Electron microscopy reveals that the flat base of the vesicular stomatitis virus is a privileged site for membrane fusion and that the glycoproteins located outside form regular arrays required at late stages of the fusion process.
Abstract
The entry of enveloped viruses into cells requires the fusion of viral and cellular membranes, driven by conformational changes in viral glycoproteins. Many studies have shown that fusion involves the cooperative action of a large number of these glycoproteins, but the underlying mechanisms are unknown. We used electron microscopy and tomography to study the low pH–induced fusion reaction catalyzed by vesicular stomatitis virus glycoprotein (G). Pre- and post-fusion crystal structures were observed on virions at high and low pH, respectively. Individual fusion events with liposomes were also visualized. Fusion appears to be driven by two successive structural rearrangements of G at different sites on the virion. Fusion is initiated at the flat base of the particle. Glycoproteins located outside the contact zone between virions and liposomes then reorganize into regular arrays. We suggest that the formation of these arrays, which have been shown to be an intrinsic property of the G ectodomain, induces membrane constraints, achieving the fusion reaction.
Introduction
Entry of enveloped viruses into host cells requires binding of the virus to one or more receptors present at the host cell surface. This binding is followed by fusion of the viral envelope with the cellular membrane. These steps are mediated by virally encoded glycoproteins, which promote both receptor recognition and membrane fusion. The membrane fusion process involves large structural rearrangements of the fusogenic glycoproteins after interaction with specific triggers (e.g., a low pH environment and/or cellular receptors). These conformational changes result in the exposure of hydrophobic motifs (so-called “fusion peptides” or “fusion loops”), which then interact with one or both of the participating membranes, resulting in their destabilization and fusion (Weissenhorn et al., 2007; Harrison, 2008). Conformational change triggered in the absence of a target membrane inactivates the fusion properties of the fusogenic glycoprotein.
Determinations of the atomic structure of the ectodomains of many viral fusion glycoproteins in their pre- and/or post-fusion states have revealed a large diversity of conformations (Skehel and Wiley, 2000; Gibbons et al., 2004; Kielian and Rey, 2006; Lamb and Jardetzky, 2007; Harrison, 2008; Roche et al., 2008; Backovic and Jardetzky, 2009), but experimental data suggest that the membrane fusion pathway is very similar for all the enveloped viruses studied to date, regardless of the organization of their fusion machinery (Chernomordik et al., 1998; Gaudin, 2000; Zaitseva et al., 2005). Fusion is generally thought to proceed via the formation of an intermediate stalk that forms a local lipid connection between the outer leaflets of the fusing membranes. Radial expansion of the stalk then induces the formation of a transient hemifusion diaphragm (i.e., a local bilayer composed of the two initial inner leaflets). The formation and enlargement of a pore within this structure results in complete fusion (Chernomordik et al., 1995; Chernomordik and Kozlov, 2005).
The stalk/pore model is largely supported by experimental results (Kemble et al., 1994; Chernomordik et al., 1998; Gaudin, 2000; Zaitseva et al., 2005). However, it remains unclear how fusion proteins catalyze the formation of these lipid intermediates during the fusion process. In particular, many studies have shown that fusion involves the cooperative action of a large number of viral proteins, interacting with and deforming the viral and target membranes (Blumenthal et al., 1996; Danieli et al., 1996; Roche and Gaudin, 2002; Leikina et al., 2004), but the underlying molecular mechanisms remain unknown.
Rhabdoviruses are enveloped viruses with a flat base and a round tip, resulting in a characteristic bullet shape (Nakai and Howatson, 1968; Brown et al., 1988; Barge et al., 1993). The two most frequently studied rhabdovirus genera are the lyssaviruses (prototype virus: rabies virus, RV) and the vesiculoviruses (prototype virus: vesicular stomatitis virus, VSV). These viruses fuse with the cell membrane after endocytosis of the viral particle and this process is triggered in the acidic environment of the vesicle (White et al., 1981; Matlin et al., 1982). Attachment and fusion are both mediated by a single viral glycoprotein, G, the only glycoprotein present in these viruses (Roche et al., 2008).
G has at least three conformational states (Clague et al., 1990; Gaudin et al., 1993): the native pre-fusion state, present at the surface of the virus at pH values above 7; the activated hydrophobic state, which interacts with the target membrane during the first step of the fusion process (Durrer et al., 1995); and the post-fusion conformation, which is structurally different from both the native and activated states (Clague et al., 1990; Gaudin et al., 1993). The different states of G are maintained in a pH-dependent equilibrium, which shifts toward the inactive state at low pH (Roche and Gaudin, 2002).
Two different structures of a thermolysin-generated VSV G ectodomain (Gth), probably corresponding to a high pH pre-fusion (Roche et al., 2007) and low pH post-fusion (Roche et al., 2006) state, have been determined by x-ray crystallography. Four distinct domains were identified in both these structures: a β-sheet–rich lateral domain, a central domain involved in trimerization, a pleckstrin homology domain (PH domain), and a fusion domain inserted into a loop of the PH domain. The fusion domain contains a membrane-interacting motif consisting of two hydrophobic loops located at the tip of an elongated three-stranded β-sheet. In striking contrast to other proteins involved in fusion, the fusion loops are not buried at an oligomeric interface in the pre-fusion conformation of G. Instead, they are exposed, pointing toward the viral membrane.
The transition from the pre- to the post-fusion structure of VSV G involves a major reorganization of the molecule (Roche et al., 2006, 2007, 2008). During this transition, the fusion domain is projected toward the target cell membrane through two structural changes: the reorganization of two hinge segments connecting the fusion domain to the PH domain, and lengthening of the central helix of the trimerization domain. Finally, the C-terminal domain associated with the transmembrane segment refolds into an α-helix that positions itself in the grooves of the trimeric core (composed of the central helices) in an antiparallel manner, to form a six-helix bundle. The post-fusion state has a typical “hairpin” structure (also found in the post-fusion states of other viral glycoproteins involved in fusion; Kielian and Rey, 2006), with the fusion loops in the vicinity of the TM domains.
In this study, we used electron microscopy (on both negatively stained and frozen samples) and tomography to analyze the structure of VSV G at the surface of the virus. We demonstrate that both crystalline structures can be observed at the surface of the virus, at different pH values. We also visualized individual fusion events between viral particles and liposomes. Fusion appears to occur preferentially at the flat base of the virion. Moreover, once the spikes on the cylindrical part of the particle have attained their post-fusion conformation, they tend to form regular arrays. At pH values below 5.5, extended helical networks of G were observed. Viral membrane tearing was observed when this helical network was formed in the absence of a target membrane. Finally, the formation of regular arrays is an intrinsic property of the G ectodomain: Gth incubated with liposomes at low pH also self-organizes to form similar networks at the surface of the bilayer.
