Abstract
A phylogenetically conserved RNA structure within the open reading frame of poliovirus and other group C enteroviruses functions as a competitive inhibitor of the antiviral endoribonuclease RNase L. Hence, we call this viral RNA the RNase L competitive inhibitor RNA (RNase L ciRNA). In this investigation we used phylogenetic information, RNA structure prediction software, site-directed mutagenesis, and RNase L activity assays to identify functionally important sequences and structures of the RNase L ciRNA. A putative loop E motif is phylogenetically conserved in the RNA structure and mutations of nucleotides within the putative loop E motif destroyed the ability of the RNA molecule to inhibit RNase L. A putative H-H kissing loop interaction is phylogenetically conserved in the RNA structure and covariant polymorphisms that maintain the Watson-Crick complementarity required for the kissing interaction provide evidence of its importance. Compensatory mutations that disrupted and then restored the putative kissing interaction confirm that it contributes to the ability of the viral RNA to inhibit RNase L. RNase L was activated late during the course of poliovirus replication in HeLa cells, as virus replication and assembly neared completion. We conclude that a putative loop E motif and an H-H kissing loop interaction are key features of the group C enterovirus RNA associated with the inhibition of RNase L.
Keywords: poliovirus, enterovirus, innate immunity, dsRNA, 2-5A, ribonuclease L, loop E motif, pseudoknot, H-H type kissing loop
Introduction
Phylogenetically conserved RNA sequences within positive-strand RNA viruses typically correlate with functionally important RNA sequence motifs and/or RNA structures. Computer algorithms like M-Fold and Kinefold can be used to identify putative secondary and tertiary structures.1–3 Furthermore, covariant polymorphisms in related RNAs that maintain Watson-Crick base pairs are considered to be good evidence for particular base pairs in related RNA structures.4 An RNA sequence within the ORF of poliovirus RNA (Fig. 1A) makes poliovirus RNA unusually resistant to cleavage by the antiviral endoribonuclease RNase L.5 Because this poliovirus RNA functions as a competitive inhibitor of the endoribonuclease domain of RNase L we call this viral RNA the RNase L competitive inhibitor RNA (RNase L ciRNA).6 The RNase L ciRNA structure is composed of two RNA sequences [poliovirus nucleotides (nts) 5742–5824 and nts 5906–5967] in the open reading frame of poliovirus RNA (Fig. 1A). An intervening RNA sequence (poliovirus nts 5825–5905) can be deleted without affecting the inhibitory action of the RNase L ciRNA (Δ in Fig. 1A indicates the position where these intervening poliovirus nts 5825–5905 are normally located relative to the ciRNA).5
Figure 1.
Ribonuclease L competitive inhibitor RNA (RNase L ciRNA) from the ORF of poliovirus. (A) Diagram of the RNA structure in the poliovirus ORF associated with the competitive inhibition of RNase L; as predicted by M-Fold and Kinefold.2,3,5,6 Putative loop E motif highlighted in green. Putative H-H type kissing loop interaction highlighted in light blue. (B) Loop E motif from X. laevis 5S rRNA (adapted from Leontis and Westhof).7
A potential loop E motif is present in the RNase L ciRNA structure (Fig. 1A, putative loop E motif nucleotides highlighted in green). Loop E motifs are common in contemporary ribosomal RNAs, and were presumably present in primordial rRNAs.7 Loop E motifs are also found in the Potato Spindle Tuber Viroid ribozyme, the RNA component of RNase P, the P7 of some group I introns, and the IRESes of hepatitis C virus and human enteroviruses.7–15 While nucleotides within loop E motifs are predicted to be unpaired using computer algorithms like M-Fold and Kinefold (Fig. 1A), atomic resolution structures revealed that six out of seven unpaired nucleotides of loop E motifs are involved in non-canonical base pairs (Fig. 1B).16,17 Loop E motifs contain a trans (locally parallel)-Hoogsteen-Hoogsteen A · A pair, a bulged G (Fig. 1A, G5761 in red), a trans-Hoogsteen U · A pair, and a sheared A · G pair (Fig. 1B). 17
Pseudoknots are another common structural feature of RNAs. Visual observation and Kinefold predict an H-H type kissing loop interaction in the RNase L ciRNA structure (Fig. 1A, complementary kissing loop nucleotides in blue).3 H-H type kissing loop pseudoknots exhibit Watson-Crick base pairing between RNA sequences in the loops of stem-loop structures. Positive-strand RNA viruses often encode RNA pseudoknots within IRES RNAs or within their open-reading frames to mediate ribosomal frameshifting.18 H-H type kissing loop interactions are present in numerous RNA structures, including the 3′ nontranslated regions of picornavirus RNA genomes.19,20
In this study we used phylogenetic information, site-directed mutagenesis and RNase L activity assays to identify features of the RNase L ciRNA that are required for the inhibition of RNase L. The data suggest that a putative loop E motif and an H-H type kissing loop interaction are key structural features of the viral RNA that are required for the inhibition of RNase L.
