Abstract
Revealing the structure and aggregation mechanism of amyloid fibrils is essential for the treatment of over 20 diseases related to protein misfolding. Coherent two dimensional (2D) infrared spectroscopy is a novel tool that provides a wealth of new insight into the structure and dynamics of biomolecular systems. Recently developed ultrafast laser sources are extending multidimensional spectroscopy into the ultraviolet (UV) region, and this opens up new opportunities for probing fibrils. In a simulation study, we show that 2DUV spectra of the backbone of a 32-residue β-amyloid (Aβ9–40) fibril associated with Alzheimer’s disease, and two intermediate prefibrillar structures carry characteristic signatures of fibril size and geometry that could be used to monitor its formation kinetics. The dependence of these signals on the fibril size and geometry is explored. We demonstrate that the dominant features of the β-amyloid fibril spectra are determined by intramolecular interactions within a single Aβ9–40, while intermolecular interactions at the “external interface” have clear signatures in the fine details of these signals.
Introduction
The formation and deposition of amyloid fibrils of misfolded proteins is associated with more than 20 neurodegenerative diseases. 1–3 Oligomeric or other prefibrillar precursors of amyloid fibrils are believed to be the main toxic species4–6 in amyloid-related diseases. Characterization of fibril structure is essential for understanding the mechanism of cell and tissue damage in amyloid-related diseases. Knowledge of the structure, kinetics and toxicity of amyloidogenic polypeptides is important for the development of effective treatments of amyloid disorders and for developing therapeutic agents against the associated human diseases. Amyloid fibrils are formed by a wide variety of peptides and proteins, and can be distinguished from other types of protein fibrils by the appearance in electron microscope (EM) images, by their dye-binding properties, and by the presence of cross-β structural motifs within the fibrils. Due to the lack of suitable probes with atomic resolution, these aggregates are not well characterized structurally and their kinetic pathways and toxicity are under active study.7–12 Numerous possible structures have been proposed and identified both experimentally13,14 and in theoretical studies.10,12 In the case of Alzheimer’s disease, fibrils are composed of β-amyloid (Aβ) peptides ranging from 39 to 42 residues, rich in β-sheet secondary structure. In most reports, 15,16 Aβ fibrils are characterized by cross-β structural motifs, indicating that they are composed of β-strands segments nearly perpendicular to the long axes of the fibrils. Tycko and co-workers have built a detailed molecular model of a 32-residue β-amyloid (Aβ9–40),13 based on solid-state NMR data, which is in agreement with atomic force microscopy and electron microscopy.
The response of proteins to sequences of femtosecond infrared (IR) pulses provides a multidimensional view into their equilibrium distribution of structures and instantaneous snapshots of fast-triggered dynamical events. Coherent multidimensional optical spectroscopic techniques use femtosecond laser pulses to excite the vibrational or electronic degrees of freedom in biomolecules and watch for correlated events taking place during two (or more) controlled time intervals. The resulting correlation data can be interpreted in terms of multipoint correlation functions that carry detailed information on structure, function, and dynamical events. 17–21 Two dimensional IR spectroscopy (2DIR) techniques provide a novel tool for studying fibril structure and aggregation mechanisms and promise to furnish much complementary structural information. 20,22–25 In joint experimental and theoretical studies, 26,27 we have investigated the sensitivity of 2DIR spectroscopy to secondary structural motifs.
New intense and stable femtosecond lasers with high repetition rates should allow femtosecond multidimensional spectroscopy to be extended into the ultraviolet (UV). Two dimensional UV (2DUV) spectroscopy offers a different observation window, with advantages over 2DIR in terms of reaching shorter pulse durations and higher quality polarization control. 28 It can be used to probe amyloid fibril structures and dynamical fluctuations as well as provide the prospect of triggering photochemical and photophysical processes, which are crucial to understand and manipulate their functionality. Multidimensional UV experiments have been reported on single bases. 29 Experimental determination of secondary structure in amyloid fibrils will require the identification of structurally ordered and disordered segments of β-strand and other segments (i.e., sheets or turns).13,30 Optimized optical pulse sequences could help distinguish the various protein structures and monitor the mechanism of formation. UV absorption and circular dichroism spectra of fibrils show signatures of distinct structural motifs. 31,32 Multidimensional UV will make it possible to study backbone and aromatic side chain (tyrosine and tryptophan) dynamics and directly monitor the formation of fibrils on membranes and how this damages the bilayer’s integrity. 31,32
In this paper we report simulations of 2DUV Spectra of the 32-residues β-amyloid (Aβ9–40) fibril and two proposed intermediate prefibrilar structures. Simulations are carried out using a QM/MM (Quantum Mechanics/Molecular Mechanics) approach we recently developed. 33 The approach has been successfuly applied in simulating multidimensional UV spectra of protein backbone transitions.28,33 The UV signals were obtained using classical MD (molecular dynamics) simulations combined with effective fluctuating QM Hamiltonians. With the same approach, we first present traditional 1DUV signals: CD (circular dichroism) and LA (linear absorption), and then turn to 2DUV signals. Spectroscopic signatures of different structural elements in Aβ9–40 amyloid fibrils are assigned using a transition charge population analysis.