Based on these observations, we suggest that the fusion of VSV with target membranes is driven by two successive structural rearrangements of G at two different sites in the virion. Glycoproteins located at the flat base of the virion initiate the fusion process by undergoing conformational changes, allowing them to interact with the target membrane. G molecules located outside the fusion site on the cylindrical part of the viral particle then self-organize, imposing membrane constraints that lead to the merging of the two membranes.
Results
Characterization of the fusion induced by VSV
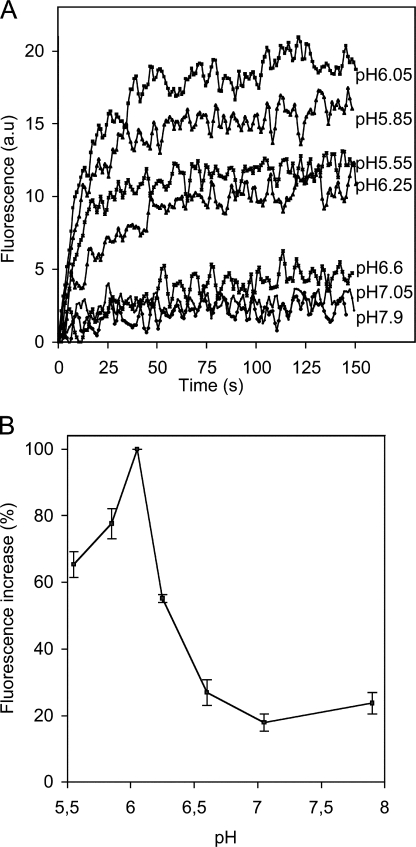
A resonance energy transfer method based on non-exchangeable fluorescent donor and acceptor probes was used to assay fusion. Fig. 1 A shows typical fusion kinetics for various pH values, at 20°C. Fusion was not observed above pH 6.5. Below this pH, fluorescence increased rapidly (t1/2 of ∼15 s) and the fusion reaction was complete within 50 s. Fusion peaked at about pH 6, decreasing thereafter with the pH (Fig. 1 B). Fusion levels at pH 5.5 were about half those at pH 6. These data are consistent with published data for VSV (Clague et al., 1990) and other rhabdoviruses (Gaudin et al., 1993, 1999). We investigated the structure of G on the surface of the virus in the absence of fusion (pH 7.5), in optimal fusion conditions (pH 6) and in conditions in which fusion is partially inhibited (pH 5.5).
Figure 1.
pH dependence of fusion between VSV and liposomes. (A) Kinetics of VSV fusion at various pH values, at 20°C, a.u. (arbitrary units). (B) pH dependence of the extent of fusion at 20°C, obtained from kinetic curves, such as those shown in A. The increase in fluorescence was maximal at pH 6.05, and this value was defined as 100%. The slight increase in fluorescence observed at pH values above 6.5 is due to virus–membrane interactions rather than membrane fusion. Error bars indicate the standard deviation (for three experiments).
Electron microscopy of VSV incubated at pH 7.5
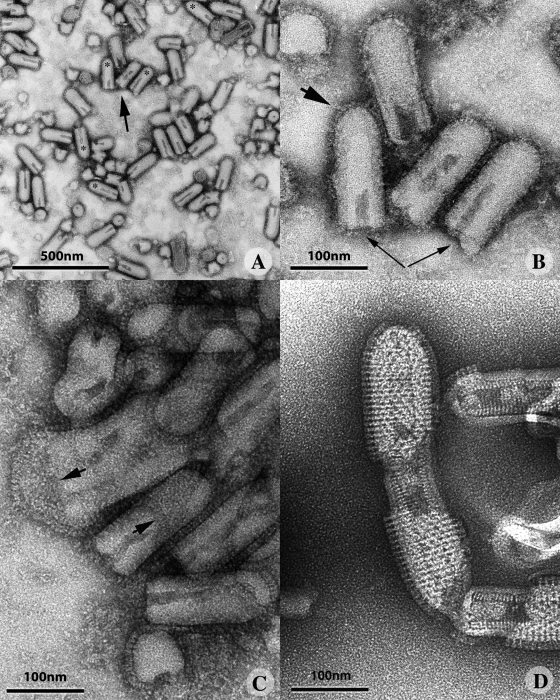
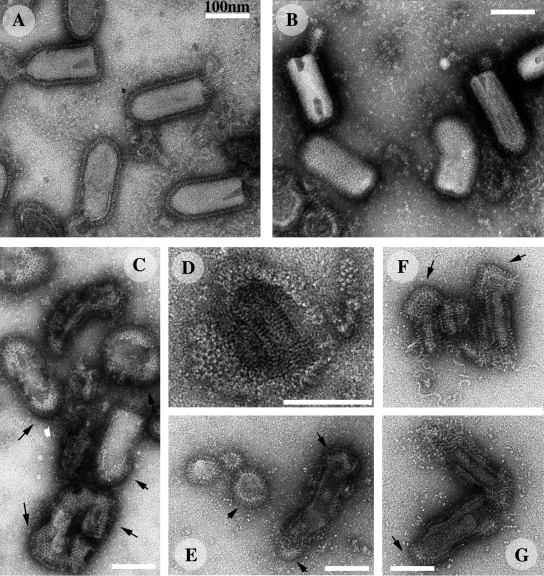
We initially used negative staining to analyze the structure of VSV G at the surface of native virions incubated at pH 7.5. Under these conditions, virions did not aggregate and were evenly distributed over the grid (Fig. 2 A). The stain did not penetrate the virion efficiently, so the RNP was not visible in most cases (Fig. 2 B). The viral particles had the characteristic bullet shape described in previous studies (Nakai and Howatson, 1968; Brown et al., 1988; Barge et al., 1993), with a flat base and a round tip. A rather continuous layer of spikes formed on the cylindrical part of the virion and on the round tip, making it difficult to distinguish between individual glycoprotein molecules (Fig. 2 B, large arrow). The width of this layer (∼8 nm) was consistent with the length of the proposed pre-fusion conformation determined by x-ray crystallography (Roche et al., 2007). The spikes exhibited a low density area close to the viral membrane, again consistent with the crystallographic model. In most cases, the domain connecting the spikes to the membrane could not be visualized by electron microscopy. Instead, the glycoprotein layer seemed to float around the virion. The oligomeric state of the glycoprotein at the surface of the virus could not be determined from electron micrographs. Spikes were often hardly visible (Fig. 2 A, asterisks; Fig. 2 B, left thin arrow) or not visible at all (Fig. 2 B, right thin arrow) on the flat base of the viral particle. Stain accumulated toward the base within many virions, indicating a non-uniform distribution of mass density along the central axis of the particle.
Figure 2.