Results
Phylogenetic conservation of the putative loop E motif and H-H type kissing loop interaction of the RNase L ciRNA in group C enteroviruses
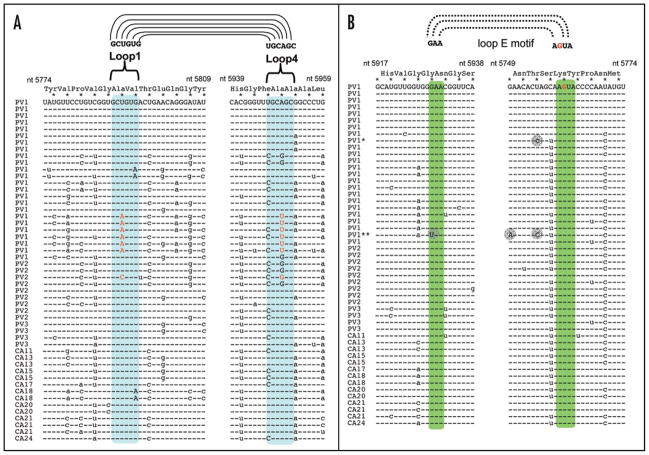
RNA sequences from poliovirus and other group C enteroviruses were aligned to examine the conservation of nucleotides in the RNase L ciRNA (Fig. 2). There are three serotypes of poliovirus [poliovirus type 1 (PV1), poliovirus type 2 (PV2), and poliovirus type 3(PV3)] as well as several serotypes of Coxsackievirus (CA11, CA13, CA15, CA17, CA18, CA20, CA21 and CA24) classified as group C enteroviruses.21 Conservation of the amino acid sequence of group C enterovirus 3C protease in this region of the virus ORF restricted the location of polymorphisms in the RNase L ciRNA predominantly to the wobble position of codons (Fig. 2, poliovirus amino acid sequence and wobble positions of codons indicated at the top of the alignments). There were polymorphisms present at many wobble positions; however, most polymorphisms maintained base pairs in the predicted ciRNA structure. Furthermore, polymorphisms at the wobble positions of the H-H type kissing loop sequences maintained the complementarity required for kissing loop interactions (Fig. 2A, kissing loop nucleotides highlighted). Importantly, several RNA sequences contained co-variant polymorphisms that maintained the complementarity required for the kissing loop interaction (Fig. 2A, co-variant kissing loop polymorphisms in red). The phylogenetic conservation of base pairs, including those in the predicted H-H type kissing loop interaction, suggests that the kissing loop interaction may be functionally important.
Figure 2.
Phylogenetic conservation of the H-H kissing loop interaction and loop E motif of RNase L ciRNA in group C enteroviruses. RNase L ciRNA sequences from 51 group C enterovirus sequences; 24 poliovirus type 1 (PV1) sequences, 9 poliovirus type 2 (PV2) sequences, 3 poliovirus type 3 (PV3) sequences, and 14 non-poliovirus group C enteroviruses, Coxsackie A viruses 11, 13, 15, 17, 18, 20, 21 and 24. (A) Stem-loop 1 and stem-loop 4 RNA sequences (nucleotides 5774–5809 and 5939–5959 of poliovirus). Sequences within putative H-H type kissing loop interaction highlighted in light blue. (B) Sequences adjacent to the loop E motif (nucleotides 5917–5938 and 5749–5774 of poliovirus). Putative loop E motif sequences highlighted in green. Nucleotide accession numbers: PV1: NC_022058, V001149, AY184119, V01150, AF538841, AF538840, AF538842, AF538843, AF462418 (PV1*), AF462419, AJ32960, AJ132961, AF405682, AF405690, AF405669, AF405666, AY560657, AF111953, AF111981, AF111961, AF111966, AF111983, AF111984 (PV1**), AY278553. PV2: AY177685, AY184220, AY278549, AY278550, AY278552, AY278551, AF448782, AF44873, M12197; PV3: AY184221, K01392, AJ293918, X04468; AF499636_CAV11, AF465511_CAV13, AF499637_CAV13, AF465512_CAV15, AF499639_CAV15, AF499639_CAV17, AF465513_CAV18, AF399640_CAV18, AF465514_CAV20, AF499642_CAV20, AF465515_CAV21, AF546702_CAV21, D00538_CAV21, D90457_CAV24.