Simulation Protocol
MD simulations were performed using the software package NAMD2.7 34 with the CHARMM2735 force field and the TIP3P water model.36 Cubic periodic boundary conditions and a 1 fs time step were used. Long-range electrostatic interactions were computed using the particle-mesh Ewald (PME)37 approach and a real space cutoff of 12 Å was used for nonbonded interactions. Langevin dynamics with a 1 ps damping coefficient were used to achieve a constant temperature. We first ran a 200 ps simulation of the amyloid fibrils in vacuum to obtain an initial periodic structure. During this simulation only the box size along the Z axis (fibril axis, see Fig. Figure 1 (A)) was allowed to vary. The amyloid fibrils were then inserted in a TIP3P water box36 containing 9236 water molecules, which were not allowed to penetrate inside the fibrils. 36 chloride ions were added to neutralize the box. Using a Nosé-Hoover Langevin piston 38 with a decay period of 200 fs and a 100 fs damping time, the system was equilibrated at 1 atm constant pressure for 500 ps, resulting in a box size of approximately 104×94×33 Å3. After equilibration, we performed 5 ns NVT simulations. Ensembles of MD geometric snapshots were recorded and extracted for the UV studies.
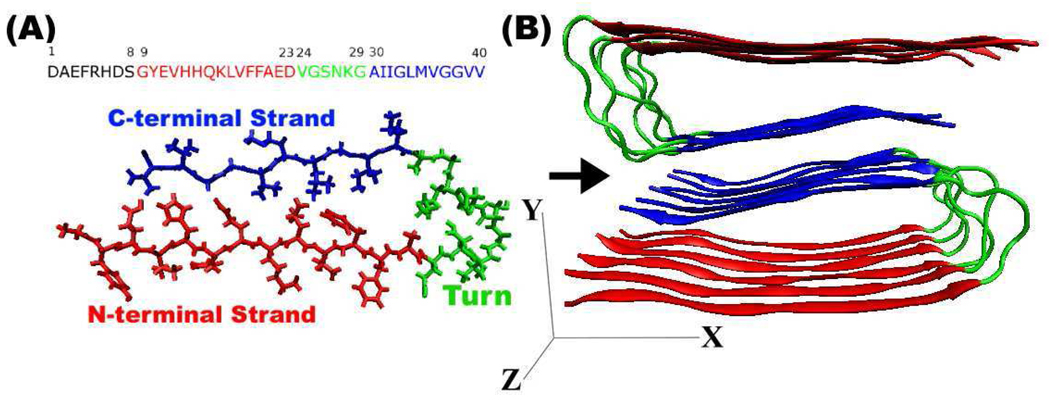
Figure 1.
(A) Aβ1–40 sequence and the structure of a single Aβ9–40 peptide, with asymmetric “U” structure. N-terminal strand (red), C-terminal strand (blue), and turn region (green), respectively. (B) Model structure of amyloid fibril made of 12 Aβ9–40 molecules. Red and blue indicate two parallel β-sheets.
QM calculations were carried out to simulate the electronic transition process. The present work focuses on the protein backbone amide n → π* (~220 nm) and π → π* (~190 nm) electronic transitions. Parameters for the transition energies of isolated peptide units, the resonant couplings, electric and magnetic dipole moments are those used in the DichroCalc package, 39 and they come from calculations on the excited states of N-methylacetamide (NMA) computed by CASSCF/SCRF (the complete-active space self-consistent-field method implemented within a self-consistent reaction field in MOLCAS.40 The electronic structure of amino side chains in proteins and the surrounding water molecules were computed at the density functional theory (DFT) level B3LYP/6-311++G** as implemented in the GAUSSIAN03 package. 41
We have then applied a computational protocol based on an efficient QM/MM algorithm called exciton Hamiltonian with electrostatic fluctuations (EHEF).33 EHEF provides an interface for reading MD simulation trajectories of standard MD packages and generating QM atom-atom charge distributions. EHEF performs charge population analysis for the MD samples. Charges contributed by localized atomic orbitals are treated as atomic partial charges, and charges arising from delocalized atomic orbitals are treated with a set of grid point charges fitted from the electrostatic potential. A set of standard atom-atom charges are generated in the “internal coordinate frame”. For a given conformation, charge distributions were deduced from the standard atom-atom charges by updating atom-atom vectors of the corresponding MD geometric structure. Using the full charge distributions we calculate the couplings between the chromophore and the explicit molecular environment. The transition energy is thus affected by the fluctuating electrostatic potential coming from the environment. We avoid repeated expensive QM calculations and obtain the fluctuating Hamiltonian at the QM level for all MD snapshots. 33 Effective fluctuating QM Hamiltonians were constructed using the matrix method in the DichroCalc core. Nonlinear optical signals of molecular systems and their complexes relevant for the resonant electronic spectroscopy have been calculated with the SPECTRON code.20 The 2DUV photon echo signal is obtained by using four impulsive coherent short laser pulses. The excitons generated by electronic peptide backbone transitions are considered as quasi-particles. 20 The 2D photon echo signals are calculated using the protocol described in ref. 28 Calculations were performed for the xxxx and xxxy polarization configuration (i.e. the k1 laser pulse with polarizations along x or y axis and the other three all along x axis). The full inhomogeneous UV spectra are obtained by averaging over 1000 MD snapshots for absorption and CD spectra, and 500 MD snapshots for 2D spectra. All calculated 2DUV spectra were normalized to 1, and then were plotted from −R to R (0 < R < 1) to enhance weak features.
