Coa3 and Cox14 form assembly intermediates with newly synthesized Cox1 and are required for association of the Mss51 translational activator with these complexes.
Abstract
Regulation of eukaryotic cytochrome oxidase assembly occurs at the level of Cox1 translation, its central mitochondria-encoded subunit. Translation of COX1 messenger RNA is coupled to complex assembly in a negative feedback loop: the translational activator Mss51 is thought to be sequestered to assembly intermediates, rendering it incompetent to promote translation. In this study, we identify Coa3 (cytochrome oxidase assembly factor 3; Yjl062w-A), a novel regulator of mitochondrial COX1 translation and cytochrome oxidase assembly. We show that Coa3 and Cox14 form assembly intermediates with newly synthesized Cox1 and are required for Mss51 association with these complexes. Mss51 exists in equilibrium between a latent, translational resting, and a committed, translation-effective, state that are represented as distinct complexes. Coa3 and Cox14 promote formation of the latent state and thus down-regulate COX1 expression. Consequently, lack of Coa3 or Cox14 function traps Mss51 in the committed state and promotes Cox1 synthesis. Our data indicate that Coa1 binding to sequestered Mss51 in complex with Cox14, Coa3, and Cox1 is essential for full inactivation.
Introduction
The inner mitochondrial membrane contains the four respiratory chain complexes that generate a proton gradient across the membrane that drives ATP synthesis by the F1Fo-ATPase. The electrons are eventually transferred to molecular oxygen by the terminal enzyme of the respiratory chain, the cytochrome oxidase (complex IV). The respiratory chain complexes are multisubunit membrane protein complexes that contain cofactors for the transport of electrons (Saraste, 1999; Hosler et al., 2006). The majority of the subunits that form the respiratory chain complexes are encoded by nuclear genes and are imported into the organelle. In addition, complexes I, III, and IV contain core subunits that are encoded by mitochondrial genes and synthesized on mitochondrial ribosomes (Fox, 1996; Herrmann and Neupert, 2003; van der Laan et al., 2006). These core subunits represent the starting points of protein complex assembly, and their expression is tightly regulated because misassembled subunits harbor the danger of generating reactive oxygen species with deleterious effects for the cell (Herrmann and Funes, 2005; Fontanesi et al., 2006). The newly synthesized proteins are probably co-translationally inserted into the inner mitochondrial membrane by the protein export machinery (Jia et al., 2003; Szyrach et al., 2003; Bonnefoy et al., 2009). To build a functional protein complex from nuclear- and mitochondria-encoded subunits and to properly insert the cofactors, a large number of assembly factors are required. In the case of the cytochrome oxidase, >20 assembly factors in the yeast Saccharomyces cerevisiae and human mediate the assembly process of 11 or 13 structural subunits, respectively (Shoubridge, 2001; Carr and Winge, 2003; DiMauro and Schon, 2003; Fontanesi et al., 2006; Fernández-Vizarra et al., 2009). Defects in the function of assembly factors compromise the activity of the respiratory chain and affect the metabolism of the cell. Thus, such defects lead to respiratory deficiency in yeast and cause severe neuromuscular disorders in human, the so-called mitochondrial encephalomyopathies (DiMauro and Schon, 2003). In fact, defects in translation of mitochondrial mRNAs are among the most common causes of mitochondrial diseases (Taylor and Turnbull, 2005; Weraarpachai et al., 2009). However, the molecular mechanisms regulating mitochondrial gene expression are ill defined.
SURF1, the human homologue of the yeast Shy1 protein, is a cytochrome oxidase assembly factor, which is important for the biogenesis of the mitochondria-encoded Cox1 protein, the core subunit of the complex. Deletion of the SHY1 gene in yeast leads to severe reduction of cytochrome oxidase complexes and growth defects on nonfermentable medium (Mashkevich et al., 1997; Nijtmans et al., 2001). In human, mutations in SURF1 are among the major causes for Leigh syndrome (Online Mendelian Inheritance in Man ID 256000), a subacute necrotizing encephalomyopathy which is commonly associated with systemic cytochrome oxidase deficiency (Tiranti et al., 1998; Zhu et al., 1998). The yeast Shy1 protein was found in association with early assembly intermediates of Cox1, and similarly, Leigh syndrome patients often accumulate aberrant forms of the cytochrome oxidase that are likely to represent intermediates of the biogenesis process (Coenen et al., 1999; Williams et al., 2004). Although the molecular function of Shy1/SURF1 is still enigmatic, recent analyses suggest a direct or indirect role in incorporation of cofactors such as copper and heme into Cox1 (Smith et al., 2005; Pierrel et al., 2007; Bundschuh et al., 2009). Moreover, Shy1 interactions included subunits involved in the regulation of COX1 expression (Mick et al., 2007; Pierrel et al., 2007).
Translation of COX1 mRNA is tightly linked to the assembly process of the Cox1 protein. Two translational activator proteins, Pet309 and Mss51, specifically recognize the COX1 mRNA and are essential for its translation (Decoster et al., 1990; Manthey and McEwen, 1995; Perez-Martinez et al., 2003; Zambrano et al., 2007). Moreover, Mss51 plays an undefined role in the assembly process (Barrientos et al., 2004; Perez-Martinez et al., 2009). Mss51 is associated with the inner mitochondrial membrane where it forms complexes with newly synthesized Cox1, Cox14, Coa1 (cytochrome oxidase assembly factor 1), and Shy1 (Mick et al., 2007; Pierrel et al., 2007; Khalimonchuk et al., 2010). In this complex, Mss51 is maintained in an inactive state and is unable to promote Cox1 expression. In the course of the Cox1 assembly process, Cox1 associates with additional subunits of the complex, leading to a release of Mss51 that is reactivated and can initiate COX1 mRNA translation. Thus, expression of Cox1 is intimately coupled to its assembly process (Perez-Martinez et al., 2003; Barrientos et al., 2004; Herrmann and Funes, 2005; Khalimonchuk and Rödel, 2005; Fontanesi et al., 2006). However, it is unclear how the assembly state of Cox1 in the membrane is monitored and what molecular roles Coa1, Cox14, and Shy1 play in regulating Mss51 activity.
In this study, we identify Coa3 as a novel regulator of Cox1 expression in mitochondria. Together with Cox14, Coa3 is required to alter the equilibrium of Mss51 from the translation-promoting state to a translationally inactive state. Coa3 is in a complex with newly synthesized Cox1, Cox14, and Mss51. A lack of Coa3 or Cox14 leads to uncontrolled expression of COX1 due to a loss of the Mss51 negative feedback regulation. Concomitantly, accumulated unassembled Cox1 is rapidly turned over in the mutants. Our findings allow novel insights into the mechanism by which assembly of cytochrome oxidase is coupled to translational regulation of Cox1. Cox14 and Coa3 are essential for Mss51 recruitment to Cox1, a first step and prerequisite for its subsequent inactivation by Coa1. Thus, we suggest that generation of the latent state of Mss51 is not solely achieved by sequestration but rather a multistep process of interactions in a defined order.
Results
Coa3 is a mitochondrial membrane protein in complex with Shy1
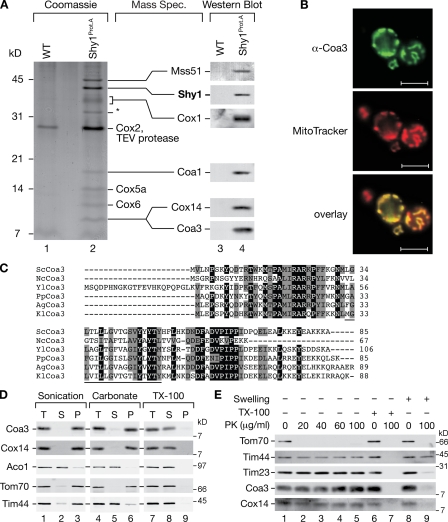
Defects in the function of assembly factors for respiratory chain complexes lead to loss of respiratory competence of cells and thus to severe human disorders. Interestingly, in the case of cytochrome oxidase, assembly is functionally coupled to the regulation of Cox1 expression, the central catalytic subunit of the complex. Defects in assembly of cytochrome oxidase can lead to down-regulation of Cox1 synthesis, preventing accumulation of unassembled Cox1. Although two molecules, Mss51 and Cox14, which participate in the regulatory process, have been determined, little is known as to how the assembly state of Cox1 is monitored at the molecular level. To identify proteins that are involved in early steps of cytochrome oxidase biogenesis and translational regulation, we isolated the assembly factor Shy1 and associated proteins from purified mitochondria. Therefore, we used a functional protein A–tagged version of Shy1 (Mick et al., 2007). After solubilization of mitochondria, Shy1 and its associated proteins were purified by IgG chromatography. Shy1 and bound proteins were released from the affinity matrix by tobacco etch virus protease treatment and analyzed by SDS-PAGE followed by Western blotting and tandem mass spectrometry (MS). In agreement with our previous analyses, subunits of cytochrome oxidase such as Cox1, Cox2, Cox5a, and Cox6 were identified (Fig. 1 A), as well as subunits of the bc1 complex (not depicted; Mick et al., 2007). As expected, we also detected Coa1, Cox14, and Mss51 in the eluate (Mick et al., 2007), proteins which have been implicated in regulation of Cox1 expression. Moreover, we identified tryptic peptides by liquid chromatography (LC)/tandem MS analysis that corresponded to a predicted novel protein with a molecular mass of 9.88 kD encoded by the open reading frame Yjl062w-A, later termed Coa3 (Fig. 1 C). The presence of Coa3 in the eluate was confirmed by Western blotting using antibodies directed against the C terminus of Coa3 (Fig. 1 A, lane 4). To confirm a mitochondrial localization of the uncharacterized Coa3, we performed immunofluorescence analyses using Coa3 antibodies. The labeling pattern of the Coa3 antibody could be superimposed with mitochondria visualized by MitoTracker staining (Fig. 1 B). These analyses confirmed a mitochondrial localization of Coa3 in agreement with a proteomic study by Reinders et al. (2006). Coa3 is highly conserved among yeast species. An in silico analysis of Coa3 did not indicate a significant probability for the presence of a cleavable N-terminal presequence. Moreover, a single segment with the potential to represent a transmembrane helix was identified, suggesting that Coa3 could represent a mitochondrial membrane protein (Fig. 1 C and Fig. S1 A).