The group C enterovirus RNA sequences in and around the putative loop E motif were particularly well conserved, including both base paired and non-base paired wobble position residues (Fig. 2B). The G 5761 predicted to be the bulged G of the putative loop E motif was absolutely conserved (Fig. 2B, bulged G5761 in red). The G5929 of the putative loop E motif is a wobble position of a glycine codon (Fig. 2B). The third base of glycine codons can be any nucleotide (GGG, GGA, GGC and GGU). Nonetheless, this G 5929 is conserved in all but one of the aligned sequences (Fig. 2B, PV1** marked with an asterisk). Notably, the PV1** sequence, in addition to the G5929 polymorphism, also has two other polymorphisms that would be inconsistent with the ciRNA structure and function (Fig. 2B, G5749A and U5753C in PV1** marked with grey circles). It remains unclear why the RNase L ciRNA is not well conserved in this one sequence; however, it appears that this PV1** virus isolate is a hyper-mutated ciRNA variant. Otherwise, the phylogenetic data indicate that the putative loop E motif and H-H type kissing loop interaction of the ciRNA are very well conserved among group C enteroviruses.
H-H kissing loop interaction contributes to the function of the RNase L ciRNA
We tested the importance of particular RNA sequences in the kissing loops of the RNase L ciRNA by assaying the ability of the ciRNA and its mutant derivatives to inhibit RNase L activity (Fig. 3). RNase L activity was measured in reactions where the cleavage of an RNA FRET probe by RNase L was detected by fluorometry (Fig. 3A and B).22 The FRET probe is a 36 base long fragment of respiratory syncytial virus intergenic RNA with a 5′ 6FAM fluorophore and a 3′ Black Hole Quencher (Fig. 3A). Incubation of the RNA FRET probe in reactions containing RNase L and 2–5A allows the activated RNase L to cleave the RNA FRET probe, thereby releasing the 6FAM fluorophore from the BHQ (Fig. 3B). Cleavage of the FRET probe is detected by measuring RFU in a fluorometer (Fig. 3D). RFUs increase over time in reactions containing 100 nM of the FRET probe, 20 nM of RNase L and 20 nM 2–5A (Fig. 3D, black line). RFUs do not increase over time of incubation when RNase L is excluded from the reaction (Fig. 3D, probe alone, grey line). When 100 nM of ciRNA is included in the reaction RFUs increase very little over time due to the ability of the ciRNA to function as a competitive inhibitor of RNase L (Fig. 3D, ciRNA, yellow line), as previously established.6 An RNA from another portion of the poliovirus ORF does not inhibit cleavage of the FRET probe by RNase L (Fig. 3D, 100 nM 2123 RNA, blue line). Thus, the FRET assay is a very specific and quantitative assay of RNase L activity. Furthermore, the RNase L ciRNA potently inhibits RNase L endoribonuclease activity in this assay, functioning as a competitive inhibitor with a Ki of 34 nM.6 A modest five-fold molar excess of RNase L ciRNA (100 nM ciRNA in reactions containing 20 nM RNase L) inhibits greater than 90% of the RNase L activity in the FRET assay (Fig. 3D).
Figure 3.
Compensatory mutagenesis of the H-H kissing loop interaction in the RNase L ciRNA. (A) Diagram of RNA FRET probe. (B) Illustration of FRET in RNase L activity assays. (C) Three complementary bases in the loop of stem-loop 1 and stem-loop 4 of the ciRNA were changed to their complementary sequences, individually and simultaneously. Mutations highlighted in grey. (D) RNase L activity measured by FRET assay, as previously established.6 Reactions containing 20 nM RNase L, 20 nM 2–5A, 100 nM FRET probe and 100 nM of the indicated viral RNAs; Poliovirus ciRNA (yellow line), loop 1 mutant ciRNA (L1M, purple line), loop 4 mutant ciRNA (L4M, brown line), PV ciRNA containing the compensatory loop 1 and loop 4 mutations (CM, azure line). A negative control reaction incubated without RNase L (probe alone, grey line). A positive control reaction containing FRET probe, RNase L and 2–5A without poliovirus ciRNA (probe + RNase L + 2–5A, black line). A control reaction containing 100 nM of an alternate fragment of the poliovirus ORF (2123, pale blue line, poliovirus nucleotides 5889–6181). RFU plotted versus time of incubation.