Results
Amyloid Fibril of Aβ1–40
The fragment Aβ9–40 is depicted in Fig. Figure 1 (A), which was taken from the Aβ1–40 conformation proposed by Tycko and co-workers. 13 Fig. Figure 1 (A) shows the structure of a single Aβ9–40 peptide, an asymmetric “U” structure composed of two β-strands as N-terminal (red) and C-terminal (blue), and a turn which links the strands (green). Using the notation of Ref., 13 our simulations are started from the STAG(+2) structure composed of 12 Aβ9–40 monomers, as shown in Fig. Figure 1 (B). The two types of β-strands form separate, parallel β-sheets in amyloid fibrils. There are two types of interfaces between different sets of side chain contacts: “internal” interfaces between the red and blue β-strands in the same Aβ9–40 peptide, and an “external” interface between two blue β-sheets or between neighboring Aβ9–40 peptides along the fibril axis.
The fragment of a 32-residue β-amyloid (Aβ9–40) fibril was used to model the Aβ1–40 amyloid fibril molecule.13 It is known that residues 1–8 are structurally disordered and are not important for fibril growth.12 Moreover, due to this fragment’s disordered nature, we assume that it does not strongly influence the UV absorption properties of the rest of the monomers. A recent experimental 2DIR study on Aβ40 has shown that its morphology at low pH much more closely resembles the long straight fibrils found in diseased brain tissue. 22 We have, therefore, used a neutral sidechain protonation state for the amino acids Glu11, Glu22 and Asp23 corresponding to a low pH. To obtain minimized structures, Tycko and co-workers 13 have used a set of restraints on the interpeptide hydrogen bonds length, backbone torsion angle and sidechains distances. However, these did not include the periodic boundary conditions. The drawback of using such periodic boundary conditions on a small aggregate (six monomers along the fibril axis) is that they induce the loss of twisting which has been observed in experiment and simulations. This twisting corresponds to a rotation over the fibril axis which induces an helical shape of the amyloid structure. The period of such helical twisting has been estimated at 50–200 nm,12 a value much greater than the dimensions of our simulation box. However, we believe that this fibril torsion will not affect strongly the UV spectral properties. We have used the hydrogen bond length and backbone torsion angle restraints employed previously. 13 Restraints are implemented using the colvars module in NAMD 2.7 program34 and include backbone torsion angle restraints for residues 10–12, 15–21, 30–32, 34–36 and 39 (ϕ = −150°, ψ = 150°, 0.01 kcal/mol-deg2 force constants) and backbone hydrogen bond distance restraints for residues 10–24 and 30–40 (2.15 Å oxygen-hydrogen distances, 10.0 kcal/mol-Å2 force constants).
Fibril formation
The mechanism of formation of amyloid fibrils involves many intermediate states. Here we study two possible intermediate prefibrillar structures. Under low pH and at elevated temperature, native lysozyme can evolve into a protofibril structure, and eventually forms an amyloid fibril. 42 In order to monitor intermediate states with UV spectra, we selected this archetypal protein, specifically hen egg-white lysozyme (PDB code 2ZYP), and a standard β-sheet protofibril protein (PDB code 1YJP) for a test study. Their X-ray crystal structures taken from the RSCB protein data bank (which were used as the starting geometries), and a segment of amyloid fibril with eight Aβ9–40 molecules are shown in the top of Fig. Figure 2. We have carried out MD simulations of these proteins in water at room temperature. The MD ensembles were used to compute UV spectra.
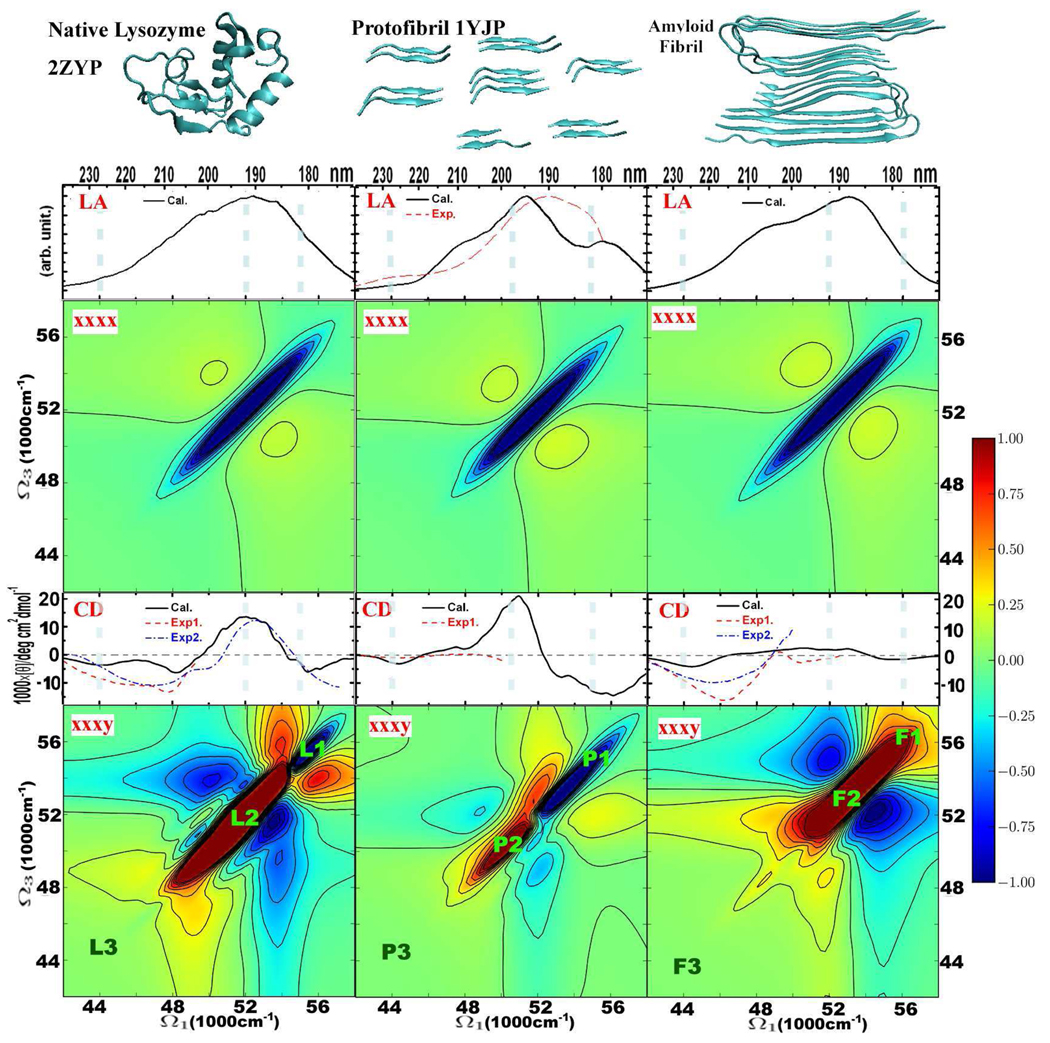
Figure 2.