Morphology of negatively stained VSV at pH 7.5, 6.0, and 5.5. (A) At pH 7.5, VSV formed a monodisperse suspension with virions that were bullet shaped when viewed from the side and circular when viewed from above. The asterisks indicate viruses with a base depleted of glycoproteins. (B) Higher magnification of the virions indicated by an arrow in A. Continuous layer of G over the surface of the virus (large arrow), with a lower density at the base of the particle (thin arrows). The right arrow indicates a virus with no spikes visible at its base. (C) VSV forms large aggregates at pH 6.0. G has a more elongated structure, making it possible to distinguish individual spikes, which are often closely packed at the apex of the viral particle (arrows). (D) At pH 5.5, the spikes form ordered helical arrays. Note that the viral particles fuse at their bases. The nucleocapsid is now clearly visible, indicating that the stain penetrated the viral particle.
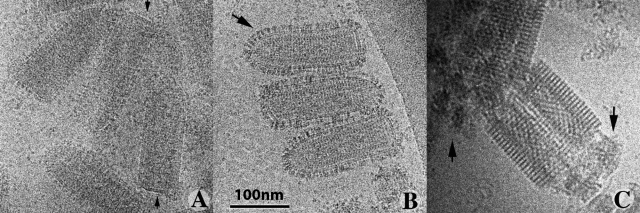
Preparation artifacts were minimized by also visualizing the viral particles in their native aqueous state, by cryo-EM. In these conditions we were able to observe the internal structure of the virions, the RNP, which displayed typical helical symmetry (Fig. 3 A). The overall organization of the spikes at the viral surface was very similar to that observed after negative staining. We also observed very few glycoproteins at the base of the virus, as after negative staining (Fig. 3 A, arrows).
Figure 3.
Morphology of VSV embedded in vitreous ice. (A) At pH 7.5, G forms a thin continuous layer around the viral particle. The arrows indicate viruses with no G visible at their base. (B) At pH 6, G elongates and individual spikes protruding from the membrane are visible. The arrow indicates an area in which the spikes are regularly organized. (C) At pH 5.5, all the spikes display a well-ordered helical organization. Note that the membrane at the base of the particle is disrupted, allowing the release of internal material (indicated by arrows). In A and B, the image is underfocused by ∼1.8 µm. In C, it is underfocused by 3 µm, to improve visualization of the helical G array.
VSV tomograms at pH 7.5
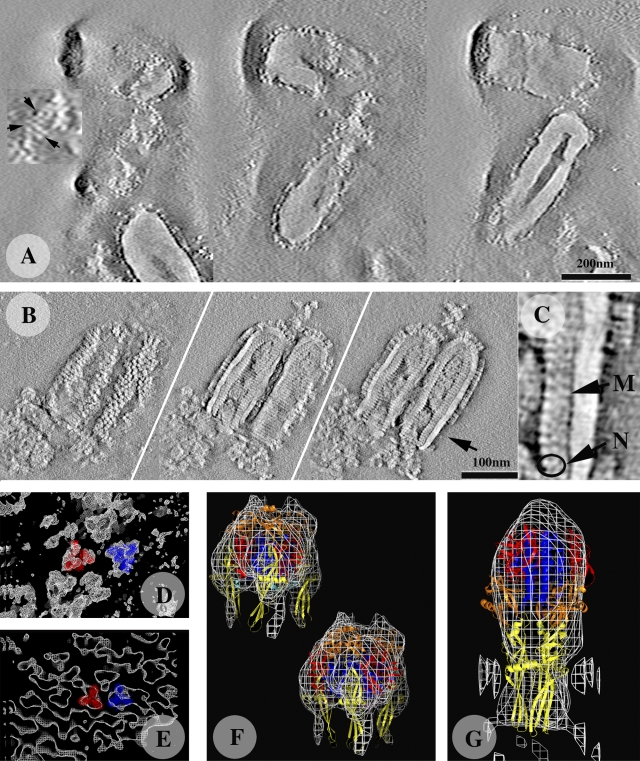
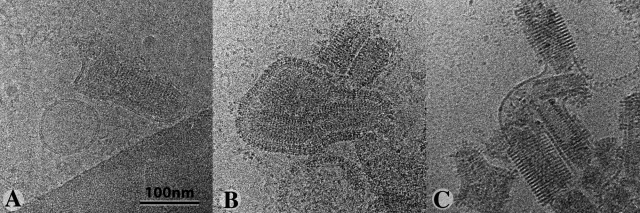
We performed electron tomography on negatively stained samples to analyze more precisely the organization of G at the surface of the virus. Image processing and tomogram construction (Fig. 4 A; Videos 1 and 2) generated a 3D reconstruction of several viral particles. VSV particles had a typical bullet shape, with a central cavity (Fig. 4 A, right frame). Nucleocapsids were not visible because the stain did not penetrate the viral particles. At the surface of the viral particle (Fig. 4 A, left frame) we observed triangular forms consisting of three dots. We also identified trimeric shapes in the calculated volume (Fig. 4 D). These volumes arose from negative staining but, as the typical tripod-like shape of the pre-fusion crystal structure including the “legs” corresponding to the fusion domains was clearly recognizable, we were able to fit G pre-fusion structures into these volumes (Fig. 4, D and F). These tripod-like structures were generally clustered (Fig. 4 A, left frame). A better fit was obtained if a monomer was inserted into the map imposing C3 symmetry (Fig. 4 F, right), rather than a crystallographic trimer (Fig. 4 F, left), suggesting that there may be some flexibility in the pre-fusion conformation of G. Finally, we observed far fewer tripods (a few tens) at the surface of the virus than expected, given the number of glycoproteins present (∼1,200 monomers, giving, potentially, 400 trimers; Thomas et al., 1985).
Figure 4.
Tomography of negatively stained VSV incubated at pH 7.5 and 5.5, and comparison of the structure of G with the corresponding x-ray crystallography model. (A and B) Three sections of the tomograms are shown (extracted from Videos 1 and 3); one at the level of the G layer (left), one at the level of the nucleocapsid (middle) and one passing through the center of the particle (right). The tilted series used to calculate the tomograms were recorded on negatively stained samples. (A) At pH 7.5, VSV is bullet shaped, with a central cavity. In some areas, the G layer contains trimeric entities (arrows in the enlargement). As the stain does not penetrate the viral particle, the nucleocapsids are not visible. (B) At pH 5.5, G shows trimeric structures that form quasi-helical arrays (left). The nucleocapsid is now visible (middle and right). In the center of the particle, a twisted material occupies the central cavity. (C) Enlargement of the central section (indicated by the arrow in the right frame in B) showing the organization of the particle beneath the membrane. The characteristic bilobed shape of N is visible and an additional domain is seen that may be attributed to M (Ge et al., 2010). (D and E) Volumes at the surface of the particle, extracted from the tomograms, revealing the presence of trimeric entities to which x-ray models of the pre-fusion (D) and post-fusion (E) structures can be manually fitted. In each case, two models (in blue and red) are displayed. (F and G) For a more quantitative fit, four trimers were isolated from the reconstructions and averaged. The x-ray models were fitted to the resulting averaged 3D reconstructions with UROX (Siebert and Navaza, unpublished program), a more user-friendly version of URO (Navaza et al., 2002). (F) For viruses incubated at pH 7.5, two fits are shown. For the fit displayed above and to the left, the crystallographic trimer, with its domains shown in color, was directly fitted in the electron microscopy reconstruction. The second fit (below and to the right) was performed with a monomer, imposing C3 symmetry. The better fit obtained with this model suggests that the trimer of G at the surface of the particle is slightly different from that in the crystalline structure. (G) At pH 5.5, the crystalline post-fusion model of G fits the tomographic model well.