Mutations were engineered into the phylogenetically conserved H-H kissing loops to assess the contribution(s) of these sequences and structure to the inhibition of RNase L (Fig. 3C and D). A three base change in loop 1 (Loop 1 mutant, L1M) and a three base change in loop 4 (Loop 4 mutant, L4M) were designed to disrupt the kissing loop interaction (Fig. 3C). Together these loop 1 and loop 4 mutations restored Watson-Crick base pairing in the kissing loops (Fig. 3C, compensatory mutant, CM). When FRET reactions contained 100 nM of loop 1 mutant ciRNA (L1M) or loop 4 mutant ciRNA (L4M) RFUs increased over time; however, the kinetics and magnitude of increase were in between those for reactions containing either wildtype ciRNA or 2123 controls (Fig. 3D). Importantly, when these loop 1 and loop 4 mutations restored Watson-Crick base pairing in the kissing loops the ciRNA with the compensatory mutations was able to inhibit RNase L to the same degree as wildtype ciRNA (Fig. 3D, ciRNA versus CM). These data indicate that the complementarity of RNA sequences in loops 1 and 4 contribute to the ability of the ciRNA to inhibit RNase L. It is most likely that Watson-Crick base pairing between sequences in loops 1 and 4 occurs intramolecularly, as diagrammed in Figure 1A; however, the data do not exclude the alternate possibility that the complementarity in loops 1 and 4 could allow for the formation of dimers or even larger oligomers of ciRNA.
Mutation of wobble positions in and around the putative loop E motif
Based on the predicted secondary structure of ciRNA (Fig. 1A) and the phylogenetic information (Fig. 2B) we targeted for mutagenesis wobble positions in and around the putative loop E motif of the ciRNA (Fig. 4A). Ten wobble position mutations were engineered into the ciRNA: 1 (G5749U); 2 (C5752U); 3 (U5755G); 4 (G5761A); 5 (C5767G); 6 (A5938U); 7 (U5935A); 8 (G5929U); 9 (U5923G) and 10 (U5920C) (Fig. 4A). These mutations were designed to create base pairs for unpaired bases or disrupt preexisting base pairs (as determined using M-Fold, data not shown). The effects of these wobble position mutations on the inhibition of RNase L was assessed in reactions containing FRET probe, RNase L and 2–5A (Fig. 4B). One hundred and 500 nM poliovirus ciRNA inhibited RNase L in reactions containing FRET probe, RNase L and 2–5A (Fig. 4B, ciRNA). Maximal amounts of RFUs were detected in reactions containing 100 and 500 nM of a negative control RNA (Fig. 4B, 2123). Four of the ten wobble position mutations in ciRNA completely disabled its ability to inhibit RNase L activity: positions 3 (U5755G), 4 (G5761A), 5 (C5767G) and 8 (G5929U) (Fig. 4). Another four wobble position mutations in ciRNA had very little effect: positions 2 (C5752U), 6 (A5938U), 9 (U5923G) and 10 (U5920C) (Fig. 4). Two mutations significantly debilitated the ciRNA, however, some residual inhibitory activity was still detected: 1(G5749U) and 7 (U5935A) (Fig. 4). These data indicated that particular phylogenetically conserved wobble positions in and around the putative loop E motif were important for the ability of the ciRNA to inhibit RNase L. Based on these data we performed more comprehensive mutagenesis of U5755, C5767, G5761 and G5929 as presented below.
Figure 4.
Mutagenesis of wobble positions within and around the loop E motif of ciRNA. (A) Wobble positions of codons with conserved RNA sequences within and around the putative loop E motif were targeted for mutagenesis (nucleotides highlighted in grey circles and numbered 1–10). 1 (G5749U); 2 (C5752U); 3 (U5755G); 4 (G5761A); 5 (C5767G); 6 (A5938U); 7 (U5935A); 8 (G5929U); 9 (U5923G) and 10 (U5920C). (B) RNase L activity measured by FRET in reactions containing 100 or 500 nM 2123 RNA, poliovirus ciRNA, or poliovirus ciRNA with the indicated mutations (1–10). RFU after 30 minutes of incubation plotted versus poliovirus RNA (PV RNA) in each reaction.