Figure 1.
Coa3 is a mitochondrial inner membrane protein facing the IMS. (A) Isolated wild-type (WT) and Shy1ProtA mitochondria were solubilized and subjected to IgG chromatography. After tobacco etch virus (TEV) protease cleavage, eluates were analyzed by SDS-PAGE, Western blotting, or peptide LC/tandem MS. The asterisk marks ADP/ATP carrier, an impurity of purification. Cytochrome bc1 complex components identified are not depicted (Mick et al., 2007). (B) Immunofluorescence analysis using MitoTracker red and Coa3-specific antibodies. Bars, 5 µm. (C) Alignment of Coa3 homologues (ClustalW 2.0.11). Black boxes indicate identical residues in at least five species; gray boxes indicate similar amino acids. Black underlining indicates the predicted transmembrane segment. Sc, S. cerevisiae; Nc, Neurospora crassa; Yl, Yarrowia lipolytica; Pp, Pichia pastoris; Ag, Ashbya gossypii; and Kl, Kluyveromyces lactis. (D and E) Membrane association and submitochondrial localization of Coa3 as described in Materials and methods. T indicates the total; S and P indicate the supernatant and pellet, respectively, after ultracentrifugation. TX-100, Triton X-100; PK, proteinase K.
To assess whether Coa3 is a mitochondrial membrane protein, we performed carbonate extraction analyses of isolated mitochondria. The soluble Aconitase (Aco1) was released from mitochondria by sonication, and the peripheral mitochondrial membrane protein Tim44 was only released efficiently by carbonate treatment (Fig. 1 D). In contrast, Coa3 was carbonate resistant and remained in the carbonate pellet similar to the integral membrane protein Tom70 (Fig. 1 D, lane 6). Thus, Coa3 behaves as an integral membrane protein. Interestingly, in contrast to previous analyses (Barrientos et al., 2004), when we assessed the fractionation pattern of Cox14 using antibodies directed against its C terminus, we found that Cox14 was similarly resistant to carbonate extraction and thus behaved as an integral membrane protein (Fig. 1 D, lane 6). In agreement with this, a single segment with the potential to span a membrane was predicted in Cox14 (Fig. S1 A). To address membrane topology of Coa3, we performed protease protection experiments. Coa3 remained stable when mitochondria were treated with increasing amounts of protease. When the mitochondrial outer membrane was disrupted osmotically, protein domains, which are exposed to the intermembrane space (IMS), become accessible to protease treatment. Under these conditions, the C termini of Coa3 and Cox14 were degraded similarly to the IMS domain of Tim23 (Fig. 1 E, lane 8 vs. lane 9), indicating that both proteins expose their C termini into the IMS. No stable fragments of Coa3 or Cox14 were detected using antibodies directed against the C termini of both proteins (Fig. S1 B). As a control, the matrix protein Tim44 remained resistant to protease treatment after swelling and became only accessible when the inner membrane was disrupted with Triton X-100 (Fig. 1 E, lanes 6–9). In summary, Coa3 and Cox14 are integral membrane proteins that expose their C termini into the IMS. Taking the prediction of a single transmembrane segment into consideration, it appears likely that the N termini of both proteins are exposed to the matrix.
Coa3 is required for cytochrome oxidase biogenesis
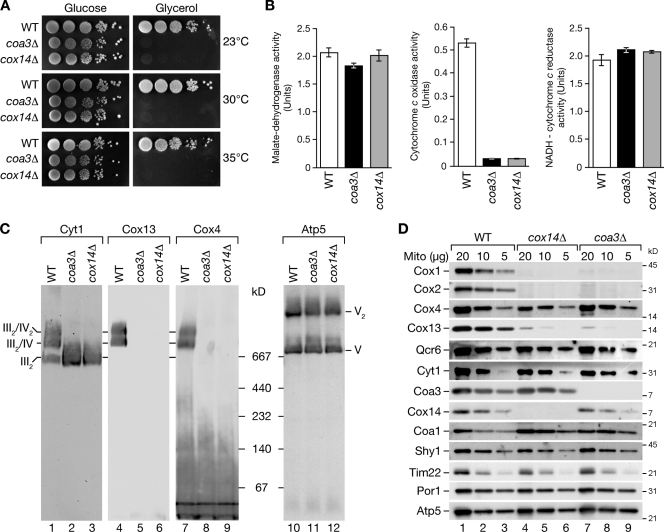
To assess the function of Coa3 in mitochondria, we deleted the COA3 open reading frame. coa3Δ mutant cells displayed a severe growth defect on nonfermentable carbon sources at all tested temperatures but displayed wild-type–like growth on fermentable medium (Fig. 2 A), in agreement with recent data from a genome-wide screen on respiratory-deficient mutants (Merz and Westermann, 2009). This growth defect was indistinguishable from the behavior of cox14Δ mutant cells, which is indicative of a defect in respiration. To determine the cause of the growth phenotype, we measured the activity of respiratory chain complexes and malate dehydrogenase as a control in isolated mitochondria. The activities of the bc1 complex (complex III) and malate dehydrogenase were similar between wild type, cox14Δ, and coa3Δ mitochondria. In contrast, cytochrome oxidase activity was drastically reduced in cox14Δ and coa3Δ mitochondria (Fig. 2 B).
Figure 2.
coa3Δ yeast cells are respiratory deficient and lack cytochrome oxidase. (A) Wild-type (WT), coa3Δ, and cox14Δ yeast cells were spotted in serial 10-fold dilutions on fermentable (glucose) and nonfermentable (glycerol) media. (B) Enzyme assays of isolated mitochondria. Means of four independent experiments (SEM; n = 4). (C) Solubilized mitochondria were separated by BN-PAGE and analyzed by Western blotting. (D) Mitochondria (Mito) were separated by SDS-PAGE and analyzed by Western blotting.
To address the reason for the lack of cytochrome oxidase activity in coa3Δ mitochondria, we analyzed respiratory chain complexes by blue native PAGE (BN-PAGE). In yeast, the respiratory chain complexes associate as supercomplexes, which can be separated by BN-PAGE upon solubilization with the mild detergent digitonin. bc1 complex forms a dimer (III2) that can associate with a single (III2IV) or two cytochrome oxidase (III2IV2) monomers. The bc1 complex was exclusively present in its dimeric form in coa3Δ and cox14Δ mitochondria and we were unable to detect cytochrome oxidase with antibodies against the nuclear-encoded subunits Cox4 and Cox13 (Fig. 2 C). Thus, we conclude that active cytochrome oxidase complex was absent from coa3Δ and cox14Δ mitochondria. To determine the basis for the lack of cytochrome oxidase, we analyzed the steady-state protein levels of selected mitochondrial proteins. In coa3Δ mitochondria, the mitochondria-encoded subunits of cytochrome oxidase, Cox1 and Cox2, were not detected, and the nuclear-encoded subunit Cox13 was dramatically reduced. In contrast, Qcr6 and Cyt1 (bc1 complex) and the nuclear-encoded Cox4 (cytochrome oxidase) were present in similar amounts compared with the wild-type control. Moreover, we assessed the levels of assembly factors for cytochrome oxidase (Fig. 2 D). Although Shy1 levels were not affected, the levels of Coa1 were slightly increased in coa3Δ mutant mitochondria. In contrast, a lack of Coa3 had no effect on levels of Cox14 and vice versa. Interestingly, the steady-state protein levels of cox14Δ mitochondria were virtually indistinguishable from coa3Δ mitochondria (Fig. 2 D, lanes 4–6 vs. lanes 7–9). Thus, a lack of selected subunits of cytochrome oxidase appears to lead to loss of respiratory competence of cox14Δ and coa3Δ cells.