Electron microscopy of VSV incubated at low pH
At low pH, massive aggregation of the virions was observed both after negative staining (Fig. 2 C) and cryo-EM (not depicted). This aggregation results from the exposure of hydrophobic regions (probably the fusion loops) at the top of the layer of spikes. We also observed some fusion between viral particles (Fig. 2, C and D). This massive aggregation and fusion made it very difficult to observe particle structure. Nevertheless, in rare cases, isolated virions (or small aggregates of a few virions) were obtained, making it possible to study particle shape (Fig. 2 D; Fig. 3, B and C).
These isolated virions no longer had the uniform layer described above: individual spikes were clearly visible, sticking out perpendicularly to the viral membrane. Their length (∼12 nm) was consistent with the post-fusion conformation determined by x-ray crystallography.
In their post-fusion conformation, the spikes tended to be organized into arrays (Fig. 2, C and D; Fig. 3, B and C), this organization being stronger at lower pH values. After the incubation of the virions at pH 5.5 for several minutes at room temperature, a regular helical network was clearly visible at the surface of the virion, extending over the entire particle in some cases (Fig. 2 D; Fig. 3 C). Once this helical network had formed, we often observed virions with a broken membrane at their base, resulting in the release of the RNP into the medium (Fig. 3 C). In the absence of this regular helical network, neither membrane tearing nor RNP release was observed, despite the post-fusion conformation of all the spikes (Fig. 3 B).
The fusion of two individual viral particles to each other always occurred at the flat bases of the particles (Fig. 2 D), consistent with the notion that this site behaves differently from the rest of the viral surface and is the preferential site for fusion.
The strong penetration of the stain into the viral particle confirmed the destabilization of the membrane at pH 5.5. In these conditions the inner structure of the virion could be visualized (Fig. 2 D).
VSV tomograms at pH 5.5
We then performed electron tomography on samples stained at pH 5.5 (Fig. 4 B; Video 3). We were able to identify volumes with an elongated shape at the surface of the virus, in which we could fit the crystalline post-fusion structure of the G ectodomain (Fig. 4, E and G).
Consistent with previous findings (Barge et al., 1993), staining revealed the presence of material within the tightly coiled helical RNP (Fig. 4 B). At some sites this material resembled uncoiled RNP in solution, whereas at others it resembled the structures formed by matrix protein (M) aggregated at low salt concentration (Gaudin et al., 1995). At the resolution of the tomogram, it is therefore difficult to draw firm conclusions concerning the nature of the central structure of the virion.
The tomograms also provided information about the organization of the viral proteins beneath the membrane. In membrane regions depleted of glycoprotein (Fig. 4 C, enlargement of a virion from the tomogram shown in Fig. 4 B) we observed another layer of protein, with punctate staining between the helical RNP—identified on the basis of the bilobed shape of the nucleoprotein (N) (Albertini et al., 2006; Green et al., 2006)— and the lipid bilayer. This additional protein probably corresponds to M protein (Fig. 4 C; Ge et al., 2010).
Fusion of VSV G with liposomes, as studied by electron microscopy
Having determined the structural characteristics of the virions, we investigated their interactions with liposomes at different pH values. At pH 7.5, virions were visualized as individual particles, by both negative staining and cryo-EM. In some cases, due to flattening and/or 2D projection artifacts, the viral particles seemed to interact with target liposomes (Fig. 5 A). However, in all cases, the viral and liposome membranes were clearly separate and no fusion event could be detected.
Figure 5.
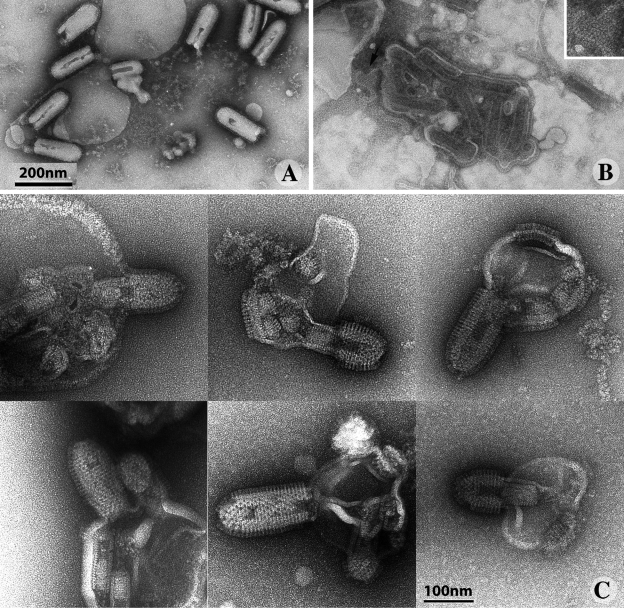
Visualization of individual fusion events between VSV and liposomes at low pH, after negative staining. (A) At pH 7.5, some viral particles interact with liposomes, but no fusion is detected. (B) At pH 6.0, viral particles and liposomes aggregate and numerous fusion events are visible. Fusion events are characterized by the presence of several nucleocapsids within a liposome, the membrane of which contains glycoproteins in the post-fusion conformation, often clustered into locally ordered arrays (arrow; magnified inset). (C) At pH 5.5 individual fusion events can be seen, demonstrating that fusion occurs at the base of the virion and that spikes located on the cylindrical part of the virus form ordered helical arrays.
At pH 6, as expected from the results of the fluorescence assay (Fig. 1), we observed massive fusion and aggregation on both negative staining and cryo-EM. This massive aggregation precluded the visualization of any individual fusion event (Fig. 5 B; Fig. 6 B).
Figure 6.
Visualization of individual fusion events between VSV and liposomes at low pH, in amorphous ice. (A) At pH 7.5, viral particles and liposomes do not fuse. (B) At pH 6.0, G shows the characteristic post-fusion conformation and numerous fusion events are observed. (C) At pH 5.5, individual fusion events can be seen. Fusion proceeds from the base of the virus, and G forms helical arrays on the cylindrical part of the viral particle.