Mutagenesis of U5755 and C5767
In order to probe the structure and function of the ciRNA more fully we substituted U5755 and C5767 with each alternate nucleotide and assessed the effects of the mutations on the inhibition of RNase L activity (Figs. 5 and 6). Substituting U5755 with any other nucleotide completely disabled the ciRNA inhibition of RNase L (Fig. 5). This unpaired, phylogenetically conserved U5755, which is present at a junction between one of the kissing stem-loops and a 3 base pair region above the putative loop E motif, is apparently important in the overall structure and function of the RNase L ciRNA. In contrast, a C5767U substitution mutation that maintains a predicted base pair did not affect the inhibition of RNase L (Fig. 6) whereas C5767G and C5767A mutations which disrupted a predicted base pair did disable the ciRNA (Fig. 6). These data are consistent with the phylogenetically conserved G-C5767 and G-U5767 base pair in ciRNA (Figs. 1A and 2B).
Figure 5.
Mutation of ciRNA U5755 prevents the inhibition of RNase L. RNase L activity in reactions containing 20 nM RNase L, 20 nM 2–5A, 100 nM FRET probe and 100 nM of the indicated viral RNAs; poliovirus ciRNA (blue line), poliovirus ciRNA U5755G (purple line), poliovirus ciRNA U5755C (brown line), poliovirus ciRNA U5755A (green line). A negative control reaction incubated without RNase L (probe alone, grey line). A positive control reaction containing FRET probe, RNase L, and 2–5A without poliovirus ciRNA (probe + RNase L + 2–5A, black line). A control reaction containing 100 nM of an alternate fragment of the poliovirus ORF (2123, yellow line, poliovirus nucleotides 5889–6181). RFU plotted versus time of incubation.
Figure 6.
A base pair involving ciRNA C5767 is required for the inhibition of RNase L. RNase L activity in reactions containing 20 nM RNase L, 20 nM 2–5A, 100 nM FRET probe and 100 nM of the indicated viral RNAs; poliovirus ciRNA (blue line), poliovirus ciRNA C5767G (purple line), polio-virus ciRNA C5767U (brown line), poliovirus ciRNA C5767A (green line). A negative control reaction incubated without RNase L (probe alone, grey line). A positive control reaction containing FRET probe, RNase L, and 2–5A without poliovirus ciRNA (probe + RNase L + 2–5A, black line). A control reaction containing 100 nM of poliovirus ORF (2123, yellow line, poliovirus nucleotides 5889–6181). RFU plotted versus time of incubation.
Phylogenetically conserved wobble positions of the putative loop E motif are required for the inhibition of RNase L
We substituted the G5761 and G5929 with each alternate nucleotide and assessed the effects of the mutations on the inhibition of RNase L activity (Fig. 7). Substituting G5761 or G5929 with any other nucleotide completely disabled the ciRNA inhibition of RNase L (Fig. 7). These data indicate that the putative loop E motif within the ciRNA is critical for the inhibition of RNase L.
Figure 7.
Wobble positions G5761 and G5929 within the loop E motif are critical for the inhibition of RNase L. RNase L activity in reactions containing 20 nM RNase L, 20 nM 2–5A, 100 nM FRET probe and 100 nM of the indicated viral RNAs. (A) poliovirus ciRNA (blue line), poliovirus ciRNA G5761A (purple line), poliovirus ciRNA G5761U (brown line), poliovirus ciRNA G5761C (green line). (B) poliovirus ciRNA (blue line), poliovirus ciRNA G5929U (purple line), poliovirus ciRNA G5929C (brown line), poliovirus ciRNA G5929A (green line). A negative control reaction incubated without RNase L (probe alone, grey line). A positive control reaction containing FRET probe, RNase L, and 2–5A without poliovirus ciRNA (probe + RNase L + 2–5A, black line). A control reaction containing 100 nM of poliovirus ORF (2123, yellow line, poliovirus nucleotides 5889–6181). RFU plotted versus time of incubation.
Mutagenesis alone cannot prove the presence of the loop E motif. Atomic structures determined by NMR or crystallography will be ultimately required to prove the presence of the loop E structure. Nonetheless, the functional importance of these wobble position mutations allowed us to engineer infectious poliovirus containing a G5761A mutation to evaluate the contribution of the ciRNA and RNase L activity in infected cells.