Coa3 associates with assembly factors and regulators of COX1 expression
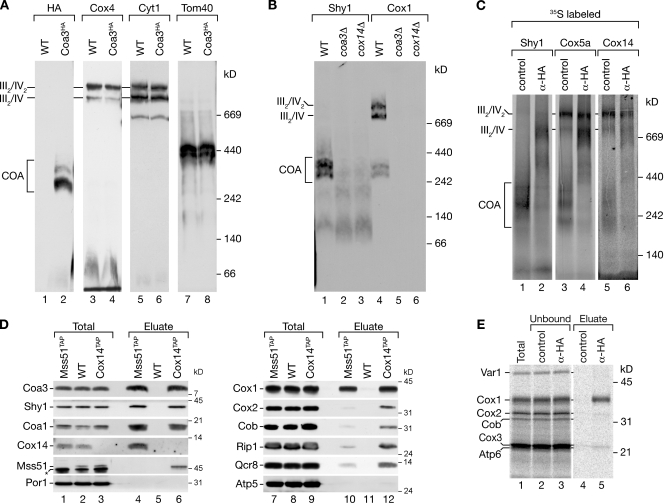
Coa3 was identified by copurification with Shy1-containing complexes. Thus, we analyzed complex formation of Coa3 by BN-PAGE using a functional C-terminally tagged version of Coa3. Coa3 was mainly detected in complexes of 250–400 kD (Fig. 3 A, lane 2). Moreover, upon longer exposure, a small fraction of Coa3 co-migrated with respiratory chain supercomplexes (Fig. S2 A). This was further supported by coisolation of Coa3 with respiratory chain supercomplexes (Fig. S2 B). The 250–400-kD complexes resembled in size cytochrome oxidase assembly intermediates formed by Shy1 and Cox1 (Fig. 3 B, lanes 1 and 4; Mick et al., 2007). These complexes were absent in coa3Δ and cox14Δ mitochondria (Fig. 3 B, lanes 2, 3, 5, and 6). This could be explained by the fact that Coa3 and Cox14 are themselves part of the complexes or the lack of Cox1 in mitochondria (Fig. 2 D). To distinguish between these possibilities, we imported Shy1, Cox14, and, as a control, the early assembling subunit Cox5a (Nijtmans et al., 1998; Mick et al., 2007; Fontanesi et al., 2008; Barrientos et al., 2009) into Coa3HA mitochondria. After solubilization, anti-HA or control antibodies were added to all samples, and analyses of complexes were performed by BN-PAGE. The anti-HA antibodies directed against Coa3 specifically shifted the 250–400-kD Shy1-, Cox14-, and Cox5a-containing complexes in size. In contrast, supercomplexes, which only contain low amounts of Coa3 (see Fig. S2 A), remained largely unaffected (Fig. 3 C). We will refer to these 250–400-kD complexes as COA complexes for cytochrome oxidase assembly intermediates. We conclude that Cox14, Shy1, and Cox5a together with Coa3 are constituents of the COA complexes in agreement with the association seen in the purification of Shy1ProtA.
Figure 3.
Coa3 associates with newly synthesized Cox1. (A) Wild-type (WT) and Coa3HA mitochondria were solubilized and analyzed by BN-PAGE and Western blotting. (B) Wild-type and mutant mitochondria were analyzed as in A. (C) Radiolabeled Shy1, Cox5a, and Cox14 were imported into isolated mitochondria containing Coa3HA. Mitochondria were solubilized and incubated with Flag (control) or HA antibodies before separation by BN-PAGE and digital autoradiography. (D) IgG chromatography of isolated mitochondria from wild type, Mss51TAP, or Cox14TAP. Samples were analyzed by SDS-PAGE and Western blotting. The asterisk indicates a cross-reactive signal detected by Mss51 antiserum. (E) In organello translation of isolated Coa3HA mitochondria was performed, and the sample was split and subjected to coimmunoprecipitations with anti-Flag (control) or anti-HA antibodies. Samples were analyzed by SDS-PAGE and digital autoradiography. (D and E) The amounts of protein loaded in the total samples correspond to 2% of the eluate.
Cox14 has been implicated in the feedback regulation of Cox1 translation through an association with the translational activator Mss51 (Perez-Martinez et al., 2003; Barrientos et al., 2004). Thus, we analyzed whether Coa3 was also in complex with Mss51 by purification of Mss51 and Cox14 complexes from mitochondria. Besides the known associated components, such as Shy1, Cox1, Cox14, and Coa1, Coa3 was efficiently copurified with Mss51 (Fig. 3 D, lanes 4 and 10). Similarly, purification of Cox14 led to coisolation of Shy1, Cox1, Mss51, Coa1, and Coa3 (Fig. 3 D). Because a fraction of Cox14 associates with respiratory chain supercomplexes (Fig. 3 C; Mick et al., 2007), additional subunits of cytochrome oxidase and bc1 complex are coisolated with Cox14 (Fig. 3 D, lane 12; and Fig. S2 C). Accordingly, Coa3 is in complex with Cox14 and Mss51. Therefore, we assessed a role of Coa3 in Cox1 biogenesis directly by analyzing whether Coa3 was associated with newly synthesized Cox1. We pulse labeled mitochondrial translation products in mitochondria that contained Coa3HA and performed immunoprecipitation experiments using antibodies against HA or control antibodies. Although control antibodies did not precipitate significant amounts of Cox1, newly synthesized Cox1 was specifically precipitated with Coa3HA (Fig. 3 E, lane 5). In summary, Coa3 interacts with newly synthesized Cox1, similar to Mss51 and Cox14 (Perez-Martinez et al., 2003; Barrientos et al., 2004). Moreover, Coa3 associates with the assembly factor Cox14 in COA complexes of 250–400 kD and is in complex with the translational regulator Mss51.
Coa3 association with the translational regulator machinery depends on Cox1
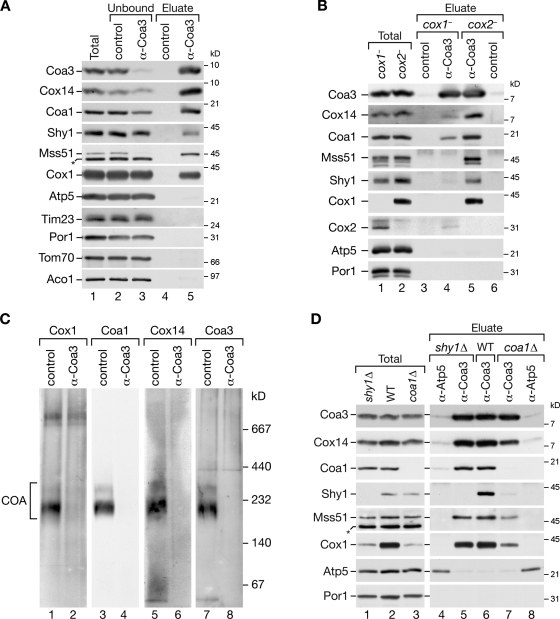
To define protein associations of Coa3 and their dynamics in more detail, we established conditions to immunoprecipitate the untagged Coa3 from mitochondrial membranes. An antibody directed against the C terminus of Coa3 efficiently immunoprecipitated the protein and was able to deplete it from digitonin-solubilized mitochondria (Fig. 4 A, lane 1 vs. lane 3). The Coa3 antibodies specifically coimmunoprecipitated significant amounts of Cox14, Coa1, Cox1, and Shy1. Similarly, Mss51 was efficiently precipitated from the mitochondrial extract. As controls, we performed precipitations with preimmune serum and checked precipitates for the presence of unrelated mitochondrial proteins. The preimmune serum did not precipitate any of the components. Control proteins such as Atp5, Porin (Por1), Tom70, Tim23, and Aco1 were not precipitated with Coa3 antibodies (Fig. 4 A, lane 4 vs. lane 5).
Figure 4.
Formation of COA complexes strictly depends on Cox1; sequestration of Mss51 is independent of Cox2, Shy1, and Coa1. (A) Coimmunoprecipitations of Coa3 from digitonin-solubilized mitochondria. After solubilization, samples were incubated with Coa3-specific or preimmune (control) antisera. Bound material was eluted and analyzed by SDS-PAGE and Western blotting. The amounts of protein loaded in the total and unbound samples correspond to 6% of the eluate. (B) Coimmunoprecipitation experiments as in A were performed in cox1− and cox2− mitochondria. (C) Isolated cox2− mitochondria were solubilized in digitonin and subjected to immunoprecipitation with Coa3 or preimmune (control) antisera. Lysates were depleted from antibody-bound complexes with protein A–Sepharose, and unbound material was separated by BN-PAGE, followed by Western blot analysis. (D) Coimmunoprecipitation experiments from wild-type (WT), shy1Δ, and coa1Δ mitochondria were performed as in A except that anti-Atp5 antiserum was used as a control. (A and D) Asterisks indicate a cross-reactive signal detected by Mss51 antiserum.
