBRA1 or ULK1 Myc-tagged proteins. 24 h after transfection, ULK1-expressing cells were nutrient starved for 2 h to stimulate ULK1 activity. Protein extracts were prepared from transfected cells and subject to IP using an anti-Myc antibody (IP Myc). An aliquot of the immunoprecipitated proteins was analyzed by WB using an anti-Myc antibody (WB Myc) to check for protein purification (right). Immunopurified ULK1 proteins were subjected to an in vitro kinase assay in the presence (+) or absence (−) of immunopurified AMBRA1 as described in Materials and methods. As negative control for kinase activity (−), the in vitro kinase assay was performed using anti-Myc–immunopurified complexes from βgal-transfected cells (βgal Myc). The reactions were resolved on SDS-PAGE and 32P-labeled proteins revealed by autoradiography (left). (D) Electrophoretic mobility hypershift of AMBRA1 upon autophagy induction. HEK293 cells were nutrient-starved for 1, 2, and 3 h. Protein extracts were prepared, loaded on a polyacrylamide gel (see Materials and methods) and analyzed by WB using an anti-AMBRA1 antibody (WB AMBRA1). (E) ULK1 phosphorylates AMBRA1. HEK293 cells were transfected with expression vectors encoding AMBRA1–Flag together with wild-type (Wt) ULK1 or K46I ULK1 Myc-tagged proteins. 48 h after transfection, protein extracts were prepared and subjected to WB using an anti-AMBRA1 antibody (WB AMBRA1). (F) HEK293 cells were transfected with an expression vector encoding Myc-tagged wild-type ULK1. 48 h after transfection, cells were lysed with HEMG and subjected to an in vitro λ-phosphatase assay as described in Materials and methods. After denaturation, protein extracts were subjected to WB using anti-AMBRA1 (WB AMBRA1; left) and anti-Myc (WB Myc; right) antibodies.
AMBRA1–BECLIN 1 complex relocalizes to the ER upon autophagy induction
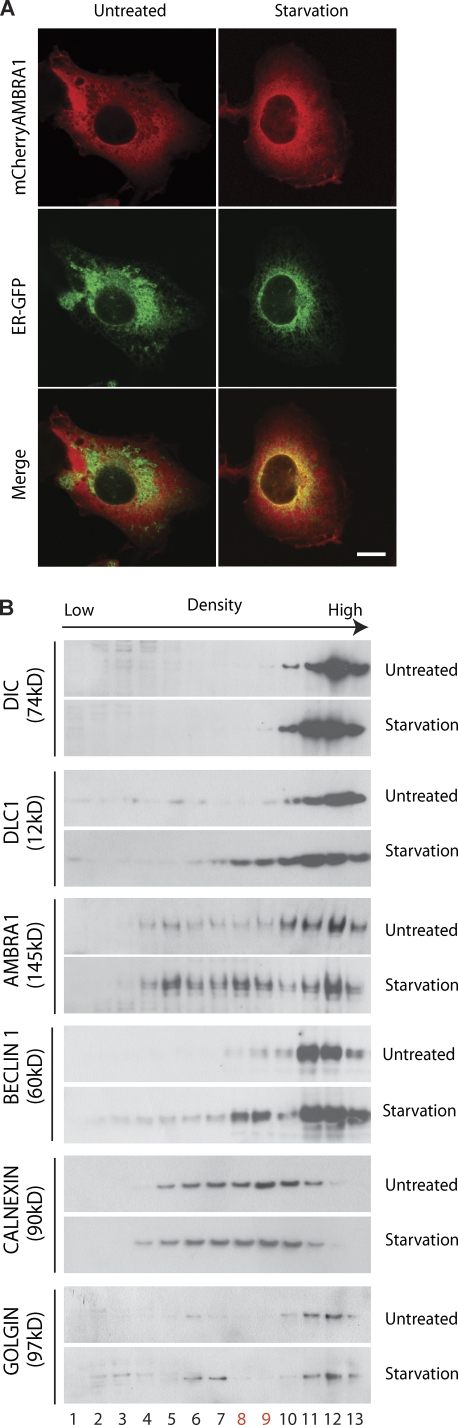
Interestingly, in parallel to the AMBRA1 dissociation from the dynein complex, autophagy induction led to an increase of AMBRA1 in the cell perinuclear region (Fig. 4 A, top). Therefore, we set out to establish which subcellular compartment AMBRA1 translocated to upon autophagy induction. By means of subcellular markers for ER, autophagosomes, mitochondria, Golgi cisternae, early endosomes, and lysosomes, a partial relocalization of AMBRA1 was detected in the ER of 2F cells induced to autophagy by nutrient starvation (Fig. 4 A; and Fig. S3, A–J). AMBRA1 translocation from the dynein complex to the ER was confirmed by a subcellular fractionation assay (Fig. 4 B).
Figure 4.
AMBRA1 translocates to the ER after autophagy induction. (A) AMBRA1 relocalizes to the ER upon autophagy induction. 2F cells expressing mCherryAMBRA1 (red) and ER-GFP (a GFP protein with an ER localization signal peptide; green) proteins were starved for 4 h or left untreated, fixed with 4% PFA followed by permeabilization with ice-cold methanol, stained with an anti-AMBRA1 antibody, and analyzed by confocal microscopy. (bottom) The images showing the merge of the two fluorescence signals are shown. Colocalization was assessed by calculating the Pearson’s correlation coefficient r of at least 10 cells analyzed in two independent experiments. (mean r: untreated, 0.42 ± 0.06; starvation, 0.62 ± 0.07) Bar, 12 µm. (B) AMBRA1 translocates to ER-enriched fractions upon autophagy induction. Postmitochondrial protein extracts from 2F cells nutrient starved for 4 h or left untreated were fractionated by discontinuous sucrose gradient sedimentation (see Materials and methods). Gradient fractions (1–13) were analyzed by WB using the indicated antibodies. The calnexin-positive fractions enriched in AMBRA1–BECLIN 1 upon starvation are indicated in red.
These results prompted us to verify whether the dynamic interaction of AMBRA1 with the dynein motor involved other components of the class III PI3-kinase complex (Xie and Klionsky, 2007). Co-IP experiments showed that both VPS34 and BECLIN 1 are associated to AMBRA1 and DLC1 before and after autophagy stimulation (Fig. 5, A and B). However, BECLIN 1–DLC1 association is mediated by AMBRA1, as demonstrated by means of the AMBRA1TAT2 mutant, which prevents this interaction (Fig. 5 C). Similarly to AMBRA1, we also detected the dynamic interaction of BECLIN 1 with DIC (Fig. 5 D). Moreover, subcellular fractionation assays and confocal analysis showed that BECLIN 1 follows the AMBRA1 translocation pattern after autophagy induction (Fig. 4 B and Fig. S3 K). Accordingly, VPS34 activity is also detectable at the sites of translocation of AMBRA1, as revealed by partial costaining with the PI3P-interacting protein GFP-DFCP1 (Fig. S3 L; Vieira et al., 2001). In particular, AMBRA1 localization is very intense in close proximity with the DFCP1-positive puncta, a hallmark of the omegasomes, which have been proposed as the autophagosome formation sites at the ER (Fig. S3 M; Vieira et al., 2001). It should be noted that a fraction of BECLIN 1, restricted to the trans-Golgi network (Kihara et al., 2001), does not colocalize with AMBRA1 (Fig. S3 K), suggesting that two pools of BECLIN 1 may be differently regulated within the cell. Notably, AMBRA1 down-regulation by RNAi impairs BECLIN 1 translocation to the ER upon autophagy induction, without altering its protein levels (Fig. 5, E–H).