At pH 5.5, consistent with the results of the fusion assay (Fig. 1), fusion was less extensive and we were able to detect individual fusion events between virions and liposomes, by both negative staining (Fig. 5 C) and cryo-EM (Fig. 6 C). The viral particle always associated with the target membrane via its flat base (21 fusion events were observed), confirming this area to be a preferential site for fusion. The helical network was also observed on the side of the virion. It seemed to impede or to delay the diffusion of glycoproteins into the liposome membrane. Indeed, even after the release of some of the RNP into the liposome, the glycoproteins often remained clustered around the virion, maintaining the shape of the viral particle.
Low pH–induced viral membrane deformation is associated with the formation of the G network
Our data suggest that viral membrane tearing resulted from the formation of a helical G network at the virion surface. We took advantage of the possibility to prevent the massive aggregation of the virions by incubating the virus at an intermediate pH (6.6) and 37°C for a few minutes before a second incubation at lower pH (Gaudin et al., 1993), to confirm this hypothesis.
Fusion was detected only at pH values below 6.5 (Fig. 1), but the incubation of virions for 15 min at 37°C and a pH as high as 6.6 resulted in most, if not all the G molecules adopting a post-fusion conformation (ectodomain length of 12 nm and trimeric head; Fig. 7 A). Thus, the structural transition of most of the virion spikes toward the post-fusion state is not sufficient to induce membrane fusion. These conditions also minimize aggregation, with the virions distributed evenly over the grid. An intact membrane and the typical bullet shape were retained by 84% of the virions (n = 531) in these conditions, comparable to 75% of the virions (n = 315) from the same preparation being intact when kept at pH 7.5.
Figure 7.
Gradual pH decrease minimizes virion aggregation, making it possible to observe individual VSV particles. (A) After incubation for 15 min at pH 6.6 and 37°C, virions are not aggregated and most of the spikes are in their post-fusion conformation (ectodomain length of 12 nm). (B) The viral preparation shown in A was subsequently incubated at pH 6.0 and 20°C for 20 min. All the spikes are in their post-fusion conformation and the virions have retained their initial shape. The arrow indicates a virion with its surface completely covered by spikes in their post-fusion conformation. (C–G) The viral preparation shown in A was subsequently incubated at pH 5.5 and 20°C for 20 min. Most of the particles are disrupted. Extensive lattices of G are visible at the surface of the virion (arrows in C, disrupted virion in D). In many cases (E–G) the particles are highly damaged, precluding the observation of large regular networks of G. Nevertheless, local regular arrays are still observed (arrows). In all frames, the bars indicate 100 nm.
After subsequent incubation at pH 6.0 and 20°C, EM observations were very similar to those after incubation at pH 6.6 (Fig. 7 B).
If the second incubation was performed at pH 5.5 and 20°C (conditions under which we previously observed the helical network), more than half of the virions were disrupted and no longer bullet shaped. Only 47% (n = 304) seemed to have retained their initial morphology. Despite the disruption of viral membranes (Fig. 7 C), many arrays of G remained visible. These arrays were less regular than those observed after direct incubation at pH 5.5.
Thus, the structural transition of all the virion spikes toward the post-fusion state is not sufficient to deform the membranes. Membrane deformation occurred only in the presence of regular arrays of glycoproteins.
Gth forms tubular structures when incubated at low pH in the presence of liposomes
In the helical G network observed at the viral surface at pH 5.5, some contacts between glycoprotein ectodomains were observed. However, the formation of this helical assembly may also be driven by interactions between the intraviral part of G and other proteins of the virus (such as the M or N proteins).
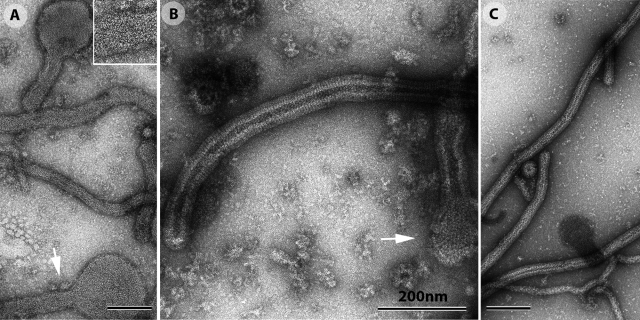
We investigated the interaction between Gth and liposomes by electron microscopy to exclude this possibility. No interaction between liposomes and proteins was observed at high pH, whereas, at low pH, Gth, in its post-fusion conformation, bound massively to the liposomes. The orientation of the molecule indicated that binding was mediated by the fusion loops located at the tip of the elongated trimer (Fig. 7).
At pH 6, the liposomes were entirely covered by Gth. Some liposomes retained their original spherical shape, but many adopted a tubular shape (Fig. 8 A). This shape change was associated with the formation of local, regular arrays of Gth (Fig. 8 A, inset).
Figure 8.
The incubation of Gth with liposomes at low pH induces the formation of tubular structures. (A) At pH 6, Gth, inserted into liposomes that were initially spherical, forms a local network (indicated by the arrow and enlarged in the top right frame), favoring the formation of tubular structures. (B) At pH 5.5, Gth forms more extensive, regular arrays at the surface of liposomes, resembling those formed by G at the surface of the virus. Spherical vesicles are nevertheless still visible (arrow). (C) At pH 5.2, only rigid tubular protein–lipid structures are observed, at the surface of which Gth displays quasi-helical symmetry. All the samples were negatively stained for electron microscopy observation.
Lowering the pH to 5.5 increased the number of tubular structures associated with the formation of an elongated, regular, helical Gth network (Fig. 8 B). At pH 5.2, most of the protein–lipid structures observed were elongated rigid tubes (Fig. 8 C), with a mean diameter of 68 nm (±8), which is smaller than the diameter of a virion (97 nm, ±3). Nevertheless, the overall pattern of interaction between Gth in such structures was very similar to that at the surface of virions at low pH. Thus, neither another viral protein nor the transmembrane domain of G is required for formation of the helical network. Thus, the G ectodomain is independently able to self-associate into a quasi-crystalline array, thereby deforming membranes.
Discussion
This electron microscopy study provides new insight into the molecular organization of VSV and the functioning of its fusion machinery.
Tomograms of negatively stained virions obtained at low pH confirmed the presence of material within the coiled nucleocapsid (Barge et al., 1993). However, due to the low resolution of the tomograms, we cannot draw firm conclusions about the nature of this material, which was not observed in the recent virus reconstruction by cryo-EM (Ge et al., 2010). However, this internal material did not display the helical symmetry imposed by Ge et al. (2010) in their reconstruction of the viral nucleocapsid.