RNase L activity in cells infected with wildtype and mutant poliovirus
Based on the phylogenetic and functional data above we engineered G5761A mutation into infectious poliovirus and monitored RNase L activity in wildtype and mutant virus-infected cells (Fig. 8). The replication of wildtype and G5761A mutant poliovirus were compared in HeLa cells expressing wildtype RNase L (Fig. 8, W12 HeLa cells) and in HeLa cells expressing a dominant negative RNase L (Fig. 8, M25 HeLa cells). One step growth analyses indicated that both wildtype and G5761A polioviruses grew with normal kinetics and magnitudes in both cell lines (Fig. 8A and B). Ribosomal RNA fragments characteristic of RNase L activity were detected 5 to 8 hours post-infection in W12 HeLa cells (Fig. 8C, asterisks and arrows) but were not evident at any time post-infection in M25 HeLa cells (Fig. 8D).23,24 Importantly, ribosomal RNA fragments characteristic of RNase L activity were not detected until virus assembly neared completion ~5 hours post-infection (Fig. 8). Notably, ribosomal RNA fragments characteristic of RNase L activity were detected 30 minutes earlier in W12 HeLa cells infected with poliovirus containing a G5761A mutation than in cells infected with wildtype poliovirus (Fig. 8C, arrows). These data indicate that RNase L was activated in poliovirus infected cells regardless of whether ciRNA was functionally intact in the viral RNA. While mutating the ciRNA may lead to RNase L activity detectable at slightly earlier times in infected cells, the difference was not profound. These data indicate that RNase L activity does not restrict the kinetics or magnitude of poliovirus replication in HeLa cells. Furthermore, G5761A mutation did not prevent the replication of poliovirus in HeLa cells. Poliovirus containing a G5761A mutation will be useful for in vivo investigations to help elucidate the contributions of RNase L and the ciRNA to viral pathogenesis.
Figure 8.
Poliovirus replication in HeLa cells expressing wildtype and dominant negative RNase L. Growth of poliovirus with a G5761A mutation was compared to the growth of wildtype poliovirus in W12 HeLa cells expressing wildtype RNase L and M25 HeLa cells expressing RNase L with a dominant-negative mutation (R667A). Virus one-step growth curve (A and B) and rRNA cleavage characteristic of RNase L activity (C and D). W12 HeLa cells (A) and M25 HeLa cells (B) were infected at an MOI of 10 PFU per cell with wildtype poliovirus (WT PV) or poliovirus encoding a G5761A mutation (G5761A PV) and titers of infectious virus were determined by plaque assay at the indicated times post-infection. Total cellular RNA from W12 HeLa cells (C) and M25 HeLa cells (D) at the indicated times post-infection was fractionated by electrophoresis in 1% agarose and detected by ethidium bromide staining and UV-light.
Discussion
In this investigation we used phylogenetic information, RNA structure prediction software, site-directed mutagenesis and RNase L activity assays to define key structural features of the group C enterovirus RNA associated with the inhibition of RNase L. An RNA structure was predicted with both M-Fold and Kinefold (Fig. 1). Phylogenetic information was consistent with the predicted structure (Fig. 2). A putative loop E motif and an H-H type kissing loop interaction appear to be key structural features of the RNA that are required for the inhibition of RNase L. Phylogenetic information (Fig. 2A) and compensatory mutagenesis (Fig. 3) indicated that the complementarity of RNA sequences were important for both the H-H kissing loop interaction and for the inhibition of RNase L activity. Likewise, wobble position mutations in and around the putative loop E motif established the functional importance of specific nucleotides in the inhibition of RNase L (Figs. 4–7). Mutagenesis alone cannot prove the presence of a loop E motif in the RNase L ciRNA. Atomic structures determined by NMR or crystallography will be ultimately required to prove the presence of a loop E structure in the RNase L ciRNA.
While RNase L becomes activated during the course of a polio-virus infection (Fig. 8C), mutations that impaired the ability of the RNA to inhibit RNase L did not prevent poliovirus replication in tissue culture cells (Fig. 8A and B). Rather, wildtype and mutant poliovirus grew indistinguishably in cells with or without RNase L activity (Fig. 8). This may be due to the activation of RNase L relatively late during the course of the poliovirus infection in vitro (Fig. 8). It is important to note that rRNA cleavage characteristic of activated RNase L was not evident until virus replication and virus assembly were nearing completion at 5 hours post-infection (Fig. 8). The activation of RNase L after virus replication in HeLa cells may limit the antiviral effect of this antiviral pathway on poliovirus in tissue culture cells.
Phylogenetic information suggests that RNase L ciRNA is functionally conserved in vivo for all of the group C enteroviruses (Fig. 2). Furthermore, OPV1 shed in the feces of a B-cell immunodeficient child indicated that RNase L ciRNA remained intact in the poliovirus genome during more than two years of infection and that OPV1-derived virus from this child was fully neurovirulent.25 Such observations suggest that the RNase L ciRNA is important during replication in the selective environment of human hosts and may contribute to the pathogenesis of paralytic poliomyelitis. Attenuating mutations elsewhere within OPV strains revert during replication in children and such OPV strains regain neurovirulence.26–28 Transmission of OPV strains from person-to-person, coupled with reversion to wildtype sequences, leads to outbreaks of paralytic poliomyelitis caused by vaccine-derived viruses.29,30 It remains to be determined how the RNase L ciRNA contributes to host-pathogen interactions in vivo and whether mutations in the RNase L ciRNA will attenuate virus replication in vivo.