Figure 5.
AMBRA1 is required for BECLIN 1 translocation to the ER after autophagy induction. (A) VPS34–AMBRA1 interaction upon autophagy induction. 2F cells infected with retroviral vectors encoding βgal Myc or AMBRA1 Myc proteins were nutrient starved for 4 h (+) or left untreated (−). Protein extracts were subjected to IP with an anti-Myc antibody (IP Myc). Purified complexes were analyzed together with the corresponding total extracts by WB using an anti-VPS34 antibody (WB VPS34). (B) BECLIN 1 interacts with DLC1. 2F cells were transiently transfected with expression plasmids encoding βgal or Flag-tagged BECLIN 1 and nutrient starved for 4 h (+) or left untreated (−). Protein extracts were subjected to IP using an anti-Flag antibody. Purified complexes were analyzed together with the corresponding total extracts by WB using anti–BECLIN 1 (WB BECLIN 1; top), anti-AMBRA1 (WB AMBRA1; middle), and anti-DLC1 antibodies (WB DLC1; bottom). (C) BECLIN 1 binding to DLC1 is mediated by AMBRA1. 2F cells were transiently transfected with expression plasmids encoding βgal or Flag-tagged BECLIN 1 together with Myc-tagged AMBRA1 full length (FL) or TAT2. Protein extracts were subjected to IP using an anti-Flag antibody. Purified complexes were analyzed together with the corresponding total extracts by WB using anti-BECLIN 1 (WB BECLIN 1; top), anti-Myc (WB AMBRA1; middle), and anti-DLC1 antibodies (WB DLC1; bottom). (D) Dynamic interaction of BECLIN 1 with the dynein motor complex during autophagy. Protein extracts from AMBRA1 expressing 2F cells were subjected to IP using an anti-DIC antibody (IP DIC) or an unrelated antibody (IP control [Ctr]). The purified complexes were analyzed together with the corresponding total extracts by WB using an anti–BECLIN 1 antibody (WB BECLIN 1). (right) The BECLIN 1–DIC interaction was also analyzed in untreated (−) or starved (+) cells. (E) 2F cells were transfected with AMBRA1 siRNA (siAMBRA1) or unrelated oligonucleotides (siCtr). 24 h after transfection, cells were starved for 4 h or left untreated, fixed, and stained with the anti–BECLIN 1 (red) and anti-ERp57 (green) antibodies. (right) The images showing the merge of the two fluorescence signals are shown. Colocalization was assessed by calculating the Pearson’s correlation coefficient r of at least 10 cells analyzed in two independent experiments (mean r: siCtr, 0.35 ± 0.17; starvation, 0.58 ± 0.19; siAMBRA1 untreated, 0.28 ± 0.06; starvation, 0.39 ± 0.03). Similar results were obtained in cells transfected with AMBRA1 siRNA1 (not depicted). Bar, 8 µm. (F) Analysis of BECLIN 1 protein levels after AMBRA1 down-regulation. AMBRA1 and BECLIN 1 expression levels in cells described in E were analyzed by WB analysis using specific antibodies. Tubulin, protein loading control. (G and H) AMBRA1 down-regulation impairs BECLIN 1 translocation to ER. 2F cells were transfected with AMBRA1 RNAi oligonucleotides (siAMBRA1) or unrelated oligonucleotides as a control (siCtr). Postmitochondrial protein extracts from 2F cells nutrient starved for 4 h or left untreated were fractionated by discontinuous sucrose gradient sedimentation (see Materials and methods). (G) Gradient fractions (1–13) were analyzed by WB using an anti–BECLIN 1 antibody. (H) Gradient fractions were also analyzed for calnexin distribution to verify similar fractionation efficiency between siCtr and siAMBRA1 cells. The calnexin-positive fractions enriched in AMBRA1–BECLIN 1 upon starvation are indicated in red.
In the light of these results, we propose that, similarly to BIM regulation during apoptosis (Puthalakath et al., 1999), the dynein motor complex might play a tethering/docking role in regulating autophagy by its association with AMBRA1 and the multimolecular BECLIN 1–VPS34 autophagosome nucleation complex.
DLC1 down-regulation induces AMBRA1-dependent autophagy
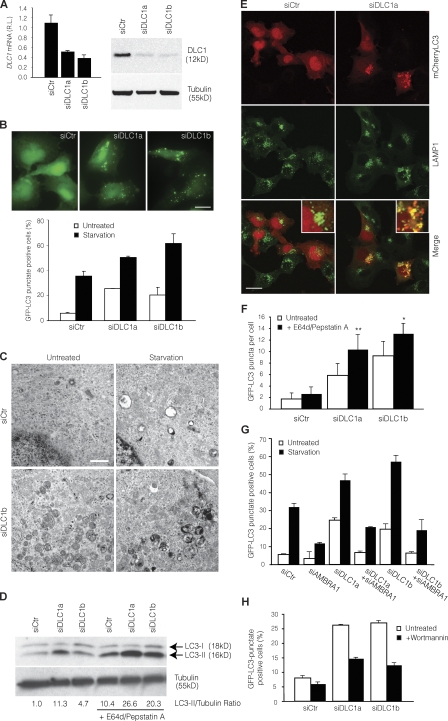
To elucidate the functional role of DLC1–AMBRA1 interaction in autophagy, we decided to test whether DLC1 modulation resulted in autophagy dysregulation. To this end, DLC1 expression was down-regulated in 2F cells by transfection of DLC1-specific siRNA oligonucleotides (Fig. 6 A), and autophagy occurrence was analyzed. We assessed both conversion of LC3-I to its cleaved and lipidated form LC3-II and its translocation to autophagic structures, two steps in autophagosome formation (Kabeya et al., 2000). We also analyzed autophagy by cell ultrastructural analysis and by measuring the increase in acidic vesicular organelles (AVOs; Paglin et al., 2001). DLC1 down-regulation led to a remarkable increase in the number of autophagosome both in basal condition and during starvation- or rapamycin-induced autophagy (Fig. 6, B–D; and Fig. S4, A–C). This effect was also obtained in different cell types, such as neural precursor cells (embryonic telencephalic naïve [ETNA] cells; Fig. S4, D–F; Cozzolino et al., 2004).
Figure 6.