Both crystal structures of Gth were observed at the surface of the virion. At pH values of 6.6 and below, all G spikes were in the elongated, post-fusion conformation. At pH 7.5, we observed only a few spikes with the tripod-like pre-fusion conformation. We cannot exclude the possibility that the oligomeric pre-fusion structure was disrupted during the preparation of the sample for observation by electron microscopy. However, the small number of G trimers observed at the surface of the virus suggests that at least one other conformation of G, different from both the structures determined by crystallography, is also present on the surface of the virion at pH values above 7. We suggest that this reflects the equilibrium between monomeric and trimeric forms of G. The existence of such an equilibrium has been demonstrated for detergent-solubilized G (Lyles et al., 1990). As previously suggested (Roche et al., 2008), the monomeric conformation of G may be an intermediate in the transition pathway.
Our data suggest that the flat base of the virion is the preferred site for fusion. This may be due to the particular curvature of the membrane at the base of the virion, a particular glycoprotein density and/or organization at this site, or both. Consistent with a role for glycoprotein density and organization, under pre-fusion conditions at pH 7.5, the flat base often seemed to be devoid of spikes, suggesting a lower abundance or a different structure and/or packing of spikes on this part of the virion. Indeed, a dense layer of protein (such as that on the cylindrical part and the round tip of the viral particle) would probably impede the formation of the initial lipid structures connecting the viral and target membranes.
Our findings demonstrate that the structural transition from the pre- to post-fusion conformation is not sufficient to drive viral fusion with a target membrane. Indeed, no fusion was detected at pH 6.6, despite most of the spikes present at the viral surface having undergone the conformational change. Indeed, lateral interactions (probably requiring a second protonation step) between G ectodomains in the post-fusion conformation were required for fusion. At pH 6, these lateral interactions, which are exclusively local (see Fig. 3 B, Fig. 5 B, and Fig. 8 A), did not deform the viral particle. However, at this pH the arrays formed by Gth induced the deformation of liposomes to form tubular structures.
At pH values of 5.5 or below, both G and Gth formed extensive helical networks on the surfaces of virions and liposomes, respectively. These networks disrupted the viral membrane and induced the formation of elongated rigid protein–lipid tubes, respectively.
Thus, the VSV fusion pathway seems to involve several distinct stages. The first stage is the association of the virion with the target membrane via its flat base. This is followed by local membrane deformations, leading to the formation of one or several stalks, and the formation of one or several initial fusion pores. Interactions between glycoproteins, in their low pH conformation, outside the contact zone, then lead to the enlargement of these initial pores, completing the fusion of the membranes. Pore enlargement is probably driven by the membrane tension induced by local reorganization of the glycoprotein network on the lateral side of the virion. This step did not require complete helical network formation: as pore enlargement requires less energy than membrane disruption, the formation of local arrays is probably sufficient. Indeed, premature formation of the helical network leads to membrane disruption, RNP release, and viral inactivation, accounting for the lower fusion efficiency at lower pH values.
A role for network formation at a late stage of the fusion process is consistent with published data for rabies virus showing that, at low temperature and at pH 6.4, fusion is arrested, probably at the stage of initial pore formation (Gaudin, 2000). In these conditions, the structural transition toward the post-fusion state is known to be blocked in rabies virus (Roche and Gaudin, 2002), precluding formation of the network outside the contact zone and preventing membrane fusion.
These observations are also consistent with the model of Kozlov and Chernomordik (2002), according to which, fusion proteins outside the contact zone generate the driving force for fusion by forming a coat around the fusion site. It remains unclear whether this mechanism can be generalized to other enveloped viruses. Nevertheless, it has been suggested that influenza hemagglutinins outside the contact zone are involved in late stages of the fusion process (Leikina et al., 2004). Furthermore, for paramyxovirus PIV5, micrographs indicate that F proteins in their post-fusion conformation tend to cluster at the surface of the virus (Ludwig et al., 2008). Finally, the spikes of class II fusion glycoproteins, in their post-fusion conformation, self-associate to form networks of various degrees of regularity, different from the icosahedral organization observed before fusion (Gibbons et al., 2003, 2004; Stiasny et al., 2004; Sánchez-San Martín et al., 2008). It is thus tempting to speculate that similar mechanisms are used by different fusion machineries.
Materials and methods
Chemicals
2-(4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine (β-BODIPY 500/510 C12-HPC) and 2-(4,4-difluoro-5,7-diphenyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-&-hexadecanoyl-sn-glycero-3-phosphocholine (β-BODIPY 530/550 C5-HPC) were purchased from Invitrogen. Phosphatidylcholine (PC; type XVI-E from egg yolk), phosphatidylethanolamine (type III from egg yolk), and gangliosides (type III from bovine brain) were supplied by Sigma-Aldrich.
Virus and cells
Wild-type VSV (Mudd-Summer strain, Indiana serotype) was propagated in BSR cells, a clone of BHK 21. Cells were grown in Eagle’s minimal essential medium supplemented with 10% calf serum. Virus particles were purified from the culture supernatant 24 h after infection. Cell debris was eliminated by a centrifugation for 30 min at 3,500 rpm in a rotor (model JA14; Beckman Coulter) at 4°C. The virus was then recovered in as a pellet by centrifugation for 3 h at 14,000 rpm in the JA14 rotor at 4°C and resuspended in TD buffer (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 25 mM Tris-HCl, pH 7.5). For the viral preparations used in fusion assays, the virus was purified by an additional centrifugation for 50 min, through 30% glycerol in 10 mM Tris HCl, pH 7.5, 50 mM NaCl, and 1 mM EDTA, at 25,000 rpm, in a rotor (model SW 28; Beckman Coulter) at 4°C. The pellet was resuspended in TD buffer and the preparation was used within 2 d.
Preparation of liposomes
We mixed 700 µg of PC, 300 µg of PE, and 100 µg of gangliosides dissolved in organic solvents, and dried the mixture under vacuum. For fusion experiments, 2.5 µg of BODIPY 500/510 C12-HPC (as the fluorescence donor) and 5 µg of BODIPY 530/550 C5-HPC (as the fluorescence acceptor) were added to the mixture before drying. The lipid film was resuspended in 1 ml of buffer (150 mM NaCl and 5 mM Tris-HCl, pH 8) and the mixture was sonicated in a water bath for 20 min.
Fusion assay
Fusion was assayed as described previously (Malinin et al., 2001; Roche and Gaudin, 2004). In brief, we mixed 10 µl of fluorescent liposomes with 980 µl of phosphate-citrate buffer at the required pH (prepared from 100 mM citric acid and 200 mM dibasic sodium phosphate solution) in the cuvette of a thermostatically controlled spectrofluorimeter (model LS50B; PerkinElmer). We then added 10 µl of virus (∼50 µg of viral protein) and continually monitored the increase in BODIPY 500/510 C12-HPC fluorescence. We used an excitation wavelength of 485 nm (slit width, 5 nm) and an emission wavelength of 518 nm (slit width, 5 nm). The mixture was stirred continually during the experiment.