The linear and 2D signals of native hen egg white lysozyme 2ZYP (left), β-sheet protofibril 1YJP (middle), and Aβ9–40 amyloid fibril (right). From top to bottom: structures, and linear absorption, 2DUV xxxx, CD, and 2DUV xxxy spectra. CD and LA simulations are based on the electronic structure at 1000 snapshots along the MD trajectory. Red dotted and blue dashed lines in CD of lysozyme represent experimental results exp144 and exp2.43 Experimental CD45 and LA46 spectra of protofibril are given by red dotted lines. Red dotted and blue dashed lines in CD of amyloid fibril represent experimental results exp144 and exp2.47 2DUV spectra are obtained by averaging over 500 MD snapshots. Red 2DUV signals are positive, and blue 2DUV signals are negative. L1-2-3, P1-2-3, and F1-2-3 in xxxy 2DUV spectra indicate the main diagonal signals, corresponding to the frequencies marked with dashed vertical line in CD and LA spectra. The 2DUV spectra normalization (R) factors from left to right: for xxxx − 0.42, 0.38, 0.20; for xxxy − 0.10, 0.44, 0.09.
The CD and LA spectra of 1000 MD snapshots were averaged to calculate the inhomogeneous spectra. The calculated CD and LA spectra of lysozyme (2ZYP), the protofibril (1YJP), and a segment of Aβ9–40 fibril (eight Aβ9–40 molecules) are compared with experiment in Fig. Figure 2. The simulated CD spectrum of lysozyme agrees with experiment. 43,44 One strong positive peak at 52000 cm−1 (190 nm), and three negative peaks at 44000 cm−1 (227 nm), 48000 cm–1 (208 nm), and 55000 cm−1 (181 nm) are observed. Good agreement is also seen with the experimental CD45 and LA46 spectra of protofibril protein. The CD spectrum shows two negative peaks at 44000 cm−1 (227 nm) and 55000 cm−1 (181 nm), and a strong positive peak at 51000 cm−1 (196 nm). The CD peak of the protofibril at 55000 cm−1 is much stronger than the 44000 cm−1 peak, which is characteristic of β-sheet proteins. 33,43 For amyloid fibrils, the negative feature observed in experiments at 44500 cm−1 (224 nm)44,47 is reproduced by the Aβ9–40 simulations. The negative 46000 cm−1 (217 nm) experimental feature is probably due to the contamination of lysozyme proteins in the fibril samples,44,47 and is not reproduced by our simulations. Conversion from native lysozyme to fibrils red-shifts the peak at 48000 cm−1.44 The negative peak of the Aβ9–40 fibril at 44000 cm−1 is much stronger than the 55000 cm−1 feature. This trend is reversed compared to typical β-sheet proteins. 33,43 The CD and LA spectra of the Aβ9–40 amyloid fibril are very different from β-sheet protofibril proteins. Although the Aβ9−40 amyloid fibril consists of β-sheet structures, the “internal” interface of Aβ9–40 molecules introduces internal couplings that change the optical signals.
We have simulated 2DUV spectra of these typical structures. Compared with 1D (CD and LA) signals, 2D spectroscopy offers many additional observation windows through varying the polarization configuration of all four pulses. 20,21,25 All-parallel xxxx (non chiral) signals are very similar for the three structures studied, only showing minor spectral shifts. The chirality-induced xxxy signals are much richer. The six intense strong peaks in lysozyme, collapse to two diagonal peaks in the protofibril, and finally to a single strong positive diagonal peak (red) together with two negative sidebands (blue) in the mature amyloid fibril. A weak negative (blue) peak (L1) is observed in lysozyme, which becomes a strong negative peak (P1) in the protofibril, and vanishes in that of Aβ9–40 amyloid fibril.