Modulation of autophagy by DLC1 down-regulation. (A) DLC1 down-regulation in GFP-LC3–expressing 2F cells using specific siRNA oligonucleotides (siDLC1a and siDLC1b). DLC1 mRNA and protein levels were analyzed by quantitative PCR (left) and WB (right). siCtr, unrelated oligo; RL, relative levels; tubulin, protein loading control. (B) Increase of basal and starvation-induced autophagy by DLC1 down-regulation. 24 h after transfection with DLC1 siRNA oligonucleotides, GFP-LC3–expressing 2F cells were starved for 4 h or left untreated, and the occurrence of autophagy was analyzed by measuring GFP-LC3 punctate–positive cells. A graph reporting data from three experiments is shown together with representative fluorescence images of siRNA oligonucleotide–transfected cells in control conditions. Bar, 20 µm. (C) Ultrastructural analysis by means of electron microscopy of ultrathin sections from 2F cells transfected with a RNAi oligonucleotides for DLC1 (siDLC1b) or an unrelated oligonucleotide (siCtr) and starved for 4 h or left untreated. Bar, 1.5 µm. (D–F) Increase of autophagosome on rate by DLC1 down-regulation. (D) After DLC1 down-regulation (siDLC1a-b), 2F cells were treated with the lysosome inhibitors E64d and pepstatin A for 4 h or left untreated, and the occurrence of autophagy was analyzed by LC3-I to LC3-II conversion. Densitometric analysis of the band density ratio of LC3-II relative to tubulin is reported with the siCtr ratio arbitrarily defined as 1.00 (Fig. S4 B). (E) mCherryLC3-expressing 2F cells transfected as in D and treated with the lysosome inhibitors E64d and pepstatin A for 4 h were stained by using the lysosome marker LAMP1 (green). Insets containing higher magnification views of the merge images are also shown. Bar, 16 µm. Colocalization was assessed by calculating the Pearson’s correlation coefficient r (mean r: siCtr, 0.24 ± 0.02; siDLC1a, 0.66 ± 0.04). (F) GFP-LC3–expressing 2F cells transfected and treated as in D were analyzed for the appearance of GFP-LC3 puncta per cell. *, P < 0.05; **, P < 0.01. (G) Autophagy induced by DLC1 down-regulation requires AMBRA1. GFP-LC3–expressing 2F cells were transfected using DLC1 and AMBRA1 siRNA oligonucleotides (siAMBRA1) either separately or in combination. 24 h after transfection, 2F cells were starved for 4 h or left untreated, and the occurrence of autophagy was analyzed by measuring GFP-LC3 punctate–positive cells. (H) Autophagy induced by DLC1 down-regulation requires PI3K activity. After DLC1 down-regulation, 2F cells were incubated with Wortmannin for 4 h and analyzed for appearance of GFP-LC3 punctate staining. Error bars indicate mean ± SD of three experiments.
Dynein is composed of heavy and intermediate chain proteins involved in the structural composition of the motor complex in combination with different light chains that are known to modulate the complex’s function (King, 2000; Höök and Vallee, 2006). To verify whether autophagy was specifically induced by DLC1 down-regulation, autophagy occurrence was analyzed in 2F cells transfected with siRNA oligonucleotides specific for other cytosolic members of the DLC family (Wilson et al., 2001), namely dynein TCTEX light chain 1, dynein ROADBLOCK light chain 1, and DLC2. Among these, only DLC2 down-regulation induced autophagy, although to a lesser extent than DLC1 (Fig. S4, G–M); accordingly, DLC2 was also able to bind AMBRA1 (Fig. S4 M). The combined down-regulation of DLC1 and DLC2 had an additive effect on autophagy induction (Fig. S4, N–P), indicating a nonredundant role for these two proteins in this context.
Because it has been shown that the dynein complex is required for autophagosome migration and fusion to lysosome (Ravikumar et al., 2005; Köchl et al., 2006; Jahreiss et al., 2008; Kimura et al., 2008), we set out to investigate whether the accumulation of autophagosomes by DLC1 depletion could have derived from an enhanced autophagic sequestration (on rate) or a reduced degradation of autophagic material (off rate; Klionsky et al., 2008). To discriminate between these possibilities, we assessed the occurrence of autophagosome–lysosome fusion in cells transfected with DLC1 siRNA oligonucleotides by monitoring the colocalization of LC3 fused to mCherry (an improved monomeric red fluorescence protein that does not lose fluorescence under acidic condition, typical of lysosomes; Shaner et al., 2004) with the lysosome markers LAMP1 and Lysotracker (unpublished data) in the presence of lysosomal protease inhibitors (E64d and pepstatin A), which prevent mCherryLC3 degradation within the lysosome (Tanida et al., 2005). We observed that upon DLC1 down-regulation, autophagosomes are capable of fusing with lysosomes and forming autophagolysosomes (Fig. 6 E). Moreover, E64d and pepstatin A enhanced the DLC1 siRNA-triggered increase of endogenous LC3-II (Fig. 6 D and Fig. S4 B) and the accumulation of GFP-LC3 puncta (Fig. 6 F). Collectively, these data imply that DLC1 inhibition increases the on rate of autophagy in cultured cells.
Next, we hypothesized that if autophagy induction by DLC1 down-regulation was caused by the release of AMBRA1 from the dynein complex, this phenomenon should be abolished by AMBRA1 inactivation. 2F cells were transfected with DLC1 siRNA oligonucleotides alone or in combination with AMBRA1 siRNA oligonucleotides and analyzed for the effect on autophagy by measuring the occurrence of GFP-LC3 puncta. As shown in Fig. 6 G, autophagy induction by DLC1 down-regulation is prevented if AMBRA1 is concomitantly inactivated, indicating that DLC1’s role in autophagy is AMBRA1 dependent. Moreover, autophagy induced by DLC1 down-regulation requires BECLIN 1–VPS34 activity, as revealed by Wortmannin-mediated PI3K inhibition (Fig. 6 H). To rule out the possibility that AMBRA1, besides its control of autophagy, could also have a role in the early endosome trafficking mediated by the dynein complex, the internalization and degradation of the EGF receptor (Driskell et al., 2007) was analyzed in cells where AMBRA1 expression had been down-regulated by siRNA oligonucleotides (Fig. S5). No significant differences were detected between AMBRA1 siRNA-transfected and control cells. Thus, AMBRA1 does not seem to play a role in early endosome trafficking of EGF receptor.
AMBRA1 mutants unable to bind DLC1 induce constitutive autophagy
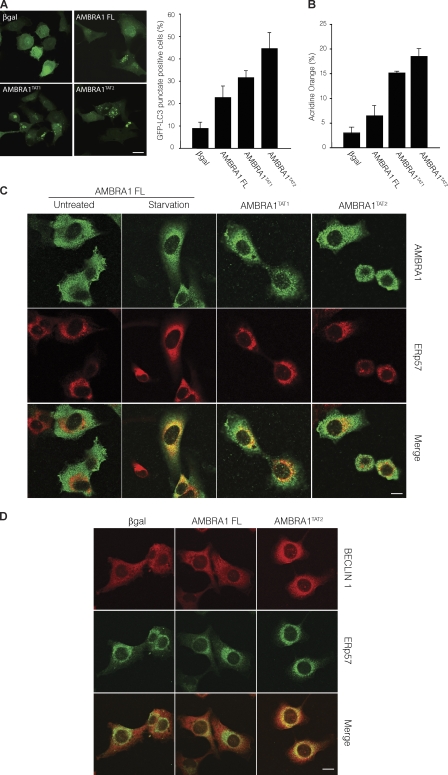
Prompted by these data, we decided to study the autophagic potential of AMBRA1 mutants lacking the DLC1-binding domain. 2F cells were transduced with retroviruses encoding the AMBRA1 wild-type (AMBRA1 FL) or the AMBRA1TAT1 and TAT2 mutants; autophagy induction was analyzed by counting GFP-LC3 punctate–positive cells or measuring the increase in AVOs. Notably, AMBRA1 defective in DLC1 binding showed a stronger ability in inducing autophagy than did the wild-type protein (Fig. 7, A and B). Moreover, AMBRA1 mutants TAT1 and TAT2 are constitutively translocated together with BECLIN 1 to the ER, even in untreated conditions (Fig. 7, C and D). These results support the hypothesis that AMBRA1 is bound in an inactive state to the dynein complex via the binding to DLC1 and that its release from the complex is an ULK1-dependent early step in AMBRA1-mediated autophagy (Fig. 8). It is important to note that, although a mutation in the DLC1-binding domain significantly increases AMBRA1 autophagic potential, ULK1 activity is required for the stimulation of autophagy by both wild-type and AMBRA1TAT mutants (Fig. S5, B–D). This result underlines how AMBRA1 release from the dynein complex is one of the ULK1-dependent regulatory steps crucial for autophagy induction, with many others that probably remain to be characterized.