Given the tight connection of the interacting factors to Cox1 assembly, we asked which of the protein interactions were dependent on the presence of Cox1. Thus, we performed immunoprecipitations from mitochondria lacking Cox1 (cox1−) and as a control of mitochondria lacking Cox2 (cox2−). In cox2 mutant mitochondria, mature cytochrome oxidase is not formed, but assembly intermediates of Cox1 accumulate (Horan et al., 2005; Mick et al., 2007). Although Coa3 was efficiently precipitated from both mitochondria, only minute amounts of Cox14, Coa1, and Mss51 were coimmunoprecipitated in cox1− mitochondria, indicating that all interactions with Coa3 largely depend on the presence of Cox1. In contrast, when Cox1 accumulated in an unassembled or partially assembled state such as in the case of cox2− mitochondria, Cox14, Coa1, Mss51, Shy1, and Cox1 were associated with Coa3 (Fig. 4 B). To assess whether proteins associate with Coa3 in distinct or similar complexes, we performed immunodepletion analyses. Therefore, we incubated mitochondrial extracts from cox2− mitochondria with Coa3 or preimmune serum coupled to protein A–Sepharose and separated complexes that remained in the unbound fraction on BN-PAGE. Although partially assembled supercomplexes that contain Cox1 (Mick et al., 2007) were not depleted by the antibody treatment (Fig. 4 C, lanes 1 and 2), the 250–400-kD COA complexes were fully depleted from the extract, and antibodies against Cox1, Coa1, Coa3, and Cox14 did not detect residual amounts of complexes in this range (Fig. 4 C). (Unfortunately, antibodies against Mss51 were not of sufficient quality to detect Mss51 complexes on BN-PAGE.) Accordingly, Coa3, Cox14, Coa1, and Cox1 are constituents of the COA complexes.
Because Coa3 was identified through its association with Shy1, we asked whether Shy1 was required for COA complex formation. Coa3 antibodies efficiently precipitated Cox14, Coa1, Cox1, and Mss51 from shy1Δ mitochondria (Fig. 4 D, lanes 4 and 5). Note that Cox1 levels are dramatically decreased in shy1Δ mitochondria (Nijtmans et al., 2001; Barrientos et al., 2002; Mick et al., 2007), whereas the coprecipitation efficiency was only slightly reduced compared with the wild-type control. Thus, Shy1 is obviously not required for complex formation between Coa3, Cox14, Coa1, Cox1, and Mss51. Previous work had linked Coa1 to Cox1 translational regulation (Mick et al., 2007; Pierrel et al., 2007). This observation and the presence of Coa1 in the COA complexes led us to assess its involvement in complex formation. When Coa3 was immunoprecipitated from coa1Δ mitochondria, the coprecipitation efficiency was slightly decreased compared with wild-type and shy1Δ mitochondria; however, Cox14, Cox1, and Mss51 were still efficiently coprecipitated with Coa3, whereas Shy1 was not recovered (Fig. 4 D, lanes 7 and 8). Thus, binding of Coa3 to Cox14, Mss51, and to unassembled Cox1 itself is independent of Coa1 and Shy1 (see Coa3 and Cox14 are required for Mss51 association with newly synthesized Cox1). However, the presence of Coa1 is a prerequisite for Shy1 recruitment into the complex.
Coa3 and Cox14 negatively regulate COX1 translation
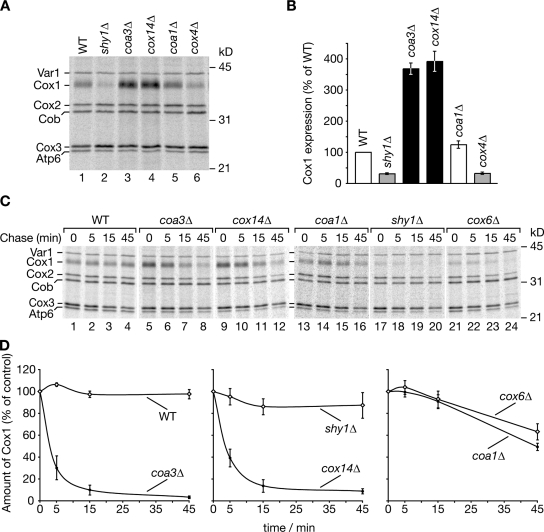
The association of Coa3 with Mss51 and Cox14 pointed toward a possible function in the COX1 translational regulation. Cox14 is considered to bind to Cox1 and to sequester Mss51 in an inactive state and thus to prevent new rounds of Cox1 translation (Barrientos et al., 2004). Therefore, we analyzed Cox1 synthesis in wild-type and mutant cells in a pulse-chase analysis. After a 5-min pulse with [35S]methionine, the synthesis of Cox1 was dramatically increased in the absence of Cox14 or Coa3 (Fig. 5 A, lanes 3 and 4). In agreement with previous in organello analyses, a lack of Coa1 resulted only in a slight increase of Cox1 synthesis (Fig. 5 A, lane 5; Mick et al., 2007). However, Cox1 translation was reduced when Shy1 or structural subunits of cytochrome oxidase, such as Cox4, were absent (Fig. 5 A, lanes 2 and 6; Barrientos et al., 2004). This reduction is probably caused by the occurring sequestration of Mss51 by Coa3 and Cox14. Thus, we conclude that both Cox14 and Coa3 negatively regulate Cox1 synthesis in mitochondria. Therefore, we tested whether overexpression of either Cox14 in coa3Δ mutant cells or of Coa3 in cox14Δ mutant cells suppressed the growth phenotype of the mutants. However, no effect on the growth phenotype was observed (unpublished data), indicating that both proteins fulfill distinct functions.
Figure 5.
coa3Δ and cox14Δ yeast cells display increased COX1 expression and reduced stability of Cox1. (A) In vivo labeling of mitochondrial translation products was performed according to Materials and methods. After 5-min pulse, cells were lysed and subjected to TCA precipitation, and labeled proteins were analyzed by SDS-PAGE and digital autoradiography. (B) Three independent experiments as in A were quantified using ImageQuant TL software (GE Healthcare). Values represent the mean ratios of Cox1/Var1, relative to wild type (WT; 100%). Error bars indicate SEM (n = 3). (C) In vivo labeling of mitochondrial translation products was performed as in A. After 5-min pulse, translations were stopped by the addition of excess unlabeled methionine and chloramphenicol. Samples were taken after 5-, 15-, and 45-min chase and analyzed as in A. (D) Four independent experiments as in C were quantified using ImageQuant TL software, and Cox1 signals were plotted against chase times. Values represent mean ratios of Cox1/Cob, relative to 0-min chase (100%). Error bars indicate SEM (n = 4).
The observation that Cox1 synthesis was drastically increased in the absence of Coa3 or Cox14 was unexpected because the steady-state levels of Cox1 were severely affected (Fig. 2 D). We speculated that the newly synthesized Cox1 was especially unstable in coa3Δ and cox14Δ mutants. To address this hypothesis, we performed pulse labeling of mitochondrial translation products in whole cells and monitored the amounts of Cox1 protein in an extended chase. Newly synthesized Cox1 remained relatively stable over a time course of 45 min in wild-type and shy1Δ cells (Fig. 5, C [lanes 1–4 and 17–20] and D; and Fig. S3 A). The lack of nuclear-encoded structural subunits such as Cox6 or the assembly factor Coa1 led to a slight decrease in Cox1 stability (Fig. 5, C [lanes 13–16 and 21–24] and D). In contrast, Cox1 was rapidly turned over in both coa3Δ and cox14Δ mitochondria (Fig. 5 C, lanes 5–12). Thus, we conclude that association of Coa3 and Cox14 to newly synthesized Cox1 directly or indirectly protects it from turnover by the quality control system (Arlt et al., 1996; Guzélin et al., 1996). In contrast to Coa3 and Cox14, Coa1 seems to play a later role because a lack of Coa1 affects Cox1 translation or stability only mildly. To support this conclusion, we generated coa1Δcox14Δ, coa1Δcoa3Δ, and cox14Δcoa3Δ double mutant strains and assessed Cox1 stability in these cells. As expected, double mutants of coa1Δ with cox14Δ or coa3Δ displayed reduced Cox1 stability similar to single coa3Δ or cox14Δ cells (Fig. S3 B).