Gth purification
We incubated 1 ml of VSV (5 mg/ml viral proteins in TD buffer) for 30 min at 37°C with 250 µl of phosphate-citrate buffer at pH 6.30 (prepared from a 0.1 M solution of citric acid and 0.2 M dibasic sodium phosphate stock solutions). G was then cleaved by adding 150 µl of 1 mg/ml thermolysin in water and incubating for 1 h at 37°C. The reaction was stopped by adding 150 µl of 25 mM EDTA, 0.9 M Tris-HCl, pH 8.7, and a protease inhibitor cocktail and incubating for 30 min at 37°C. The “shaved” viral particles were pelleted by centrifugation at 13,000 g for 1 h. The supernatant (S1) was retained and the pellet was resuspended in 100 µl of 50 mM Tris-HCl, pH 8.7. The resulting suspension was centrifuged for 30 min at 13,000 g and the supernatant was mixed with S1. The pooled supernatants were overlaid on a 20% sucrose cushion in 5 mM Tris-HCl, pH 8.7, and centrifuged in a rotor (model SW55; Beckman Coulter) for 1 h at 40,000 rpm (∼100,000 g). The supernatants (∼2 ml) were diluted in 20 ml of 5 mM Tris-HCl, pH 8.7, and loaded onto a DEAE-Trisacryl column (equilibrated in 5 mM Tris-HCl buffer, pH 8.7). Gth was eluted in 200 mM NaCl and 5 mM Tris-HCl, pH 8.7.
Electron microscopy
Sample preparation.
Purified virions were diluted in 100 mM NaCl in either 50 mM Tris-HCl, pH 7.5, or 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 6.0 or 5.5. A gradual decrease in pH was achieved by incubating virions for 15 min in 100 mM NaCl, 50 mM MOPS, pH 6.6, at 37°C and then dialyzing for 20 min, at room temperature, against a buffer of the appropriate pH (6 or 5.5).
Negative staining.
Samples were adsorbed onto airglow discharge carbon-coated grids and stained with sodium phospho-tungstic acid adjusted to the sample pH (7.5, 6.0, and 5.5). For samples maintained at pH 5.5, no major difference was observed between the results obtained with phospho-tungstic acid (pH 5.5) and uranyl acetate (UA; pH 4.8) staining. UA was therefore used in these conditions, even for the recording of the tomograms. Images were recorded in an electron microscope (model CM12; Philips) operated at 80 kV, with a nominal magnification of 35,000.
Electron cryomicroscopy.
We dispensed 3 µl of the studied suspension on a holey carbon grid. The drop was blotted and the grid was plunged into liquid nitrogen–cooled ethane and transferred to a liquid nitrogen–cooled holder (model 626; Gatan). Samples were observed in an electron microscope (model CM12; Philips) operating at 80 kV. Images were recorded on image plates (SO163; Kodak) developed for 12 min in full-strength Kodak D19. Data were recorded under minimal dose conditions, at a magnification of 35,000, with an underfocus of 1.5–3 µm.
Tomographic data collection.
Tomograms were recorded with a transmission electron microscope (Technai 200; FEI) equipped with a field emission gun operating at 200 kV. Images were recorded with a 2048 × 2048-pixel CCD camera (Gatan) at a magnification of 29,000 (5.2 nm-pixel), with a defocus of 1.5–3 µm, corresponding to first zeros at (2.0 nm)−1 − (3.0 nm)−1. Data were recorded with the microscope software, under low-dose conditions.
Image processing
Projections in a tilt series were mutually aligned and tomograms were calculated by the weighted back-projection method, using the FEI Inspect3D package, according to the protocol outlined in the manual. 3D reconstructions were averaged with Imagic (van Heel et al., 1996). The atomic models of G were visually fitted to our reconstructions with Pymol (DeLano Scientific LLC). Quantitative fits were performed in reciprocal space, with UROX software (Siebert and Navaza, unpublished program), a more user-friendly version of URO (Navaza et al., 2002). The quality of the fit was evaluated by calculating correlation coefficients and R factors. Two fits were tested for each tomogram: one with the crystallographic trimer without the imposition of symmetry and the other with the crystallographic protomer and the imposition of C3 symmetry. For data with a resolution of up to 1/(40Å), the correlation coefficients and R factors were equal to (0.88 and 0.38), (0.94 and 0.37), (0.79 and 0.44), and (0.83 and 0.43) for the post-fusion conformation without symmetry, the post-fusion conformation with imposed C3 symmetry, the pre-fusion conformation without symmetry, and the pre-fusion conformation with imposed C3 symmetry, respectively. The mean root standard deviations between the models obtained with the trimer and the monomer, with the imposition of C3 symmetry, were 15 and 22Å, for the post- and pre-fusion structures, respectively.
Online supplemental material
Videos 1 and 2 are 3D reconstructions of electron tomograms (negative stain) of VSV at pH 7.5. Sections of the tomogram presented in Video 1 are used in Fig. 4 A. Video 3 is a tomogram of VSV at pH 5.5. Sections of this tomogram are used in Fig. 4 B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006116/DC1.
Acknowledgments
We thank Drs. M. Kozlov and B. Delmas for careful reading of the manuscript.
This project was supported by the CNRS, INRA, and grants from the ANR, including postdoctoral fellowships to S. Libersou (ANR-06-BLAN-0103-01) and A. Albertini (ANR-08-BLAN-0256), and from Région Ile-de-France (SESAME 2006, #1-07-579/R).