Figure 7.
AMBRA1 interaction with DLC1 regulates its proautophagic function. (A and B) AMBRA1 mutants defective for DLC1 interaction have an increased autophagic potential. GFP-LC3–expressing 2F cells were transduced with retroviral vectors encoding AMBRA1 wild-type (AMBRA1 FL) or TAT1 and TAT2 mutants (AMBRA1TAT1 and TAT2) and analyzed for the appearance of GFP-LC3 punctate staining (A) or for AVO formation by FACS measurement of Acridine orange staining (B). Values represent the mean ± SD of three experiments. (C) 2F cells infected with retroviral vectors encoding Myc-tagged FL, TAT1, or TAT2 AMBRA1 proteins were fixed and stained with the anti-Myc (AMBRA1; green) and anti-ERp57 (red) antibodies. AMBRA1-FL–overexpressing cells were analyzed either in control or starvation conditions. (bottom) The images showing the merge of the two fluorescence signals are shown. Colocalization was assessed by calculating the Pearson’s correlation coefficient r of at least 10 cells analyzed in two independent experiments (mean r: AMBRA1 FL untreated, 0.30 ± 0.10; AMBRA1 FL starvation, 0.52 ± 0.12; AMBRA1TAT1 untreated, 0.54 ± 0.04; AMBRA1TAT2 untreated, 0.53 ± 0.01). A βgal retroviral vector was used as a negative control. (D) 2F cells infected with retroviral vectors encoding Myc-tagged βgal, FL AMBRA1, or TAT2 AMBRA1 proteins were fixed and stained with the anti–BECLIN 1 (red) and anti-ERp57 (green) antibodies. (bottom) The images showing the merge of the two fluorescence signals are shown. Colocalization was assessed by calculating the Pearson’s correlation coefficient r (mean r: βgal, 0.37 ± 0.03; AMBRA1 FL, 0.44 ± 0.03; AMBRA1TAT2, 0.54 ± 0.05). Bars, 8 µm.
Figure 8.
Proposed model of AMBRA1 dynamic interaction with the dynein motor complex during autophagy induction. AMBRA1 is bound in a presumably inactive state to the dynein complex via binding to DLC1. After autophagy induction, AMBRA1 is released from the dynein complex upon ULK1-dependent phosphorylation and translocates to the ER together with BECLIN 1–VPS34 to allow autophagosome formation. The dynein motor complex consists of a Head domain (dynein heavy chain) and a Base domain (DIC, DLC1, or DLC2 and other factors such as TCTEX1 and ROADBLOCK1).
Discussion
DLC1 is a component of the microtubule-based molecular dynein motor complex. As such, it is involved in cell division, vesicular trafficking, and ciliary/flagellar motility (King, 2000). However, DLC1 also interacts with proteins that are not directly associated with dynein- or microtubule-dependent roles, such as factors involved in apoptosis (Puthalakath et al., 1999), enzyme regulation (Jaffrey and Snyder, 1996), mitochondrial dynamics (Varadi et al., 2004), and viral pathogenesis (Raux et al., 2000). In particular, the proapoptotic BH3-only protein BIM is tethered by DLC1 on the dynein complex (Puthalakath et al., 1999), from which it is released upon induction of apoptosis by means of both DLC1 and BIM phosphorylation (triggered by p21-activated kinase 1 [Vadlamudi et al., 2004] and JNK [Lei and Davis, 2003], respectively). In this study, we show that DLC1 may also play a role in regulating autophagy through its interaction with the proautophagic protein AMBRA1. In addition to its role in carrying the autophagosome toward the lysosome (Ravikumar et al., 2005; Fass et al., 2006; Köchl et al., 2006; Jahreiss et al., 2008; Kimura et al., 2008), our results imply an alternative, more regulative role for the dynein motor complex in mediating autophagy initiation. Because microtubule dynamics are obviously linked to cell cycle regulation and cell motility, we are tempted to speculate that AMBRA1 release from microtubules may also signal to the motor complex the initiation of the autophagy process, i.e., modulating the global cytoskeleton response and rearrangement observed during autophagy.
Despite much progress in the field of autophagy, a critical question remains unanswered, i.e., how ULK1 activation and the recruitment of the ULK1–ATG13–FIP200 complex may turn out in autophagosome formation. In this study, we show that ULK1 phosphorylates AMBRA1, regulating AMBRA1–DLC1 dissociation from the dynein complex. Also, we report that AMBRA1–DLC1, upon autophagy induction, translocates from the dynein complex to the ER. Autophagosome formation is known to require PI3P (Xie and Klionsky, 2007) and is believed to occur near the ER (Juhasz and Neufeld, 2006). Recent evidence points to the existence of a PI3P-enriched membrane compartment, the omegasome, in dynamic equilibrium with the ER, which provides a platform for accumulation of autophagosomal proteins, expansion of autophagosomal membranes, and the emergence of fully formed autophagosomes (Axe et al., 2008; Hayashi-Nishino et al., 2009). Supporting this hypothesis is the fact that the antiapoptotic factors BCL2 and BCLXL require ER localization to regulate autophagy by BECLIN 1 binding (Pattingre et al., 2005).
We previously showed that AMBRA1 is required for an effective BECLIN 1–VPS34 complex formation (Fimia et al., 2007). Now, our findings indicate that a preassembled complex, containing AMBRA1, BECLIN 1, and VPS34, translocates from the AMBRA1-docking site on the dynein motor complex to the omegasome compartment of the ER, where autophagosomes originate. An important goal for the future will be the identification of the factors responsible of docking the AMBRA1–BECLIN 1 complex on the omegasome compartment, with DFCP1 (or other PI3P-binding proteins) or ATG14, which are found to localize at the ER (Axe et al., 2008; Matsunaga et al., 2009), being good candidates to play such a role.
AMBRA1 is strongly expressed in the developing brain and in adult brain compartments, such as hippocampus, cerebellum, and striatum, which are all severely affected in neurodegenerative conditions. ULK proteins also play roles in neurons. Indeed, ULK1 regulates growth cones, filopodia extension, and axon branching; expression of the dominant-negative forms of ULK1 and ULK2 suppresses neurite extension of cerebellar granular neurons (Tomoda et al., 1999; Zhou et al., 2007; Mizushima, 2010). These roles have not yet been clearly related to ULK1 proautophagic functions. However, our findings that ULK1 is a direct regulator of AMBRA1 and that AMBRA1 plays a positive role in neuron morphogenesis might imply the existence of a common pathway for these factors in neurogenesis, based on autophagy regulation.
Finally, unraveling the mechanism of autophagy initiation in higher eukaryotes is relevant for designing new molecules for interfering with this process, and moreover, the use of specific drugs able to deregulate AMBRA1–DLC1 interaction may prove highly useful in the therapeutic strategy to fight neurodegeneration.