Coa3 and Cox14 are required for Mss51 association with newly synthesized Cox1
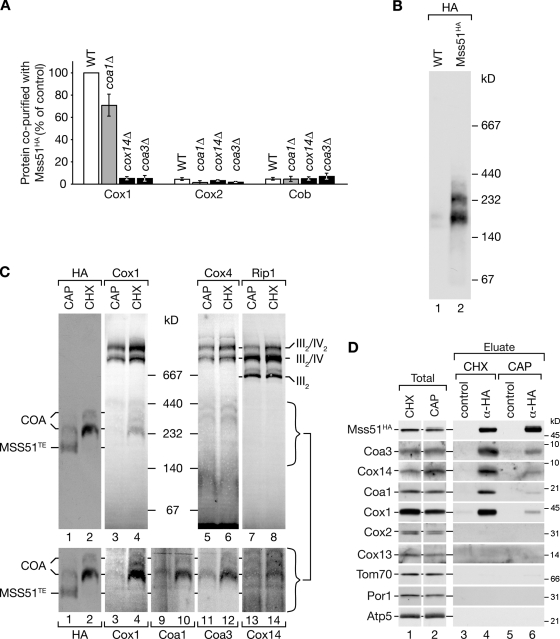
Mss51 is required for translation of COX1 mRNA. Recruitment of Mss51 to unassembled Cox1 in complex with Cox14 is thought to be essential to inactivate Mss51 (Perez-Martinez et al., 2003; Barrientos et al., 2004). Thus, we assessed Mss51 binding to newly synthesized Cox1 in wild-type and mutant mitochondria by coimmunoprecipitation. Although newly synthesized Cox1 was efficiently bound by Mss51 in wild-type mitochondria, Cox1 binding was severely reduced and close to background levels in coa3Δ and cox14Δ mitochondria (Fig. 6 A). In contrast, in coa1Δ mitochondria, a mild but obvious reduction of Cox1 binding to Mss51 was observed. Thus, recruitment of Mss51 to newly synthesized Cox1 depends on the presence of Coa3 as well as Cox14 but not on Coa1.
Figure 6.
Mss51 complexes are dynamic in nature. (A) In organello labeling of mitochondrial translation products was performed in the indicated strains, expressing Mss51HA. Coimmunoprecipitation experiments were analyzed as in Fig. 3 E. Autoradiograph signals were quantified using ImageQuant TL software. The ratios of signals for Cox1, Cox2, and Cob between eluate and total were calculated and normalized to the Cox1 eluate/total ratio in wild type (WT; 100%). Means of three independent experiments (SEM; n = 3) are shown. (B) Mss51HA and wild-type mitochondria were solubilized and analyzed by BN-PAGE and Western blotting using anti-HA antibodies. (C) Mitochondria isolated from Mss51HA yeast cells treated with chloramphenicol (CAP) or cycloheximide (CHX) were analyzed as in B. Brackets indicate the molecular mass range of the Mss51 complexes. Lanes 1–4 in the bottom panels are cropped from the top panels and displayed with increased contrast for Cox1. (D) Solubilized mitochondria from pretreated cells were subjected to coimmunoprecipitation with anti-HA and control antibodies and analyzed by SDS-PAGE and Western blotting.
To assess Mss51 complexes in mitochondria at steady-state, we chromosomally integrated a HA-tagged version replacing the wild-type Mss51. Mss51HA was detected in three distinct complexes in BN-PAGE analyses of digitonin-solubilized mitochondria (Fig. 6 B). This pattern resembles previous observations for an Mss51MYC construct (Khalimonchuk et al., 2010). However, the apparent molecular weights of the complexes observed in this study are different in size. Thus, we investigated whether Coa3 and other assembly factors were constituents of Mss51 complexes and asked how they are linked to mitochondrial translation. Therefore, mitochondria were isolated from cells treated with the translational inhibitors chloramphenicol or cycloheximide. Chloramphenicol inhibits mitochondrial translation, whereas cycloheximide treatment stops supply of nuclear-encoded cytochrome oxidase subunits and leads to accumulation of unassembled Cox1 (Fig. 6 C, lanes 3 and 4). In the latter case, Mss51 was found exclusively in 250- and 300-kD complexes, and, according to the sequestration model, Mss51 should be inactivated through association with unassembled Cox1 into COA complexes. In support of this, we detected Cox1, Coa1, Coa3, and Cox14 in complexes that co-migrated with the 250- and 300-kD COA complexes (Fig. 6 C), suggesting that in complex with these four proteins, Mss51 is in the latent, translational resting state. In contrast, chloramphenicol treatment shifted the distribution of Mss51 to a 180-kD complex, whereas only minute amounts of the 250-kD complex were detected. In this study, recruitment of unassembled Cox1 to imported subunits releases Mss51 into a smaller translation-effective complex of 180 kD (MSS51TE; translation effective), with Mss51 being the only constituent identified so far (Fig. 6 C). To confirm physical associations, we performed coimmunoprecipitation of Mss51HA. In agreement with the results of the BN-PAGE analyses, Mss51 was associated with Coa3, Cox14, Cox1, and Coa1 only when assembly intermediates accumulated (Fig. 6 D, lane 4), whereas the interactions were mostly lost when mitochondrial translation was blocked (Fig. 6 D, lane 6). In summary, by changing conditions for the function of Mss51 between its committed, translation-stimulating situation, and the latent, translation-inactive state, we were able to assign the low molecular weight complex of Mss51 as the translation-promoting and larger complexes with Cox1, Coa1, Cox14, and Coa3 as the inactive forms. The analysis of untreated mitochondria (Fig. 6 B) shows that at steady-state, all forms of Mss51 are present, suggesting that they are in a dynamic equilibrium. Treatment with chloramphenicol or cycloheximide shifts the equilibrium to provide a view of the two extreme states of Mss51.
Lack of Coa3 or Cox14 shifts the equilibrium of Mss51 complexes to the translation-committed state
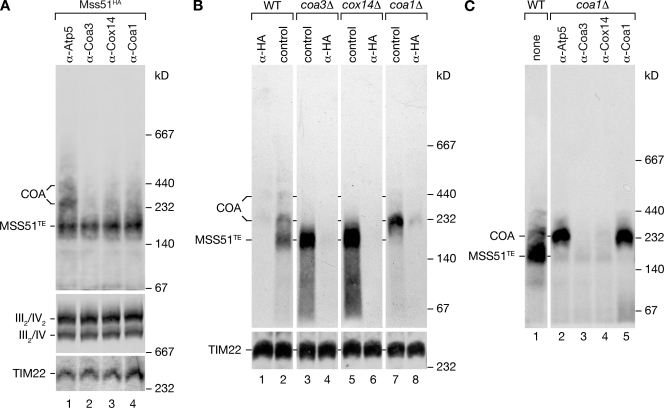
To provide direct evidence that at steady-state Cox14, Coa1, and Coa3 are together in the 250- and 300-kD complexes with Mss51, we performed antibody depletion BN-PAGE analyses. Antibodies directed against Coa3, Cox14, and Coa1 completely depleted the 250- and 300-kD complexes from solubilized mitochondria (Fig. 7 A, lanes 2–4). In contrast, the MSS51TE complex was not affected by the treatment. As a control, we used antibodies directed against Atp5 that did not affect Mss51 complexes (Fig. 7 A, lane 1). Thus, Cox14, Coa3, and Coa1 are specific constituents of the Mss51 complexes representing its latent state.
Figure 7.
Lack of Coa1 stalls Mss51 in a 250-kD complex. (A) Mss51HA mitochondria were solubilized, and samples were split and incubated with indicated antibodies. Antibodies with bound material were precipitated with protein A–Sepharose, and unbound material was analyzed by BN-PAGE and Western blotting using anti-HA antibodies. Respiratory chain supercomplexes and carrier translocase (TIM22) were detected with anti-Cox1 or anti-Tim54 antibodies, respectively. (B) Mitochondria from the indicated mutants expressing Mss51HA were analyzed as in A. (C) Antibody depletion experiments were performed in coa1Δ mitochondria as in A. For comparison, untreated wild-type (WT) mitochondria were analyzed in lane 1.