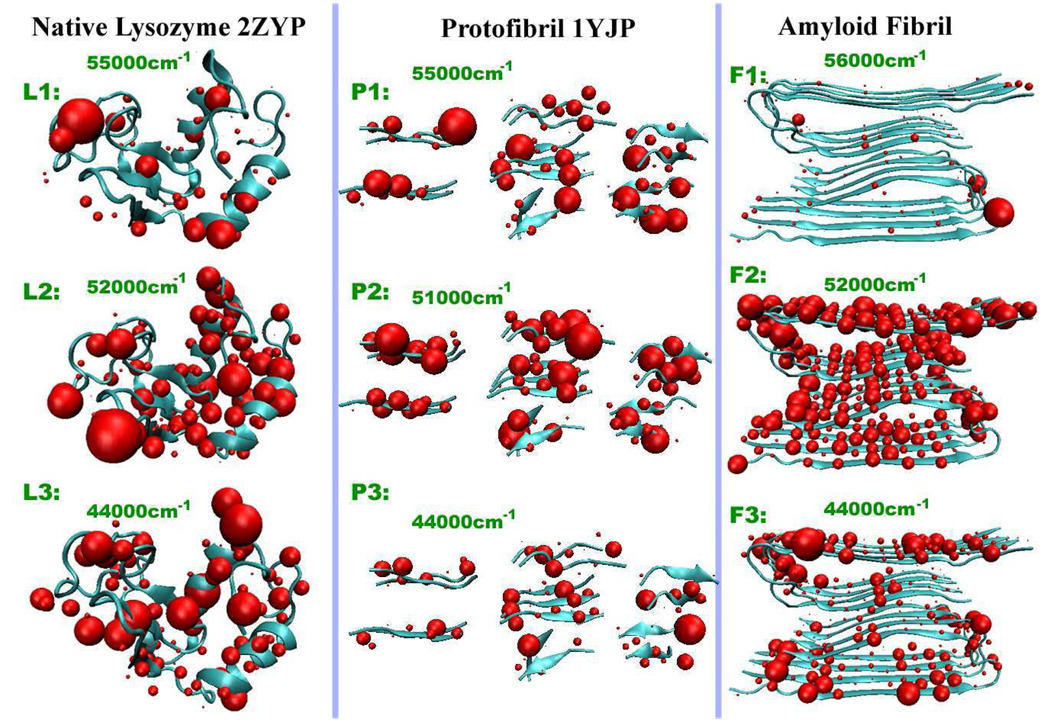
To assign the spectral features, we have computed the transition populations, which are the squares of the exciton wave function in the basis of local amide excitations. Transition populations associated with the three peaks marked with a dashed vertical line, corresponding to xxxy 2DUV peaks L1-2-3, P1-2-3, F1-2-3, are plotted in Fig. Figure 3. The CD peaks at 51000–52000 cm−1 and the xxxy 2DUV peaks L2, P2, F2 are induced by electronic transitions from all peptide groups, which are similar for lysozyme, the protofibril, and the fibril. The CD signals at 55000 cm−1 and the negative (blue) xxxy 2DUV signal L1 in lysozyme are attributable to the random coil regions of the structure. The CD peaks at 55000 cm−1 and the negative (blue) xxxy 2DUV signal P1 are very strong in the protofibril 1YJP protein, and can be ascribed to the delocalized transition populations at the β-sheet structures. In contrast, the transition population in the β-sheet structures of the Aβ9–40 fibril is small and localized at 56000 cm−1, which explains why we see a weak negative CD peak and no negative (blue) 2DUV xxxy signals at F1. The helix part in lysozyme leads to strong transitions (L3 in Fig. Figure 3) and CD peaks at 44000 cm−1, while the pure sheet structures in protofibril 1YJP contribute to the weak transitions P3 in Fig. Figure 3 and small CD peaks at 44000 cm−1. The Aβ9–40 fibril has many turn regions and internal interfaces between two β-strands, giving strong CD peaks at 44000 cm−1. These simulations demonstrate the sensitivity of UV signals to the internal interactions between sidechains and the protein backbone.
Figure 3.
Transition populations of native lysozyme 2ZYP, protofibril 1YJP, Aβ9–40 fibril averaged over 500 MD snapshots. L1-2-3, P1-2-3, and F1-2-3 correspond to the xxxy 2DUV peaks marked in Fig. Figure 2.
Fibril structural motifs
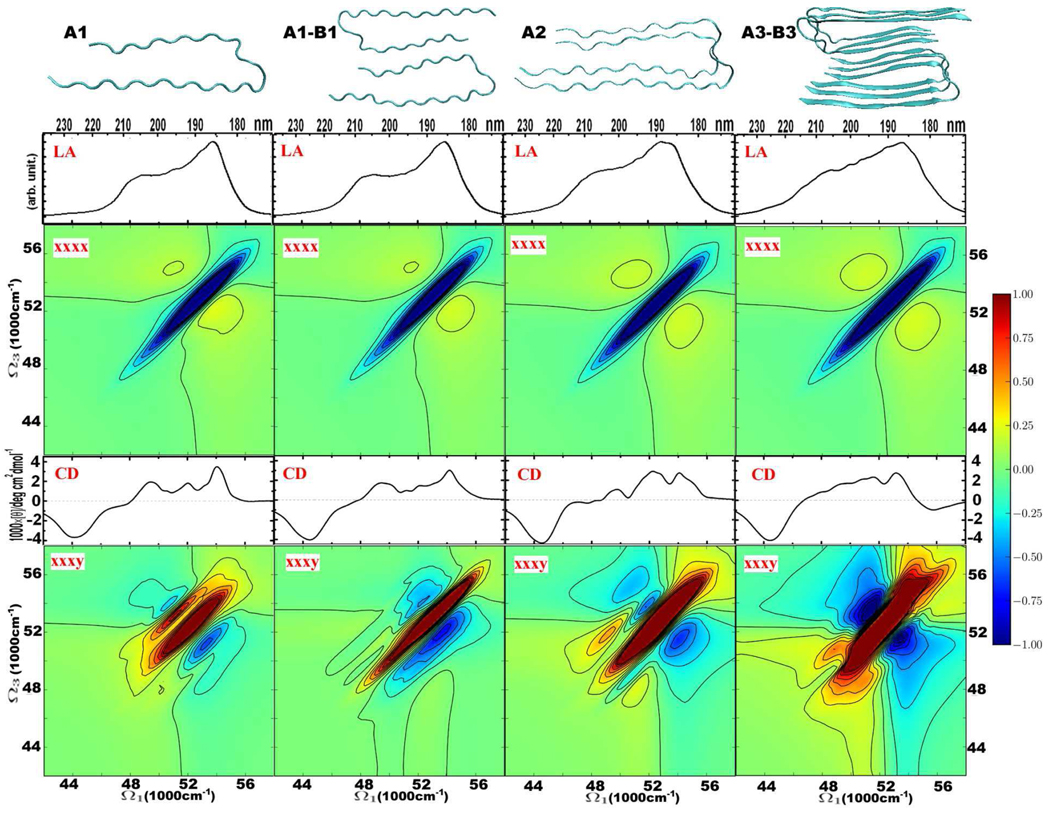
Based on the ensemble of MD snapshots of the 12-Aβ9–40 amyloid fibril, we have taken several segments to study the variation of spectra with amyloid size. In Fig. Figure 4, we show the structures of fibril segments consisting of one (A1), two (molecules A and B perpendicular to fibril axis: A1-B1), two (molecules A and A along fibril axis: A2), and six Aβ9–40 molecules (A3-B3), together with the simulated 2DUV spectra. The LA and xxxx 2DUV signals show how two well-separated peaks in the smallest fibril merge into a single broad feature as the system size increases. This is represented by the rise of the elongated diagonal peak in the xxxx signal. The CD and xxxy 2D UV signal show much stronger size dependence: the positive (red) peak evolves into a stronger and wide diagonal band with increasing size. Simulations of these four small Aβ9–40 