Materials and methods
Yeast two-hybrid screening
pGBKT7-AMBRA1 was generated by cloning the C-terminal region (aa 533–1,269) of AMBRA1 into the EcoRI and SalI sites of pGBKT7 (Takara Bio Inc.). A human brain cDNA library cloned in pACT2 (Takara Bio Inc.) was screened by cotransformation with the pGBKT7-AMBRA1 into AH109 yeast strain. Positive clones were selected based on their growth on Trp, Leu, Ade, and His dropout media (Takara Bio Inc.) containing 5 mM 3-amino-1,2,4-triazole (3AT; Sigma-Aldrich). Recovery of the plasmids and β-galactosidase (βgal) assay were performed following the manufacturer’s directions (Matchmaker two-hybrid system protocol; Takara Bio Inc.).
Cell culture
The human fibrosarcoma 2F, HEK293, and ETNA cells were cultured in DME (Sigma-Aldrich) supplemented with 10% FCS (Sigma-Aldrich), 2 mM l-glutamine, and 1% penicillin/streptomycin solution at 37°C (2F and HEK293) or 33°C (ETNA) under 5% CO2. For autophagy induction, cells were treated with 0.2–2 µM rapamycin (Sigma-Aldrich) or cultured for 4 h in Earle’s balanced salt solution (Invitrogen). When indicated, cells were incubated in complete or starvation medium in the presence of 1 or 10 µM nocodazole (Sigma-Aldrich), 10 µg/ml E64d together with 10 µg/ml pepstatin A (Sigma-Aldrich), 200 nM Wortmannin (Sigma-Aldrich), 10 mM 3-methyl adenine, and 10 µM JNK inhibitor SP600125 (Sigma-Aldrich) for 4 h before the assay. For EGFR internalization assay, cells were grown overnight in DME plus 0.1% FCS and treated with 100 ng/ml EGF (PeproTech) for 10, 20, and 40 min before analysis. For lysosome staining, 50 nM Lysotracker green (Invitrogen) was added to cells 30 min before analysis. 2F and 293 cells were transiently transfected with expression vectors using Lipofectamine 2000 (Invitrogen) as indicated by the supplier. 2F cells stably expressing ER-GFP protein were obtained by culturing pCMV/Myc/ER-GFP–transfected cells in the presence of 0.4 mg/ml G418 (Invitrogen) for 2 wk.
cDNA cloning
For retroviral expression, all constructs were cloned in pCLPCX vector. BECLIN 1, GFP-LC3, GFP-p40Phox, and FL, F1, F2, and F3 AMBRA1 vectors were previously described (Fimia et al., 2007). GFP-DFCP1 plasmid was provided by N.T. Ktistakis (Babraham Institute, Cambridge, England, UK; Axe et al., 2008). ULK1 constructs were provided by S.A. Tooze (London Research Institute, London, England, UK; Chan et al., 2009). The human DLC1 was cloned in frame with HA tag. AMBRA1 mutants TAT1 and TAT2 were generated by using the site-directed mutagenesis kit (Agilent Technologies) and cloned in frame with a Myc or Flag tag. mCherryLC3 was generated by replacing, in pCLPCX-GFP-LC3 plasmid, the GFP sequence with mCherry amplified by PCR from the pmCherry-N1 vector (Takara Bio Inc.). mCherryAMBRA1 was generated by subcloning mCherry sequence at the 5′ end of AMBRA1 in pCLPCX-AMBRA1. pCMV Myc ER-GFP was purchased from Invitrogen. The sequences of all PCR-amplified cDNAs were verified by DNA sequencing analysis.
Retrovirus generation and infection
15 µg retroviral vectors was cotransfected with 5 µg expression plasmid for the vesicular stomatitis virus G protein into 293 gp/bsr cell line by using the calcium phosphate method. 48 h later, the supernatant containing the retroviral particles was recovered and supplemented with 4 µg/ml polybrene. 2F or ETNA cells were infected by incubation with retroviral containing supernatant for 6–8 h.
Antibodies
The following primary antibodies were used in this study: rabbit anti-Myc tag (Millipore), mouse anti-HA tag (Sigma-Aldrich), mouse anti-Flag tag (WB and EM analyses; Sigma-Aldrich), mouse anti-DLC1 (BD), mouse anti-DIC (Santa Cruz Biotechnology, Inc.), rabbit anti-ULK1 (Sigma-Aldrich), rabbit and goat anti–BECLIN 1 (WB and immunofluorescence analyses, respectively; Santa Cruz Biotechnology, Inc.), rabbit anti–BECLIN 1 (WB analysis; Cell Signaling Technology), rabbit anti-VPS34 (Invitrogen), rabbit anti-LC3 (Cell Signaling Technology), rabbit anti-AMBRA1 (WB analysis; Strategic Diagnostic, Inc.), rabbit anti-AMBRA1 (IF and EM analysis; Covalab), rabbit anti-AMBRA1 CT (IP and EM analysis; ProSci, Inc.), mouse anti-ERp57 (Stressgen), mouse anti-LAMP1 and anti-EEA1 (Abcam), mouse anti-GOLGIN (Invitrogen), mouse anti–complex V α subunit (Invitrogen), mouse anti–α-tubulin (Sigma-Aldrich), mouse anti-EGFR (Millipore), and rabbit anti-calreticulin (Stressgen).
Autophagy assays
Autophagy was measured as described previously (Fimia et al., 2007). In brief, starvation was induced by incubating cells in Earle’s balanced salt solution medium (Sigma-Aldrich) for 4–5 h. For immunodetection of LC3 puncta, cells were grown on coverslips and fixed with 4% PFA in PBS, washed three times, and directly examined by confocal microscopy. The results indicate the percentage of GFP-LC3–positive cells with GFP-LC3 punctate dots or the numbers of GFP-LC3 punctate dots per cell. A minimum of 50–100 cells per sample was counted for triplicate samples per condition per experiment. To quantify the development of AVOs, cells were detached by trypsin digestion, washed with PBS, stained with 1 µg/ml Acridine orange (Sigma-Aldrich) for 15 min, and analyzed using a flow cytometer (FACScan; BD) and CellQuest software (BD). Autophagy experiments in vivo were performed using C57BL/6 mice. 5-mo-old male mice from the same littermate were kept without food but with water for 24 and 48 h or fed ad libitum before sacrifice and tissue analysis.
For electron microscopy, cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 45 min at 4°C, rinsed in cacodylate buffer, postfixed in 1% OsO4 in cacodylate buffer, dehydrated, and embedded in epon. Ultrathin sections were briefly contrasted with uranyl acetate and photographed with an electron microscope (CM900; Carl Zeiss, Inc.).