What is the role of Coa3 or Cox14 in Mss51 regulation? To address this question, we analyzed Mss51 complexes in mutant mitochondria by BN-PAGE. A lack of Coa3 or Cox14 shifted the equilibrium of Mss51 complexes toward the MSS51TE complex. The larger 250- and 300-kD COA complexes were absent in these mitochondria at steady-state (Fig. 7 B, lanes 3 and 5). Using anti-HA antibodies directed against Mss51HA for depletion analyses, we demonstrated the authenticity of the Mss51 complexes. The altered complex pattern of Mss51 in cox14Δ and coa3Δ mitochondria is in full agreement with the increased translation phenotype observed in the mutants (Fig. 5 B). Interestingly, analysis of coa1Δ mitochondria revealed the presence of Mss51 primarily in a 250-kD complex, which accumulates compared with the wild-type situation (Fig. 7 B, lane 7). In these mitochondria, MSS51TE was not detected. It appears likely that, despite a lack of an obvious size difference on BN-PAGE, the observed 250-kD complex in coa1Δ is similar to the 250-kD complex of wild-type mitochondria with the exception that Coa1 is absent. coa1Δ mitochondria displayed wild type–like synthesis of Cox1 but significantly less than coa3Δ or cox14Δ mitochondria (Fig. 5 B). Thus, we concluded that in the absence of Coa1, Mss51 is present in the 250-kD complex but not fully inactivated and therefore is able to promote translation closely resembling the wild-type situation. To demonstrate that the 250-kD complex in coa1Δ in fact contained Coa3, Cox14, and Mss51, we performed antibody depletion BN-PAGE analyses. Incubation with antibodies directed against Coa3 or Cox14 specifically depleted the 250-kD complex from the extract, whereas antibodies directed against Atp5 or Coa1 had no effect (Fig. 7 C). In summary, the 250-kD Mss51 complex in coa1Δ mitochondria consists of Coa3, Cox14, and sequestered Mss51. It is likely that this complex is of transient nature in wild type and that binding of Coa1 allows subsequent steps of assembly such as Shy1 binding. Thus, the 250-kD complex accumulates in the absence of Coa1. Despite the presence of Coa3 and Cox14, Mss51 is not fully inactivated because COX1 mRNA translation occurs in these mitochondria. Thus, we conclude that Coa1 is required for full inactivation of Mss51.
Discussion
In mitochondria, translation occurs on membrane-bound ribosomes and thus in proximity to respiratory chain complexes. This spatial coupling is a prerequisite to link regulation of COX1 mRNA translation to the assembly state of the Cox1 protein. The current model for the regulatory circuit of COX1 mRNA translation suggests that Cox14 recruits Mss51 to Cox1 by a yet unknown mechanism (Perez-Martinez et al., 2003; Barrientos et al., 2004). Thus, physical sequestration is considered to lead to inactivation of Mss51 and to block COX1 mRNA translation. Coa3 is a novel Cox1-associated factor that negatively regulates Cox1 expression. We find that, similar to Cox14, Coa3 is an integral inner mitochondrial membrane protein exposed to the IMS. The topology of Cox14 has been a matter of debate. In agreement with the data presented in this study, initially Cox14 had been found to represent a membrane-spanning protein (Glerum et al., 1995), but a later study suggested a matrix localization of Cox14 (Barrientos et al., 2004).
A minor fraction of Coa3 and Cox14 appears to be associated with respiratory chain supercomplexes, similar to Shy1 (Mick et al., 2007). The function of this pool is still not clear (Stuart, 2008). It is currently speculated that the supercomplex-associated assembly factors could play a role in repair processes of mature complexes or alternatively associate with not fully assembled intermediates that already bind to complex III.
Newly synthesized Cox1 is associated with Cox14 and Coa3, even in the absence of Coa1 or Shy1. Moreover, Mss51 binding to newly synthesized Cox1 requires Coa3 and Cox14, and at the same time Mss51 binding to Cox14 and Coa3 depends on Cox1 but not on Coa1 or Shy1. Thus, the 12 times membrane-spanning Cox1 protein is apparently recognized by two transmembrane proteins: Cox14 and Coa3. These observations extend a recent study, which showed that the Cox14 interaction with Mss51 requires Cox1 (Perez-Martinez et al., 2009). Loss of Cox14 or Coa3 function leads to similar defects in the biogenesis of Cox1: Cox1 expression is significantly increased, and the newly synthesized Cox1 is rapidly turned over. How can these opposing effects be explained? Mss51 interacts with the 5′ untranslated region of the COX1 mRNA and promotes translation (Decoster et al., 1990; Perez-Martinez et al., 2003; Barrientos et al., 2004). Cox14 was believed to represent the sole link between Cox1 and Mss51 and to mediate inactivation by physical sequestration of Mss51 (Perez-Martinez et al., 2003; Barrientos et al., 2004). Loss of Cox14 would release Mss51 in its active state to stimulate Cox1 expression. In this study, we show that the mechanism of the regulatory circuit is more complex than anticipated and possibly not just a topological phenomenon. First, Cox14 and Coa3 are not membrane-associated proteins, fully exposed to the matrix, which only serve as bridging proteins to sequester Mss51; they are integral membrane proteins with small domains exposed to the IMS as well as the matrix. Second, both membrane proteins, Cox14 and Coa3, are required to bind to the unassembled Cox1 and to communicate back for translational regulation. Despite the fact that coa3Δ and cox14Δ mutant mitochondria display virtually indistinguishable phenotypes, we did not observe crosswise suppression of the mutant growth phenotype upon overexpression of Coa3 or Cox14, indicating that they fulfill independent functions. In contrast to what is found in cox14Δ and coa3Δ, loss of Coa1 causes only a slight increase in translation and destabilization of Cox1 (Fig. 5; Mick et al., 2007). The analysis of Cox1 stability in double mutants supports the conclusion that Coa1 acts downstream of Cox14 and Coa3. In agreement with this observation, a 250-kD complex consisting of Mss51, Coa3, Cox14, and Cox1 is formed in the absence of Coa1. In contrast, recruitment of Shy1 depends on the presence of Coa1 in the complex.
How is the regulation of translation linked to complex assembly? Assembly of Cox1 requires factors that chaperone the compilation of nuclear-encoded subunits with Cox1, insertion of cofactors, and addition of mitochondrial-encoded Cox2 and Cox3 (Herrmann and Funes, 2005; Fontanesi et al., 2006). The process appears to occur through distinct assembly steps, and defects in this process can lead to the accumulation of assembly intermediates (Nijtmans et al., 2001; Williams et al., 2004; Stiburek et al., 2005; Mick et al., 2007).
Mss51 is present in different complexes that we assign to its functional state. Under conditions that inactivate Mss51 through accumulation of Cox1 assembly intermediates (loss of supply of nuclear-encoded subunits) Mss51 is present in 250- and 300-kD complexes. In contrast, when assembly intermediates are depleted through chloramphenicol treatment, Mss51 is solely present in the translation-effective state (MSS51TE). We have been unable to identify assembly factors or structural subunits of the cytochrome oxidase in the MSS51TE complex by Western blotting. Thus, it is tempting to speculate that this complex is an oligomeric form of Mss51 or contains subunits that are not yet known. At steady-state, all complexes are present in mitochondria, indicating that they are in dynamic equilibrium, which can be observed by analyzing Mss51 complexes under different growth conditions of yeast (unpublished data). We clearly show that Mss51, Shy1, Cox1, Coa1, Cox14, and Coa3 are found in the 250- and 300-kD COA complexes, whereas only Mss51 is detected in the MSS51TE complex. This finding partially differs from a recent study by Khalimonchuk et al. (2010). However, our findings with regard to Cox1 synthesis and complex composition in mutants agree well with our characterization of the complexes. (a) Shy1 is not required for Mss51, Cox1, Coa1, Cox14, and Coa3 complex formation, and shy1Δ cells display reduced Cox1 synthesis compared with wild-type mitochondria. (b) Coa1 is not required for Mss51, Cox1, Cox14, and Coa3 complex formation, and coa1Δ cells display slightly increased Cox1 synthesis compared with wild-type mitochondria. (c) Cox14 and Coa3 are required for interaction of Mss51 with Cox1, Shy1, and Coa1. In the absence of either Cox14 or Coa3, Mss51 is exclusively present in the MSS51TE complex. In agreement, coa3Δ and cox14Δ cells display drastically increased Cox1 synthesis and a constitutively active Mss51. Obviously, a lack of Coa3 or Cox14 shifts the dynamic equilibrium of Mss51 toward the translation-effective state of Mss51.
Based on our findings, we propose a model in which inactivation of Mss51 is a sequential process. Initially, Mss51 is recruited to a Cox1, Cox14, Coa3 complex, leading to stabilization of Cox1. In this 250-kD form, which accumulates in coa1Δ mitochondria, Mss51 is not fully inactivated. Association of Coa1 is required to shift Mss51 to the latent, translation-inactive state and further stabilize Cox1. This step appears to precede Shy1 association. Thus, we suggest that inactivation of individual Mss51 molecules for translation is not necessarily caused by a single physical sequestration step. Our data rather suggest that inactivation is a multistep process at the inner membrane that leads to a sequential inactivation of the Mss51 protein. The equilibrium between these states is regulated through specific factors at distinct stages of Cox1 biogenesis, and Coa3 and Cox14 are the initial factors in the process.