segments have reflected the overall patterns of 2DUV spectra of the fibril with eight Aβ9–40 molecules (right panel of Fig. Figure 2), suggesting the internal interactions within the Aβ9–40 molecule dictate the electronic and photonic properties of the fibrils. On the other hand, the fine details of every major UV feature in the CD and xxxy 2DUV spectra are quite distinctive for these four Aβ9–40 fibrils. Shifting of CD peaks is observed as the system size varies. The positive signals on the diagonal of the xxxy 2DUV spectrum (two red peaks) in the A1 system (a single Aβ9–40 molecule) manifest as a single narrow red peak in the A2 system (two molecules perpendicular to fibril axis), a stronger red peak in the A1-B1 system (two molecules along fibril axis), and a even stronger red peak with extensions to the off-diagonal region in the A3-B3 system (six Aβ9–40 molecules). The two negative (blue) sidebands in the xxxy 2DUV spectrum are symmetric in the A3-B3 system, which agrees with those observed in the segment of eight Aβ9–40 molecules (right panel of Fig. Figure 2). However, these two negative sidebands are actually asymmetric in different ways in the smaller segments A1, A2, and A1-B1. Therefore, simulation of these specific fine details in high resolution will help to track the aggregation process of amyloid fibrils.
Figure 4.
Variation of CD, LA and 2DUV spectra with the number of structural elements in Aβ9–40 amyloid fibrils. From top to bottom: structures, and absorption, 2DUV xxxx, CD, and 2DUV xxxy spectra. Linear and 2D signals are based on 1000 and 500 MD snapshots, respectively. Red 2DUV signals are positive, and blue 2DUV signals are negative. The 2DUV spectra normalization (R) factors from left to right: for xxxx − 0.38, 0.41, 0.36, 0.33; for xxxy − 0.42, 0.24, 0.18, 0.11.
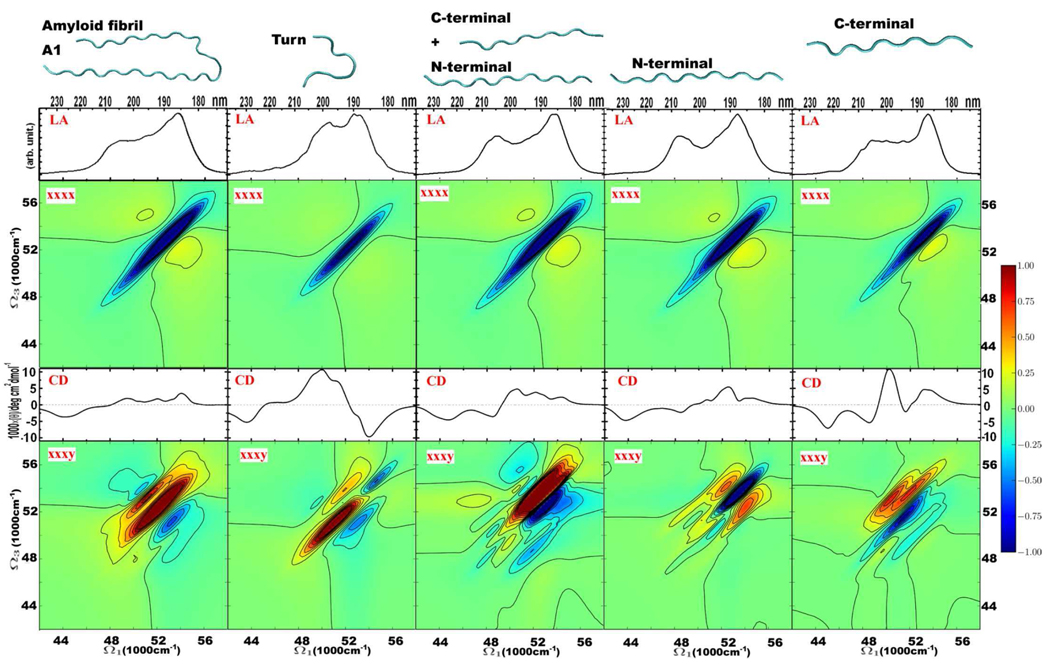
We used additional model systems to examine further the intramolecular couplings at the “internal” interface of the Aβ9–40 molecule. One Aβ9–40 molecule, and its isolated turn, the N-terminal plus C-terminal strands, the N-terminal strand in isolation, and the C-terminal strand, together with their corresponding UV spectra are drawn in Fig. Figure 5. The CD, LA and 2DUV spectra of the fragments are quite different from the entire molecule. Compared to the entire Aβ9–40 molecule, the turn fragment has stronger negative CD and xxxy 2D UV signals at around 56000 cm−1; the fragment comprising the N-terminal plus C-terminal strands, the isolated N-terminal strand, and the isolated C-terminal strand all give two negative CD peaks below 52000 cm−1. The xxxy 2DUV signals of a single strand are very different to those of Aβ9–40, while those of the turn and the N-terminal plus C-terminal strands start to exhibit the Aβ9–40 feature, implying that the couplings at the “internal” interface of the Aβ9–40 molecule are very important for the electronic structures and the induced optical response. We conclude that regarding the UV spectroscopy of the protein backbone, it is more appropriate to characterize Aβ9–40 amyloid fibril as an asymmetric “U”-shape protein, instead of an extended β-sheet fibril.