IP and WB assays
In IP experiments, cells or tissues were lysed in HEMG buffer (25 mM Hepes, pH 8.0, 100 mM NaCl, 25 mM MgCl2, 0.5% Triton X-100, 0.1 mM EDTA, and 10% glycerol) plus protease and phosphatase inhibitors (protease inhibitor cocktail plus 1 mM sodium fluoride, 1 mM sodium orthovanadate, and 1 mM sodium molibdate; Sigma-Aldrich). For analysis of the BECLIN 1–dynein complex, IP was performed with a different lysis buffer (40 mM Hepes, pH 7.4, 2 mM EDTA, 10 mM glycerophosphate, and 0.3% CHAPS plus protease and phosphatase inhibitors) and a wash buffer (40 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM EDTA, 10 mM glycerophosphate, and 0.3% CHAPS) as described previously (Hosokawa et al., 2009). 1–3 mg lysates was incubated at 4°C for 30 min. After a centrifugation at 4°C for 10 min at 13,000 g to remove insoluble debris, equal amounts of protein were incubated with 20 µl monoclonal anti-cMyc or anti-Flag antibody conjugated with protein A agarose beads (Takara Bio Inc. and Sigma-Aldrich, respectively) with rotation at 4°C for 4 h or with 30 µl monoclonal anti-HA antibody conjugated with protein A agarose (Sigma-Aldrich) or 2 µg anti-DIC antibody overnight at 4°C, or 2 µg anti-AMBRA1 antibody (CT; ProSci, Inc.) followed by 60-min incubation with 30 µl protein A/G–Sepharose beads (Roche). The beads were collected by centrifugation and washed four times with HEMG buffer. Proteins bound to the beads were eluted with 50 µl SDS-PAGE sample buffer and boiled at 95°C for 10 min. WB analyses were performed using 5% vol/vol of the whole extracts and 30% vol/vol of eluted proteins. In AMBRA–tubulin interaction experiments, immunocomplexes were eluted by incubation with a Flag peptide (Sigma-Aldrich) for 30 min at RT to prevent the presence of immunoglobulins in the gel.
Proteins were separated on NuPAGE Bis-Tris gel (Invitrogen) and electroblotted onto nitrocellulose (Protran; Schleicher & Schuell) or PVDF (Millipore) membranes. Blots were incubated with primary antibodies in 5% nonfat dry milk in TBS plus 0.1% Tween-20 overnight at 4°C. Detection was achieved using horseradish peroxidase–conjugated secondary antibody (Bio-Rad Laboratories) and visualized with ECL plus (GE Healthcare). Note that endogenous AMBRA1 usually migrates as a doublet band, whereas the overexpressed AMBRA1 shows additional lower molecular weight bands.
To better visualize AMBRA1 mobility shift induced by phosphorylation, protein samples were resolved on SDS-PAGE gels (Anderson et al., 1973). For the λ-phosphatase assay, cells were lysed in HEMG and incubated with 400 U λ-phosphatase (New England BioLabs, Inc.) for 45 min at 30°C. Reactions were stopped by addition of 4× SDS sample buffer and boiled for 10 min.
Confocal and immunogold analysis
For confocal analysis, cells were grown on coverslips and fixed with 4% PFA in PBS followed by permeabilization with 0.1% Triton X-100 in PBS or, when indicated, fixed with ice-cold methanol or with 4% PFA in PBS followed by permeabilization with ice-cold methanol. Primary antibodies were incubated for 1 h at RT and visualized by means of Cy3- and Cy2-conjugated secondary antibodies. (Jackson ImmunoResearch Laboratories, Inc.). Coverslips were mounted in antifade (SlowFade; Invitrogen) and examined under a confocal microscope (TCS SP2; Leica) equipped with a 63× 1.40–0.60 NA HCX Plan Apo oil λBL objective at RT. For colocalization analysis, both confocal (Leica) and ImageJ software (National Institutes of Health) were used. Regarding the confocal software analysis, the local intensity distribution of both fluorophores across a line scan drawn on a single optical slice was plotted in a graph. Regarding ImageJ software analysis, the pixels of two 8-bit images (red and green channels of each image) are considered colocalized if their intensities are higher than the threshold of their channels (set at 50) and if the ratio of their intensity is higher than the ratio setting value (set at 50%). Colocalization was assessed by calculating the Pearson’s correlation coefficient r of at least 10 cells analyzed in two independent experiments. The Pearson’s correlation coefficient was expressed as mean ± SD. The fluorescence intensity of each fluorochrome was simultaneously analyzed and plotted.
For electron microscopy, 2F cells were fixed in 2% freshly depolymerised PFA and 0.2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 1 h at 4°C. Samples were rinsed in buffer, partially dehydrated, and embedded in London Resin white (LR White; Agar Scientific Ltd.). As shown in Fig. S3 J, to preserve membrane structures, the fixed cells were rinsed in the same buffer, treated with 0.25% tannic acid in buffer for 1 h at 4°C, and rinsed in 50 mM ammonium chloride in buffer. The samples were postfixed for 30 min in 2% uranyl acetate in buffer, partially dehydrated, and embedded under UV light at −20°C in London Resin gold (LR White; Agar Scientific Ltd.). Ultrathin sections were processed for the immunogold technique. Grids were preincubated with 10% normal goat serum in 10 mM PBS containing 1% BSA and 0.13% NaN3 (medium A) for 15 min at RT. Sections were incubated with anti-AMBRA1 or anti-Flag antibodies diluted in medium A overnight at 4°C. After rinsing in medium A containing 0.01% Tween-20 (Merck), sections were incubated with the appropriate secondary antibodies conjugated to 15-nm colloidal gold (British Biocell International) diluted 1:30 in medium A containing fish gelatin for 1 h at RT. After incubation in medium A for 15 min at RT, a second immunolabeling was performed using a rabbit polyclonal anti-DLC1 or anti-calreticulin, as primary antibodies and goat anti–rabbit IgG conjugated to 5-nm colloidal gold as secondary antibody. Grids were thoroughly rinsed in distilled water, contrasted with aqueous 2% uranyl acetate for 20 min, and photographed in an electron microscope (EM 900 Carl Zeiss, Inc.).
RNAi
RNAi was performed using the following oligonucleotides from Invitrogen: DLC1, DLC2, ROADBLOCK1, TCTEX1, and ULK1 oligo. AMBRA1 siRNA oligonucleotides were described previously (Fimia et al., 2007). 2 × 105 cells/well were transfected with 100 pmol siRNA in 6-well plates by Lipofectamine 2000 (Invitrogen) as indicated by the supplier. Transfection was repeated on two consecutive days to increase transfection efficiency. RNA decrease was checked by real-time PCR and WB analysis 48 h after transfection.
Real-time PCR
RNA was prepared with Trizol reagent (Invitrogen). cDNA synthesis was generated using the reverse transcription kit (Promega) according to manufacturer recommendations. Real-time PCR reactions were performed with the LightCycler (Roche). The LightCycler FastStart DNA Master SYBR green I (Roche) was used to produce fluorescent-labeled PCR products during repetitive cycling of the amplification reaction as previously described (Fimia et al., 2007). Primer sets for all amplicons were designed using the Primer Express software system (version 1.0; Applied Biosystems): AMBRA1 (forward), 5′-AACCCTCCACTGCGAGTTGA-3′ and (reverse) 5′-TCTACCTGTTCCGTGGTTCTCC-3′; L34 (forward), 5′-GTCCCGAACCCCTGGTAATAGA-3′ and (reverse) 5′-GGCCCTGCTGACATGTTTCTT-3′; DLC1 (forward), 5′-CCCCCACCTCAGGTAACCAT-3′ and (reverse) 5′-GCCTGAGTAGCGCACTCCAC-3′; DLC2 (forward), 5′-GACTCGCCTCCGTGAAGTGTC-3′ and (reverse) 5′-GGCGCAGTCAACGGCAT-3′; ROADBLOCK1 (forward), 5′-GAGCCAGAAGGGAGTGCAGG-3′ and (reverse) 5′-TGAGGCTGGCATACTGGGTG-3′; TCTEX1 (forward), 5′-GGGACAGCTCTACTGACGGGA-3′ and (reverse) 5′-AAGGCCATAGGCTGGACTGC-3′; and ULK1 (forward), 5′-AAGGCCATAGGCTGGACTGC-3′ and (reverse) 5′-AAGGCCATAGGCTGGACTGC-3′. L34 mRNA level was used as an internal control. β-Actin and GAPDH levels were used as additional controls to confirm significant decreases.