Materials and methods
Yeast strains and mitochondrial isolation
S. cerevisiae strains in this study are derivatives of YPH499 (Sikorski and Hieter, 1989), except for Mss51TAP and Cox14TAP (BY4741; Thermo Fisher Scientific; Mick et al., 2007) and cox1− and cox2− (Netter et al., 1982) strains. Deletions and tagging of COA3, COX14, and MSS51 were performed by introducing HISMX6 or TRP1 cassettes amplified via PCR with site-specific 5′ extensions for homologous recombination (Longtine et al., 1998; Knop et al., 1999). Yeast cells were grown at 30°C in rich medium containing 1% yeast extract and 2% peptone or on synthetic medium lacking histidine. As a carbon source, 3% glycerol, 2% galactose, or 2% sucrose for strains affected in respiration was used. For growth tests, yeast cells from liquid cultures were adjusted to an OD600 of 0.3, and serial 10-fold dilutions were spotted onto agar plates containing rich medium with glucose or glycerol as carbon source. Plates were incubated at the indicated temperatures for up to 72 h.
Pretreatment of yeast cells with translation inhibitors was performed as follows. Cells were grown at 30°C on rich medium containing 3% glycerol to mid–log phase and supplemented with 2 mg/ml chloramphenicol (Osman et al., 2007) or 150 µg/ml cycloheximide in ethanol as solvent. Yeast cells were then grown for an additional 3 h, before mitochondria were isolated.
Immunofluorescence assay
Yeast cells were grown in rich, nonfermentable medium at 30°C to mid–log phase. Mitochondria were stained by incubation of intact cells with 0.5 µg/ml MitoTracker orange CMTMRos (Invitrogen) for 30 min at 30°C, followed by extensive washing with medium. Cells were fixed by adding formaldehyde to a final concentration of 3.7%. Immunofluorescence microscopy was performed according to Girzalsky et al. (1999). Rabbit antisera against Coa3 were used at dilutions of 1:200. For detection, Alexa Fluor 488–conjugated goat anti–rabbit IgG (Invitrogen) was used in a dilution of 1:200. Images were prepared using a microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with a Plan-Fluar 100× NA 1.45 oil objective lens (Carl Zeiss, Inc.). Images were recorded with a monochrome camera (AxioCam MRm camera; Carl Zeiss, Inc.) and processed with Axiovision 4.5 software (Carl Zeiss, Inc.).
Protein localization analysis
Analysis was performed essentially as previously described (Mick et al., 2007). In brief, isolated mitochondria were sonicated using a Branson sonifier (model 450; 3 × 30 s; 40% duty cycle) in 10 mM MOPS, pH 7.2, and 500 mM NaCl or were extracted with 0.1 M carbonate, pH 10.8, or 1% Triton X-100 buffer containing 150 mM NaCl. Subsequently, samples were subjected to ultracentrifugation at 55,000 rpm, 4°C for 45 min, in a TLA-55 rotor (Beckman Coulter). Submitochondrial localization was assessed by proteinase K treatment of intact, swollen (10 mM MOPS, pH 7.2) or Triton X-100–lysed mitochondria. After TCA precipitation, samples were analyzed by SDS-PAGE and Western blotting.
Determination of enzymatic activities in mitochondria
Malate dehydrogenase activity was determined by following the oxaloacetate-dependent oxidation of NADH at 340 nm. The assay buffer contained 100 mM potassium phosphate, pH 7.5, 0.1 mM NADH, and 0.2 mM oxaloacetate. Cytochrome oxidase and NADH–cytochrome reductase activities were assessed spectrophotometrically by measuring absorbance at 550 nm in buffers containing 40 mM potassium phosphate, pH 7.5, and 0.02% cytochrome c (Sigma-Aldrich; dithionite reduced for oxidase activity). Reactions were started by the addition of Triton X-100–lysed mitochondria. For cytochrome reductase activity, nonlysed mitochondria were added, and buffer was supplemented with 0.5 mM NADH and 0.1 mM KCN (Tzagoloff et al., 1975). Concentrations of reduced/oxidized cytochrome c were determined using the extinction coefficient at 550 nm of 21.84 mM−1cm−1.
Import of radiolabeled precursor proteins, antibody shift, antibody depletion, and coimmunoprecipitation assays
In vitro import of radiolabeled precursor proteins was performed as described in Mick et al. (2007). In brief, in vitro translation was performed in reticulocyte lysate (Promega) using open reading frames cloned into pGEM-4Z under control of the SP6 promotor (Promega). Import and assembly of the radiolabeled preproteins into isolated mitochondria was performed essentially as described previously (Wiedemann et al., 2006). Therefore, isolated mitochondria were incubated with radiolabeled precursor proteins at 25°C in import buffer (3% BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM KH2PO4, 5 mM methionine, 10 mM MOPS-KOH, pH 7.2, 2 mM ATP, and 2 mM NADH) and subjected to a proteinase K treatment after the import reaction. For antibody shift or depletion experiments, mitochondria were lysed in BN-PAGE solubilization buffer containing 1% digitonin. For antibody shift experiments, samples were split in 50-µg mitochondria aliquots, 2 µg of antibodies against HA (Roche) or Flag (Sigma-Aldrich) epitopes was added, and samples were incubated for 30 min at 4°C with gentle agitation before separation by BN-PAGE. For depletion experiments, samples were split and incubated with antibodies coupled to protein A– or protein G–Sepharose (GE Healthcare) for up to 90 min. Unbound material was directly used for BN-PAGE analysis. Coimmunoprecipitation experiments were performed according to Hutu et al. (2008), with the exception that lysis buffers contained 60 mM NaCl.
Labeling of mitochondrial translation products (in vivo and in organello)
In vivo labeling of mitochondrial translation products was performed in whole cells grown on rich medium with 2% galactose as carbon source as described previously (Barrientos et al., 2002). In brief, 0.25 OD600 of cells were harvested, washed once with buffer A (40 mM KPi, pH 6.0, and 2% galactose), and resuspended in 500 µl of buffer A. After 10-min incubation at 30°C, cycloheximide was added to a final concentration of 150 µg/ml, and labeling reactions were started by the addition of 20 µCi [35S]methionine. After 5-min pulse, reactions were stopped by adding 10 mM unlabeled methionine and 100 µg/ml chloramphenicol. Samples were further incubated, and proteins were extracted by alkaline treatment and precipitated with 10% TCA. In isolated mitochondria, translation products were labeled for 20 min for coimmunoprecipitations in Mss51HA mitochondria and 30 min in Coa3HA mitochondria with [35S]methionine, essentially as described previously (Westermann et al., 2001).
Miscellaneous
Protein complex isolations via IgG chromatography and LC/tandem MS analyses were performed as previously published (Frazier et al., 2006; Mick et al., 2007). Standard techniques were used for SDS-PAGE and Western blotting to polyvinylidene fluoride membranes. Antibodies against the bc1 holocomplex were provided by B. Trumpower (Dartmouth Medical School, Hanover, NH). Detection of antibody–protein complexes was performed by enhanced chemiluminescence (GE Healthcare) or fluorescence detection using an image scanner (FLA-9000; Fujifilm). BN-PAGE protocols followed published procedures (Dekker et al., 1997). Mitochondria were isolated essentially as previously described (Meisinger et al., 2006).
Online supplemental material
Fig. S1 shows a schematic presentation of Cox14 and Coa3 and lower gel sections of Western blot panels displayed in Fig. 1 E. Fig. S2 shows the association of Coa3 with respiratory chain supercomplexes by BN-PAGE and affinity purification. Fig. S3 shows an analysis of the stability of newly synthesized Cox1 in double mutant cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007026/DC1.
Acknowledgments
We thank M. Balleininger, I. Perschil, and M. Wissel for expert technical assistance and Drs. T. Fox, A. Chacinska, M. van der Laan, C. Meisinger, N. Pfanner, and N. Wiedemann for helpful discussions. We are indebted to Dr. B. Trumpower for antibodies against the bc1 complex.
This work was supported by the Deutsche Forschungsgemeinschaft, the Ministerium für Innovation, Wissenschaft, Forschung und Technologie des Landes Nordrhein-Westfalen, the Bundesministerium für Bildung und Forschung, and the Niedersächsisches VW Vorab.