Figure 5.
Structures, CD, LA and 2DUV spectra of an Aβ9–40 molecule and its different fragments. From top to bottom: structures, and absorption, 2DUV xxxx, CD, and 2DUV xxxy spectra. Linear and 2D signals are based on 1000 and 500 MD snapshots, respectively. Red 2DUV signals are positive, and blue 2DUV signals are negative. The 2DUV spectra normalization (R) factors from left to right: for xxxx − 0.38, 0.82, 0.40, 0.43, 0.56; for xxxy − 0.42, 0.62, 0.42, 0.51, 1.11.
Conclusions
Using a generalized QM/MM approach, we have simulated the 2DUV spectra of Aβ9–40 amyloid fibrils and related proteins. We found that the spectra are sensitive to the secondary structure, and they carry structural signatures. Chiral xxxy 2DUV signals could monitor the amyloid structure formation at different stages. A simulated fibrillation trajectory could be used to study spectral signatures of all intermediate structural configurations so that the fibrillation could be followed with high resolution. The size and geometry dependencies have been investigated. The overall 2DUV patterns do not vary much with increasing numbers of Aβ9–40 molecules. The spectrum of a single Aβ9–40 molecule already resembles the 2DUV features seen for Aβ9–40 amyloid fibrils. However, the specific fine details of most 2DUV features do vary with the number of Aβ9–40 molecules involved and the way molecules interact with each other. Detailed 2DUV signals of amyloid fibrillar systems of various sizes will aid in tracking the real time growth of fibrils. Fragments of one Aβ9–40 molecule show very different electronic and optical properties than that of fibrils. The electronic structures and patterns of spectral features of the Aβ9–40 amyloid fibril are mainly determined by the “U”-like structure of the Aβ9–40 molecule. Our simulations explore the fine details of fibril structure, which should help in identifying various intermediate toxic structures and the aggregated stages of fibril formation.
Acknowledgement
We gratefully acknowledge the support of the National Institutes of Health (Grand GM059230 and GM091364), and the National Science Foundation (Grant CHE-0745892). JDH thanks the Leverhulme Trust for a Research Fellowship. BMB was the grateful recipient of an Early-Stage Researcher Short Visit award from the Collaborative Computational Project for Biomolecular Simulation. We thank Prof. Yi Luo and Daniel Healion for helpful discussions.