Bidimensional gel electrophoresis analysis
Proteins were precipitated using a clean up kit (Ettan 2-D; GE Healthcare) following the manufacturer’s instructions and were subsequently resuspended in urea buffer (7 M urea, 2 M thiourea, 2% CHAPS, 1% sulfobetaine SB3-10, 1% amidosulfobetaine ASB14, and 50 mM DTT).
For the first dimension of protein separation, isoelectric focusing was performed using 7-cm immobilized nonlinear pH gradient strips, 3–10, and linear pH gradient strips, 4–7 (GE Healthcare), on an electrophoresis unit (IPGphor II; GE Healthcare). 30 µg proteins was loaded by passive in-gel rehydration for 12 h then run using a program in which the voltage was set for 90 min at 30 V, 30 min at 100 V, 30 min at 200 V, voltage gradient 15 min up to 500 V, 30 min at 500 V, voltage gradient 15 min up to 1,000 V, 30 min at 1,000 V, voltage gradient 60 min up to 5,000 V, and 180 min at 5,000 V.
Before the second dimension of electrophoresis, IPG gel strips were equilibrated for 15 min at RT in 1% DTT to reduce the proteins, and sulfhydryl groups were subsequently derivatized using 4% iodoacetamide (both solutions were prepared in 50 mM Tris, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and 2% bromophenol blue) for 15 min. Strips were transferred to 1.0-mm-thick 8%, 10%, and 15% (wt/vol) polyacrylamide minigels for AMBRA1, DIC, and DLC1 detection, respectively, and the second-dimension gels were run at 120 V for 2 h. The 2D gels were transferred to nitrocellulose filters as described for WB.
In vitro mixed beads kinase assay
HEK293 cells were transiently transfected with Myc-tagged wild-type or K46I ULK1 or with Myc-tagged AMBRA1 cDNAs, and the ULK1-transfected ones were nutrient starved for 2 h. Protein extracts were subjected to IP with an anti-Myc antibody, and a mixed beads kinase assay was performed as described previously (Chan et al., 2009). In brief, wild-type Myc-ULKI– and K46I Myc-ULK1–immunopurified proteins were washed four times with HEMG buffer and once with kinase reaction buffer (20 mM Hepes, pH 7.5, 20 mM MgCl2, 25 mM β-glycerophosphate, 2 mM DTT, and 100 µM sodium orthovanadate) before being combined together with Myc-AMBRA1–immunopurified proteins and incubated in a 20-µl final volume of kinase reaction buffer containing 5 µCi γ-[32P]ATP (PerkinElmer) at 30°C for 30 min. Reactions were stopped by addition of 4× SDS sample buffer and boiled for 10 min. [32P]-labeled reaction products were resolved on SDS-PAGE and analyzed by autoradiography using a phosphoimager scanner (Typhoon; GE Healthcare).
Velocity sedimentation by sucrose gradient
2F cells were treated with 5 µM taxol (Sigma-Aldrich) for 4 h. Cell lysates were suspended in a buffer containing 0.25 M sucrose, 10 mM Hepes, and 1 mM EDTA plus a protease inhibitor cocktail. Cell suspension was homogenized by 100 strokes in a dounce potter homogenizer and centrifuged for 10 min at 600 g to obtain a postnuclear supernatant. The postnuclear supernatant was recentrifuged for 15 min at 11,000 g to obtain a postmitochondrial supernatant. The postmitochondrial supernatant was layered onto a discontinuous four-step gradient consisting of 2 ml each of 2.0 M, 1.3 M, 1.0 M, and 0.6 M sucrose in 10 mM Hepes. Centrifugation was performed using a rotor (SW41 Ti; Beckman Coulter) at 27,000 g for 18 h, and 0.4-ml fractions were manually collected and checked for density.
Statistical analysis
All experiments were performed at least three times. Excel (Microsoft) was used for statistical analysis. Statistical significance was determined using the Student’s t test. P ≤ 0.05 was considered significant.
Online supplemental material
Fig. S1 shows the colocalization of AMBRA1 and DLC1 in normal and starvation conditions by immunogold analysis. Fig. S2 shows that AMBRA1 is phosphorylated during autophagy in an ULK1-dependent manner both in vitro and in vivo. In particular, ULK1 K46I dominant negative prevents AMBRA1 phosphorylation after autophagy induction, and the dissociation of AMBRA1 from the dynein motor complex during autophagy is independent of JNK and PI3K activity. Fig. S3 shows a detailed analysis of AMBRA1 localization in different subcellular compartments, thus demonstrating the translocation of AMBRA1 to the ER after autophagy induction. It also shows that AMBRA1 partially colocalizes with the PI3P-binding protein DFCP1 upon autophagy induction. Fig. S4 shows the modulation of autophagy levels by down-regulation of different DLCs. Moreover, the interaction between AMBRA1 and DLC2 is reported. Fig. S5 shows that AMBRA1 is not required for the endosomal sorting of EGF receptor and that ULK1 is required for the activity of constitutive proautophagic AMBRA1 mutants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002100/DC1.
Acknowledgments
We thank the Animal Facility (STA) of the University of Rome Tor Vergata for the mouse work, M. Acuña Villa and M.W. Bennett for editorial and secretarial work, and R. Laricchia, C. Antinozzi, T. Vescovo, V. Cianfanelli, F. Florenzano, and P. Mattioli for research assistance and help with image processing. We are indebted to S.A. Tooze, N.T. Ktistakis, and S. Hirotsune (Osaka University, Osaka, Japan) for kindly providing us with ULK1-, DFCP1- and DIC-encoding constructs. A special thank you also goes to F. Totti for his strenuous support.
This work was supported in part by grants from the Telethon Foundation, Advanced Imaging Research Center, Italian Ministry of University and Research (to F. Cecconi and M. Piacentini), Ricerca Corrente and Ricerca Finalizzata from the Italian Ministry of Health (to F. Cecconi, M. Piacentini, and G.M. Fimia), the European Union Apo-Sys Consortium (to M. Piacentini), and by Compagnia di San Paolo (to F. Cecconi and M. Piacentini).