Ribonuclease L is an antiviral endoribonuclease in the interferon-regulated, dsRNA-activated 2′–5′ oligoadenylate synthetase/ribonuclease L pathway (reviewed in refs. 31 and 32). When RNase L is activated during the course of a viral infection, the antiviral endoribonuclease cleaves viral RNAs at single-stranded UA and UU dinucleotides.33,34 Hepatitis C virus RNA is cleaved efficiently by RNase L,35 providing the selective pressure for a genome with a paucity of UA and UU dinucleotides.36 Experiments in mice indicate that interferon-mediated antiviral pathways like the 2′–5′ OAS/RNase L pathway restrict poliovirus pathogenesis and tissue tropism in vivo.37 Furthermore, type I interferon signaling decreases in nonpermissive host cells when they are explanted and grown in tissue culture, leading to decreased expression of 2′-5′ OAS (which is required for the antiviral activation of RNase L), converting the host cells into permissive environments for poliovirus replication.38 Because the integrity of the ciRNA was maintained during more than two years of OPV1 replication in the gut a of child, we would expect RNase L and the RNase L ciRNA to be biologically significant in the undefined host cells that support poliovirus replication in the gut. The RNase L ciRNA may be important in other host tissues important for poliovirus pathogenesis such as motor neurons. Poliovirus encoding the G5761A mutation (Fig. 8) will be useful for future in vivo investigations to elucidate the biological significance of RNase L and the ciRNA in particular tissues.
RNase L and the group C enterovirus RNA associated with the inhibition of RNase L likely share evolutionarily ancient origins. RNase L is related to Ire1p, a host protein involved in the ER stress response.39,40 The presence of ancient RNA structural motifs like the putative loop E motif and H-H type kissing loop interaction are consistent with the possibility that the ciRNA may have evolutionarily ancient origins yet the RNase L ciRNA has only been found in group C enteroviruses (Fig. 2) and is not evident in other related picornaviruses (data not shown). Therefore, the evolutionary origins of the RNase L ciRNA remain obscure. The ability of a contemporary viral RNA to directly inhibit an evolutionarily ancient antiviral endoribonuclease provides an intriguing glimpse into the evolution of host-virus interactions.
Materials and Methods
RNase L
RNase L was expressed and purified as previously described, with modifications.41–43 Briefly, untagged recombinant human RNase L in a baculovirus vector was expressed in SF21 insect cells. Suspension cultures of SF21 cells were grown in SFM 900 insect cell medium (Invitrogen) supplemented with 10% fetal bovine serum to a cell density of 1.5 × 106 to 2.0 × 106 cells/ml. Cells were infected at a multiplicity of infection (MOI) of 5 PFU per cell at 27°C for 72 h before being harvested. The cell pellets were washed with chilled phosphate-buffered saline (PBS), resuspended in lysis buffer (20 mM HEPES, pH 7.4, 5 mM MgCl2, 50 mM KCl, 1 mM EDTA, 10% glycerol, 14 mM 2-mercaptoethanol [2-ME], 100 μM ATP, 20 μg/ml leupeptin, 20 μg/ml pepstatin, 50 μM phenyl-methylsulfonyl fluoride [PMSF]), and disrupted with a French press. Supernatants were collected after centrifugation at 100,000 ×g and incubated for 1 hour at 4°C with CL6B Blue Sepharose affinity resin (Amersham Bioscience). The protein-bound affinity resin was packed in an HR16/24 column and washed with buffer A (20 mM HEPES, pH 7.4, 5 mM MgCl2, 50 mM KCl, 1 mM EDTA, 10% glycerol, 7 mM 2-ME, 100 μM ATP, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 50 μM PMSF). The bound RNase L was eluted with a 0 to 100% linear gradient of buffer B (20 mM HEPES, pH 7.4, 5 mM MgCl2, 1.0 M KCl, 1 mM EDTA, 10% glycerol, 7 mM 2-ME, 100 μM ATP, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 50 μM PMSF) over a period of 1 h at a flow rate of 1 ml/min. The peak fractions containing RNase L were pooled, dialyzed against four changes of buffer A, loaded onto a MonoQ HR 10/10 column, and eluted with a 0 to 50% buffer B gradient over a period of 60 min at a flow rate of 1 ml/min. The purity of RNase L was >90%, as judged by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by Coomassie blue staining.