References
- 1.Tycko R. Curr. Opin. Struct. Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Caughey B, Lansbury PT. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 3.Lester-Coll NE, Riviera EJ, Soscia SJ, Doiron K, Wands JR, de la Monte SM. J Alzheimers Dis. 2006;9:13–33. doi: 10.3233/jad-2006-9102. [DOI] [PubMed] [Google Scholar]
- 4.Glabe C, Kayed R. Neurology. 2006;66:S74–S78. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Annu. Rev. Biochem. 2006;75:333–336. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira ST, Vieira MNN, Felice FGD. IUBMB Life. 2007;59:332–345. doi: 10.1080/15216540701283882. [DOI] [PubMed] [Google Scholar]
- 7.Rishton GM. Nat. Chem. Biol. 2008;4:159–160. doi: 10.1038/nchembio0308-159. [DOI] [PubMed] [Google Scholar]
- 8.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BCH, Prusiner SB, Weissman J, Shoichet BK. Nat. Chem. Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiti F, Dobson CM. Nat. Chem. Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 10.Fawzi NL, Yap E-H, Okabe Y, Kohlstedt K, Brown S, Head-Gordon T. Acc. Chem. Res. 2008;41:1037–1047. doi: 10.1021/ar800062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankarq GM, Li S, Mehta T, N.E. Shepardson AG-M, Smith I, Brett F, Farrell M, Rowan M, Lemere C, Regan C, Walsh D, Sabatini B, Selkoe D. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchete N-V, Tycko R, Hummer G. J. Mol. Biol. 2005;353:804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 13.Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daggett V. Acc. Chem. Res. 2006;39:594–602. doi: 10.1021/ar0500719. [DOI] [PubMed] [Google Scholar]
- 15.Sunde M, Blake CCF. Q. Rev. Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 16.Eanes ED, Glenner G. J. Histochem. Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 17.Chung HS, Ganim Z, Jones KC, Tokmakoff A. Proc. Natl. Acad. Sci. USA. 2007;104:14237–14242. doi: 10.1073/pnas.0700959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho M. Chem. Rev. 2008;108:1331–1418. doi: 10.1021/cr078377b. [DOI] [PubMed] [Google Scholar]
- 19.Mukamel S. Principles of Nonlinear Optical Spectroscopy. New York: Oxford University Press; 1995. [Google Scholar]
- 20.Abramavicius D, Palmieri B, Voronine DV, Šanda F, Mukamel S. Chem. Rev. 2009;109:2350–2408. doi: 10.1021/cr800268n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukamel S, Abramavicius D, Yang L, Zhuang W, Schweigert IV, Voronine D. Acc. Chem. Res. 2009;42:553–562. doi: 10.1021/ar800258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Proc. Natl. Acad. Sci. USA. 2008;105:7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strasfeld D, Ling Y, Shim S, Zanni M. J. Am. Chem. Soc. 2008;130:6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim S-H, Strasfeld DB, Ling YL, Zanni MT. Proc. Natl. Acad. Sci. USA. 2007;104:14197–14202. doi: 10.1073/pnas.0700804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang W, Abramavicius D, Voronine DV, Mukamel S. Proc. Natl. Acad. Sci. USA. 2007;104:14233–14236. doi: 10.1073/pnas.0700392104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta N, Maekawa H, Zhuang W, Mukamel S, Tobias D, Ge N-H. J. Phys. Chem. B. 2009;113:12037–12049. doi: 10.1021/jp901504r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagchi S, Falvo C, Mukamel S, Hochstrasser R. J. Phys. Chem. 2009;113:11260–11273. doi: 10.1021/jp900245s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abramavicius D, Jiang J, Bulheller BM, Hirst JD, Mukamel S. J. Am. Chem. Soc. 2010;132:7769. doi: 10.1021/ja101968g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng C, Matsika S, Weinacht T. Opt. Express. 2009;17:18788–18793. doi: 10.1364/OE.17.018788. [DOI] [PubMed] [Google Scholar]
- 30.Antzutkin ON, Balbach JJ, Tycko R. Biophys. J. 2003;84:3326–3335. doi: 10.1016/S0006-3495(03)70057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shashilov VA, Lednev IK. J. Am. Chem. Soc. 2008;130:309–317. doi: 10.1021/ja076225s. [DOI] [PubMed] [Google Scholar]
- 32.Sikirzhytski V, Topilina NI, Higashiya S, Welch JT, Lednev IK. J. Am. Chem. Soc. 2008;130:5852. doi: 10.1021/ja8006275. [DOI] [PubMed] [Google Scholar]
- 33.Jiang J, Abramavicius D, Bulheller BM, Hirst JD, Mukamel S. J. Phys. Chem. B. 2010;114:8270. doi: 10.1021/jp101980a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips J, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel R, Kalé L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKerell AD, Jr, et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 37.Essmann U, Perera L, Berkowitz ML, Darden T, Pedersen LG. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 38.Feller SE, Zhang Y, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 39.Besley NA, Hirst JD. J. Am. Chem. Soc. 1999;121:9636–9644. [Google Scholar]
- 40.Aquilante F, Vico LD, Ferré N, Ghigo G, Malmqvist P-A, Neogrády P, Pedersen TB, Pitonák M, Reiher M, Roos BO, Serrano-Andrés L, Urban M, Veryazov V, Lindh R. J. Comput. Chem. 2009;31:224–247. doi: 10.1002/jcc.21318. [DOI] [PubMed] [Google Scholar]
- 41.Frisch MJ, et al. Gaussian 03, Revision C.02. Wallingford, CT: Gaussian, Inc.; 2004. 2004. [Google Scholar]
- 42.Cao A, Hu D, Lai L. Protein Sci. 2004;2:319–324. doi: 10.1110/ps.03183404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bulheller BM, Miles AJ, Wallace BA, Hirst JD. J. Phys. Chem. B. 2008;112:1866–1874. doi: 10.1021/jp077462k. [DOI] [PubMed] [Google Scholar]
- 44.Kalhor HR, Kamizi M, Akbari J, Heydari A. Biomacromolecules. 2009;10:2468–2475. doi: 10.1021/bm900428q. [DOI] [PubMed] [Google Scholar]
- 45.Huang THJ, Yang D-S, Plaskos NP, Go S, Yip CM, Fraser PE, Chakrabartty A. J. Mol. Biol. 2000;297:73–87. doi: 10.1006/jmbi.2000.3559. [DOI] [PubMed] [Google Scholar]
- 46.Bulheller BM, Rodger A, Hicks MR, Dafforn TR, Serpell LC, Marshall K, Bromley EHC, King PJS, Channon KJ, Woolfson DN, Hirst JD. J. Am. Chem. Soc. 2009;131:13305–13314. doi: 10.1021/ja902662e. [DOI] [PubMed] [Google Scholar]
- 47.Hirota-Nakaoka N, Hasegawa K, Naiki H, Goto Y. J. Biochem. 2003;134:159–164. doi: 10.1093/jb/mvg124. [DOI] [PubMed] [Google Scholar]