Footnotes
Abbreviations used in this paper:
- AVO
- acidic vesicular organelle
- βgal
- β-galactosidase
- DIC
- dynein intermediate chain
- DLC
- dynein light chain
- ETNA
- embryonic telencephalic naïve
- IP
- immunoprecipitation
- PI3K
- phosphatidylinositol 3-OH kinase
- PI3P
- phosphatidylinositol 3-phosphate
- WB
- Western blot
References
- Anderson C.W., Baum P.R., Gesteland R.F. 1973. Processing of adenovirus 2-induced proteins. J. Virol. 12:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K.S., Cooper S., Dessing M., Yates S., Buder A. 1998. Interaction of p59fyn kinase with the dynein light chain, Tctex-1, and colocalization during cytokinesis. J. Immunol. 161:1728–1737 [PubMed] [Google Scholar]
- Cecconi F., Levine B. 2008. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 15:344–357 10.1016/j.devcel.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F., Piacentini M., Fimia G.M. 2008. The involvement of cell death and survival in neural tube defects: a distinct role for apoptosis and autophagy? Cell Death Differ. 15:1170–1177 10.1038/cdd.2008.64 [DOI] [PubMed] [Google Scholar]
- Chan E.Y., Longatti A., McKnight N.C., Tooze S.A. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29:157–171 10.1128/MCB.01082-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D.J. 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 19:668–681 10.1091/mbc.E07-08-0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M., Ferraro E., Ferri A., Rigamonti D., Quondamatteo F., Ding H., Xu Z.S., Ferrari F., Angelini D.F., Rotilio G., et al. 2004. Apoptosome inactivation rescues proneural and neural cells from neurodegeneration. Cell Death Differ. 11:1179–1191 10.1038/sj.cdd.4401476 [DOI] [PubMed] [Google Scholar]
- Crépieux P., Kwon H., Leclerc N., Spencer W., Richard S., Lin R., Hiscott J. 1997. I kappaB alpha physically interacts with a cytoskeleton-associated protein through its signal response domain. Mol. Cell. Biol. 17:7375–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell O.J., Mironov A., Allan V.J., Woodman P.G. 2007. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat. Cell Biol. 9:113–120 10.1038/ncb1525 [DOI] [PubMed] [Google Scholar]
- Fass E., Shvets E., Degani I., Hirschberg K., Elazar Z. 2006. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J. Biol. Chem. 281:36303–36316 10.1074/jbc.M607031200 [DOI] [PubMed] [Google Scholar]
- Fimia G.M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., et al. 2007. Ambra1 regulates autophagy and development of the nervous system. Nature. 447:1121–1125 [DOI] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Herzig R.P., Andersson U., Scarpulla R.C. 2000. Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and drosophila. J. Cell Sci. 113:4263–4273 [DOI] [PubMed] [Google Scholar]
- Höök P., Vallee R.B. 2006. The dynein family at a glance. J. Cell Sci. 119:4369–4371 10.1242/jcs.03176 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20:1981–1991 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey S.R., Snyder S.H. 1996. PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science. 274:774–777 10.1126/science.274.5288.774 [DOI] [PubMed] [Google Scholar]
- Jahreiss L., Menzies F.M., Rubinsztein D.C. 2008. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 9:574–587 10.1111/j.1600-0854.2008.00701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G., Neufeld T.P. 2006. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 4:e36 10.1371/journal.pbio.0040036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F.J., Tavassoli K., Van den Bemd G.J., Chang G.T., Horsthemke B., Möröy T., Lüdecke H.J. 2003. Nuclear interaction of the dynein light chain LC8a with the TRPS1 transcription factor suppresses the transcriptional repression activity of TRPS1. Hum. Mol. Genet. 12:1349–1358 10.1093/hmg/ddg145 [DOI] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 19:2039–2050 10.1091/mbc.E07-10-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. 2001. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2:330–335 10.1093/embo-reports/kve061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2008. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33:109–122 10.1247/csf.08005 [DOI] [PubMed] [Google Scholar]
- King S.M. 2000. The dynein microtubule motor. Biochim. Biophys. Acta. 1496:60–75 10.1016/S0167-4889(00)00009-4 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchl R., Hu X.W., Chan E.Y., Tooze S.A. 2006. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 7:129–145 10.1111/j.1600-0854.2005.00368.x [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H., Yan J., Seki N., Yamanouchi Y., Suzuki Y., Takano T., Muramatsu M., Shirasawa T. 1998. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics. 51:76–85 10.1006/geno.1998.5340 [DOI] [PubMed] [Google Scholar]
- Lei K., Davis R.J. 2003. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA. 100:2432–2437 10.1073/pnas.0438011100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K.W., Pfister K.K. 2007. Methods to study the interactions of the dynein light chains and intermediate chains. Methods Mol. Biol. 392:85–95 10.1007/978-1-59745-490-2_6 [DOI] [PubMed] [Google Scholar]
- Lo K.W., Naisbitt S., Fan J.S., Sheng M., Zhang M. 2001. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J. Biol. Chem. 276:14059–14066 10.1074/jbc.M104701200 [DOI] [PubMed] [Google Scholar]
- Machado R.D., Rudarakanchana N., Atkinson C., Flanagan J.A., Harrison R., Morrell N.W., Trembath R.C. 2003. Functional interaction between BMPR-II and Tctex-1, a light chain of Dynein, is isoform-specific and disrupted by mutations underlying primary pulmonary hypertension. Hum. Mol. Genet. 12:3277–3286 10.1093/hmg/ddg365 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11:385–396 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22:132–139 [DOI] [PubMed] [Google Scholar]
- Paglin S., Hollister T., Delohery T., Hackett N., McMahill M., Sphicas E., Domingo D., Yahalom J. 2001. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 61:439–444 [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 122:927–939 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Puthalakath H., Huang D.C., O’Reilly L.A., King S.M., Strasser A. 1999. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell. 3:287–296 10.1016/S1097-2765(00)80456-6 [DOI] [PubMed] [Google Scholar]
- Puthalakath H., Villunger A., O’Reilly L.A., Beaumont J.G., Coultas L., Cheney R.E., Huang D.C., Strasser A. 2001. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 293:1829–1832 10.1126/science.1062257 [DOI] [PubMed] [Google Scholar]
- Raux H., Flamand A., Blondel D. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74:10212–10216 10.1128/JVI.74.21.10212-10216.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O’Kane C.J., Brown S.D., Rubinsztein D.C. 2005. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 37:771–776 10.1038/ng1591 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. 2008. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 105:19211–19216 10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. 2007. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581:2156–2161 10.1016/j.febslet.2007.01.096 [DOI] [PubMed] [Google Scholar]
- Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 1:84–91 10.4161/auto.1.2.1697 [DOI] [PubMed] [Google Scholar]
- Tomoda T., Bhatt R.S., Kuroyanagi H., Shirasawa T., Hatten M.E. 1999. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 24:833–846 10.1016/S0896-6273(00)81031-4 [DOI] [PubMed] [Google Scholar]
- Vadlamudi R.K., Bagheri-Yarmand R., Yang Z., Balasenthil S., Nguyen D., Sahin A.A., den Hollander P., Kumar R. 2004. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 5:575–585 10.1016/j.ccr.2004.05.022 [DOI] [PubMed] [Google Scholar]
- Varadi A., Johnson-Cadwell L.I., Cirulli V., Yoon Y., Allan V.J., Rutter G.A. 2004. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 117:4389–4400 10.1242/jcs.01299 [DOI] [PubMed] [Google Scholar]
- Vieira O.V., Botelho R.J., Rameh L., Brachmann S.M., Matsuo T., Davidson H.W., Schreiber A., Backer J.M., Cantley L.C., Grinstein S. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–25 10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.J., Salata M.W., Susalka S.J., Pfister K.K. 2001. Light chains of mammalian cytoplasmic dynein: identification and characterization of a family of LC8 light chains. Cell Motil. Cytoskeleton. 49:229–240 10.1002/cm.1036 [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Wang Q.J., Li X., Yan Y., Backer J.M., Chait B.T., Heintz N., Yue Z. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11:468–476 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Babu J.R., da Silva S., Shu Q., Graef I.A., Oliver T., Tomoda T., Tani T., Wooten M.W., Wang F. 2007. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl. Acad. Sci. USA. 104:5842–5847 10.1073/pnas.0701402104 [DOI] [PMC free article] [PubMed] [Google Scholar]