Footnotes
Abbreviations used in this paper:
- BN-PAGE
- blue native PAGE
- IMS
- intermembrane space
- LC
- liquid chromatography
- MS
- mass spectrometry
References
- Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. 1996. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 85:875–885 10.1016/S0092-8674(00)81271-4 [DOI] [PubMed] [Google Scholar]
- Barrientos A., Korr D., Tzagoloff A. 2002. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J. 21:43–52 10.1093/emboj/21.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Zambrano A., Tzagoloff A. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472–3482 10.1038/sj.emboj.7600358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Gouget K., Horn D., Soto I.C., Fontanesi F. 2009. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta. 1793:97–107 10.1016/j.bbamcr.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy N., Fiumera H.L., Dujardin G., Fox T.D. 2009. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta. 1793:60–70 10.1016/j.bbamcr.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh F.A., Hannappel A., Anderka O., Ludwig B. 2009. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284:25735–25741 10.1074/jbc.M109.040295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr H.S., Winge D.R. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36:309–316 10.1021/ar0200807 [DOI] [PubMed] [Google Scholar]
- Coenen M.J., van den Heuvel L.P., Nijtmans L.G., Morava E., Marquardt I., Girschick H.J., Trijbels F.J., Grivell L.A., Smeitink J.A. 1999. SURFEIT-1 gene analysis and two-dimensional blue native gel electrophoresis in cytochrome c oxidase deficiency. Biochem. Biophys. Res. Commun. 265:339–344 10.1006/bbrc.1999.1662 [DOI] [PubMed] [Google Scholar]
- Decoster E., Simon M., Hatat D., Faye G. 1990. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet. 224:111–118 10.1007/BF00259457 [DOI] [PubMed] [Google Scholar]
- Dekker P.J., Martin F., Maarse A.C., Bömer U., Müller H., Guiard B., Meijer M., Rassow J., Pfanner N. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408–5419 10.1093/emboj/16.17.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S., Schon E.A. 2003. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348:2656–2668 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- Fernández-Vizarra E., Tiranti V., Zeviani M. 2009. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 1793:200–211 10.1016/j.bbamcr.2008.05.028 [DOI] [PubMed] [Google Scholar]
- Fontanesi F., Soto I.C., Horn D., Barrientos A. 2006. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 291:C1129–C1147 10.1152/ajpcell.00233.2006 [DOI] [PubMed] [Google Scholar]
- Fontanesi F., Jin C., Tzagoloff A., Barrientos A. 2008. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 17:775–788 10.1093/hmg/ddm349 [DOI] [PubMed] [Google Scholar]
- Fox T.D. 1996. Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia. 52:1130–1135 10.1007/BF01952112 [DOI] [PubMed] [Google Scholar]
- Frazier A.E., Taylor R.D., Mick D.U., Warscheid B., Stoepel N., Meyer H.E., Ryan M.T., Guiard B., Rehling P. 2006. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 172:553–564 10.1083/jcb.200505060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girzalsky W., Rehling P., Stein K., Kipper J., Blank L., Kunau W.H., Erdmann R. 1999. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2–dependent protein import into peroxisomes. J. Cell Biol. 144:1151–1162 10.1083/jcb.144.6.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum D.M., Koerner T.J., Tzagoloff A. 1995. Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J. Biol. Chem. 270:15585–15590 10.1074/jbc.270.26.15585 [DOI] [PubMed] [Google Scholar]
- Guzélin E., Rep M., Grivell L.A. 1996. Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 381:42–46 10.1016/0014-5793(96)00074-9 [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Funes S. 2005. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene. 354:43–52 10.1016/j.gene.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Neupert W. 2003. Protein insertion into the inner membrane of mitochondria. IUBMB Life. 55:219–225 10.1080/1521654031000123349 [DOI] [PubMed] [Google Scholar]
- Horan S., Bourges I., Taanman J.W., Meunier B. 2005. Analysis of COX2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem. J. 390:703–708 10.1042/BJ20050598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler J.P., Ferguson-Miller S., Mills D.A. 2006. Energy transduction: proton transfer through the respiratory complexes. Annu. Rev. Biochem. 75:165–187 10.1146/annurev.biochem.75.062003.101730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutu D.P., Guiard B., Chacinska A., Becker D., Pfanner N., Rehling P., van der Laan M. 2008. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol. Biol. Cell. 19:2642–2649 10.1091/mbc.E07-12-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R.A. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22:6438–6447 10.1093/emboj/cdg624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O., Rödel G. 2005. Biogenesis of cytochrome c oxidase. Mitochondrion. 5:363–388 10.1016/j.mito.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Bestwick M., Meunier B., Watts T.C., Winge D.R. 2010. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 30:1004–1017 10.1128/MCB.00640-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15:963–972 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Manthey G.M., McEwen J.E. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14:4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashkevich G., Repetto B., Glerum D.M., Jin C., Tzagoloff A. 1997. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J. Biol. Chem. 272:14356–14364 10.1074/jbc.272.22.14356 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfanner N., Truscott K.N. 2006. Isolation of yeast mitochondria. Methods Mol. Biol. 313:33–39 [DOI] [PubMed] [Google Scholar]
- Merz S., Westermann B. 2009. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 10:R95 10.1186/gb-2009-10-9-r95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick D.U., Wagner K., van der Laan M., Frazier A.E., Perschil I., Pawlas M., Meyer H.E., Warscheid B., Rehling P. 2007. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347–4358 10.1038/sj.emboj.7601862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netter P., Carignani G., Jacq C., Groudinsky O., Clavilier L., Slonimski P.P. 1982. The cytochrome oxidase subunit I split gene in Saccharomyces cerevisiae: genetic and physical studies of the mtDNA segment encompassing the ‘cytochrome b-homologous’ intron. Mol. Gen. Genet. 188:51–59 10.1007/BF00332995 [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., Taanman J.W., Muijsers A.O., Speijer D., Van den Bogert C. 1998. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 254:389–394 10.1046/j.1432-1327.1998.2540389.x [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., Artal Sanz M., Bucko M., Farhoud M.H., Feenstra M., Hakkaart G.A., Zeviani M., Grivell L.A. 2001. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 498:46–51 10.1016/S0014-5793(01)02447-4 [DOI] [PubMed] [Google Scholar]
- Osman C., Wilmes C., Tatsuta T., Langer T. 2007. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol. Biol. Cell. 18:627–635 10.1091/mbc.E06-09-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X., Broadley S.A., Fox T.D. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22:5951–5961 10.1093/emboj/cdg566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X., Butler C.A., Shingu-Vazquez M., Fox T.D. 2009. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell. 20:4371–4380 10.1091/mbc.E09-06-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F., Bestwick M.L., Cobine P.A., Khalimonchuk O., Cricco J.A., Winge D.R. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26:4335–4346 10.1038/sj.emboj.7601861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Zahedi R.P., Pfanner N., Meisinger C., Sickmann A. 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5:1543–1554 10.1021/pr050477f [DOI] [PubMed] [Google Scholar]
- Saraste M. 1999. Oxidative phosphorylation at the fin de siècle. Science. 283:1488–1493 10.1126/science.283.5407.1488 [DOI] [PubMed] [Google Scholar]
- Shoubridge E.A. 2001. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106:46–52 10.1002/ajmg.1378 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Gray J., Mitchell L., Antholine W.E., Hosler J.P. 2005. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 280:17652–17656 10.1074/jbc.C500061200 [DOI] [PubMed] [Google Scholar]
- Stiburek L., Vesela K., Hansikova H., Pecina P., Tesarova M., Cerna L., Houstek J., Zeman J. 2005. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 392:625–632 10.1042/BJ20050807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart R.A. 2008. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40:411–417 10.1007/s10863-008-9168-4 [DOI] [PubMed] [Google Scholar]
- Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J.M. 2003. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22:6448–6457 10.1093/emboj/cdg623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.W., Turnbull D.M. 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6:389–402 10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V., Hoertnagel K., Carrozzo R., Galimberti C., Munaro M., Granatiero M., Zelante L., Gasparini P., Marzella R., Rocchi M., et al. 1998. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63:1609–1621 10.1086/302150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R.B. 1975. Assembly of the mitochondrial membrane system: isolation of nuclear and cytoplasmic mutants of Saccharomyces cerevisiae with specific defects in mitochondrial functions. J. Bacteriol. 122:826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Rissler M., Rehling P. 2006. Mitochondrial preprotein translocases as dynamic molecular machines. FEMS Yeast Res. 6:849–861 10.1111/j.1567-1364.2006.00134.x [DOI] [PubMed] [Google Scholar]
- Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E., Lochmüller H., Chevrette M., Kaufman B.A., Horvath R., Shoubridge E.A. 2009. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 41:833–837 10.1038/ng.390 [DOI] [PubMed] [Google Scholar]
- Westermann B., Herrmann J.M., Neupert W. 2001. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol. 65:429–438 10.1016/S0091-679X(01)65025-8 [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Pfanner N., Rehling P. 2006. Import of precursor proteins into isolated yeast mitochondria. Methods Mol. Biol. 313:373–383 [DOI] [PubMed] [Google Scholar]
- Williams S.L., Valnot I., Rustin P., Taanman J.W. 2004. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 279:7462–7469 10.1074/jbc.M309232200 [DOI] [PubMed] [Google Scholar]
- Zambrano A., Fontanesi F., Solans A., de Oliveira R.L., Fox T.D., Tzagoloff A., Barrientos A. 2007. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 18:523–535 10.1091/mbc.E06-09-0803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A.P., Newbold R.F., Wang J., Chevrette M., et al. 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20:337–343 10.1038/3804 [DOI] [PubMed] [Google Scholar]