Footnotes
Abbreviations used in this paper:
- G
- glycoprotein
- Gth
- thermolysin-generated VSV G ectodomain
- VSV
- vesicular stomatitis virus
References
- Albertini A.A., Wernimont A.K., Muziol T., Ravelli R.B., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 313:360–363 10.1126/science.1125280 [DOI] [PubMed] [Google Scholar]
- Backovic M., Jardetzky T.S. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189–196 10.1016/j.sbi.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barge A., Gaudin Y., Coulon P., Ruigrok R.W. 1993. Vesicular stomatitis virus M protein may be inside the ribonucleocapsid coil. J. Virol. 67:7246–7253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R., Sarkar D.P., Durell S., Howard D.E., Morris S.J. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63–71 10.1083/jcb.135.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.C., Newcomb W.W., Lawrenz-Smith S. 1988. pH-dependent accumulation of the vesicular stomatitis virus glycoprotein at the ends of intact virions. Virology. 167:625–629 [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. 2005. Membrane hemifusion: crossing a chasm in two leaps. Cell. 123:375–382 10.1016/j.cell.2005.10.015 [DOI] [PubMed] [Google Scholar]
- Chernomordik L., Kozlov M.M., Zimmerberg J. 1995. Lipids in biological membrane fusion. J. Membr. Biol. 146:1–14 [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Frolov V.A., Leikina E., Bronk P., Zimmerberg J. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369–1382 10.1083/jcb.140.6.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M.J., Schoch C., Zech L., Blumenthal R. 1990. Gating kinetics of pH-activated membrane fusion of vesicular stomatitis virus with cells: stopped-flow measurements by dequenching of octadecylrhodamine fluorescence. Biochemistry. 29:1303–1308 10.1021/bi00457a028 [DOI] [PubMed] [Google Scholar]
- Danieli T., Pelletier S.L., Henis Y.I., White J.M. 1996. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 133:559–569 10.1083/jcb.133.3.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrer P., Gaudin Y., Ruigrok R.W., Graf R., Brunner J. 1995. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 270:17575–17581 10.1074/jbc.270.29.17575 [DOI] [PubMed] [Google Scholar]
- Gaudin Y. 2000. Rabies virus-induced membrane fusion pathway. J. Cell Biol. 150:601–612 10.1083/jcb.150.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y., Ruigrok R.W., Knossow M., Flamand A. 1993. Low-pH conformational changes of rabies virus glycoprotein and their role in membrane fusion. J. Virol. 67:1365–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y., Barge A., Ebel C., Ruigrok R.W. 1995. Aggregation of VSV M protein is reversible and mediated by nucleation sites: implications for viral assembly. Virology. 206:28–37 10.1016/S0042-6822(95)80016-6 [DOI] [PubMed] [Google Scholar]
- Gaudin Y., de Kinkelin P., Benmansour A. 1999. Mutations in the glycoprotein of viral haemorrhagic septicaemia virus that affect virulence for fish and the pH threshold for membrane fusion. J. Gen. Virol. 80:1221–1229 [DOI] [PubMed] [Google Scholar]
- Ge P., Tsao J., Schein S., Green T.J., Luo M., Zhou Z.H. 2010. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 327:689–693 10.1126/science.1181766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons D.L., Erk I., Reilly B., Navaza J., Kielian M., Rey F.A., Lepault J. 2003. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell. 114:573–583 10.1016/S0092-8674(03)00683-4 [DOI] [PubMed] [Google Scholar]
- Gibbons D.L., Vaney M.C., Roussel A., Vigouroux A., Reilly B., Lepault J., Kielian M., Rey F.A. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 427:320–325 10.1038/nature02239 [DOI] [PubMed] [Google Scholar]
- Green T.J., Zhang X., Wertz G.W., Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 313:357–360 10.1126/science.1126953 [DOI] [PubMed] [Google Scholar]
- Harrison S.C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 10.1038/nsmb.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble G.W., Danieli T., White J.M. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 76:383–391 10.1016/0092-8674(94)90344-1 [DOI] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76 10.1038/nrmicro1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M.M., Chernomordik L.V. 2002. The protein coat in membrane fusion: lessons from fission. Traffic. 3:256–267 10.1034/j.1600-0854.2002.030403.x [DOI] [PubMed] [Google Scholar]
- Lamb R.A., Jardetzky T.S. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436 10.1016/j.sbi.2007.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E., Mittal A., Cho M.S., Melikov K., Kozlov M.M., Chernomordik L.V. 2004. Influenza hemagglutinins outside of the contact zone are necessary for fusion pore expansion. J. Biol. Chem. 279:26526–26532 10.1074/jbc.M401883200 [DOI] [PubMed] [Google Scholar]
- Ludwig K., Schade B., Böttcher C., Korte T., Ohlwein N., Baljinnyam B., Veit M., Herrmann A. 2008. Electron cryomicroscopy reveals different F1+F2 protein States in intact parainfluenza virions. J. Virol. 82:3775–3781 10.1128/JVI.02154-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles D.S., Varela V.A., Parce J.W. 1990. Dynamic nature of the quaternary structure of the vesicular stomatitis virus envelope glycoprotein. Biochemistry. 29:2442–2449 10.1021/bi00462a002 [DOI] [PubMed] [Google Scholar]
- Malinin V.S., Haque M.E., Lentz B.R. 2001. The rate of lipid transfer during fusion depends on the structure of fluorescent lipid probes: a new chain-labeled lipid transfer probe pair. Biochemistry. 40:8292–8299 10.1021/bi010570r [DOI] [PubMed] [Google Scholar]
- Matlin K.S., Reggio H., Helenius A., Simons K. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 156:609–631 10.1016/0022-2836(82)90269-8 [DOI] [PubMed] [Google Scholar]
- Nakai T., Howatson A.F. 1968. The fine structure of vesicular stomatitis virus. Virology. 35:268–281 10.1016/0042-6822(68)90267-5 [DOI] [PubMed] [Google Scholar]
- Navaza J., Lepault J., Rey F.A., Alvarez-Rúa C., Borge J. 2002. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D Biol. Crystallogr. 58:1820–1825 10.1107/S0907444902013707 [DOI] [PubMed] [Google Scholar]
- Roche S., Gaudin Y. 2002. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 297:128–135 10.1006/viro.2002.1429 [DOI] [PubMed] [Google Scholar]
- Roche S., Gaudin Y. 2004. Evidence that rabies virus forms different kinds of fusion machines with different pH thresholds for fusion. J. Virol. 78:8746–8752 10.1128/JVI.78.16.8746-8752.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Bressanelli S., Rey F.A., Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 313:187–191 10.1126/science.1127683 [DOI] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 315:843–848 10.1126/science.1135710 [DOI] [PubMed] [Google Scholar]
- Roche S., Albertini A.A., Lepault J., Bressanelli S., Gaudin Y. 2008. Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell. Mol. Life Sci. 65:1716–1728 10.1007/s00018-008-7534-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-San Martín C., Sosa H., Kielian M. 2008. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe. 4:600–608 10.1016/j.chom.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- Stiasny K., Bressanelli S., Lepault J., Rey F.A., Heinz F.X. 2004. Characterization of a membrane-associated trimeric low-pH-induced Form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J. Virol. 78:3178–3183 10.1128/JVI.78.6.3178-3183.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Newcomb W.W., Brown J.C., Wall J.S., Hainfeld J.F., Trus B.L., Steven A.C. 1985. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J. Virol. 54:598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Harauz G., Orlova E.V., Schmidt R., Schatz M. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17–24 10.1006/jsbi.1996.0004 [DOI] [PubMed] [Google Scholar]
- Weissenhorn W., Hinz A., Gaudin Y. 2007. Virus membrane fusion. FEBS Lett. 581:2150–2155 10.1016/j.febslet.2007.01.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Matlin K., Helenius A. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674–679 10.1083/jcb.89.3.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva E., Mittal A., Griffin D.E., Chernomordik L.V. 2005. Class II fusion protein of alphaviruses drives membrane fusion through the same pathway as class I proteins. J. Cell Biol. 169:167–177 10.1083/jcb.200412059 [DOI] [PMC free article] [PubMed] [Google Scholar]