2–5A
Enzymatic synthesis of 2–5A from ATP was done with recombinant porcine 2′–5′ oligoadenylate synthetase (pOAS1) (a generous gift of Rune Hartmann, Aarhus, Denmark) activated with poly(I):poly(C) conjugated to CL6B agarose (Ag-pI:pC). Briefly, poly(I):poly(C) was conjugated with CL6B agarose beads (Amersham Biosciences) using per-iodate conjugation chemistry and Ag-poly(I):poly(C) was incubated with 0.4 mg/ml of purified pOAS1 at 25°C in 10 mM Hepes pH 7.5 containing 1.5 mM magnesium acetate, 20% glycerol, 50 mM KCl and 7 mM β-mercaptoethanol for 1 h. pOAS1 immobilized on Ag-pI:pC beads was washed three times with buffer (10 mM Hepes pH 7.5 containing 1.5 mM magnesium acetate, 20% glycerol, 50 mM KCl and 7 mM β-mercaptoethanol) and re-suspended in the reaction mixture (10 mM ATP in 1.5 mM magnesium acetate, 20% glycerol, 50 mM KCl and 7 mM β-mercaptoethanol) for 20 h. The unfractionated 2–5A mixture was harvested by centrifugation at 3,000 ×g for 15 min. The unfractionated 2–5A mix was separated from pOAS1 using Centriprep™ (Millipore) molecular weight cut off of 3,000 Da. The 2–5A was analyzed by injecting 50 μl of 1:10 diluted stock of 2–5A mix on Dionex P100 4 × 250 analytical column (Dionex Inc.,) interfaced with System gold HPLC connected to a 32 Karat work station in 25 mM Tris-HCl pH 7.5 and eluted with a gradient of 0–750 mM NaCl in 25 mM Tris-HCl in 1 hour. The large scale purification was done on mono Q column HR10/10 (Amersham Biosciences) interfaced with FPLC™ (Amersham Biosciences) and characterized by HPLC.
Viral RNA
cDNA encoding the poliovirus RNA inhibitor of RNase L (illustrated in Fig. 1A) was amplified by PCR using a 5′ primer with a T7 transcription promoter (underlined) (5′ GAAAT TAATA CGACT CACTA TAGGA TCGTG AACAC TAGCA AGTAC CCCAA TATG 3′) and a 3′ reverse primer (5′ GGATC GCTTC AGGGC CGCTG CAAAC CCGTG TGAACC 3′). Mutations in poliovirus ciRNA were engineered into the cDNA by PCR using the following primers: Mutant 1 (G5749U) GAAAT TAATA CGACT CACTA TAGGA TCGTT5749 AACAC TAGCA AGTAC CCCAA TATG 3′, Mutant 2 (C5752U) GAAAT TAATA CGACT CACTA TAGGA TCGTG AAT5752AC TAGCA AGTAC CCCAA TATG 3′, Mutant 3 (U5755G) GAAAT TAATA CGACT CACTA TAGGA TCGTG AACAC5755AGCA AGTAC CCCAA TATG 3′, Mutant 4 (G5761A) 5′ GAAAT TAATA CGACT CACTA TAGGA TCGTG AACAC TAGCA AA5761TAC CCCAA 3′, Mutant 5 (C5765G) 5′ GAAAT TAATA CGACT CACTA TAGGA TCGTG AACAC TAGCA AGTAC CCG5765AA TATG 3′, Mutant 6 (A5938U) 5′ GGATC GCTTC AGGGC CGCTG CAAAC CCGTG A5938GAAC CGTTC CCACC AACAT GCATC C 3′, Mutant 7 (U5935A) 5′ GGATC GCTTC AGGGC CGCTG CAAAC CCGTG TGAT5935C CGTTC CCACC AACAT GCATC C 3′, Mutant 8 (G5929U) 5′ GGATC GCTTC AGGGC CGCTG CAAAC CCGTG TGAAC CGTTA5929 CCACC AACAT GCATC C 3′, Mutant 9 (U5923G) 5′ GGATC GCTTC AGGGC CGCTG CAAAC CCGTG TGAAC CGTTC CCACC5923ACAT GCATC C 3′, Mutant 10 (U5920C) 5′ GGATC GCTTC AGGGC CGCTG CAAAC CCGTG TGAAC CGTTC CCACC5920T GCATC C 3′. In addition to the mutants described U5755, C5767, G5761 and G5929 within poliovirus ciRNA were substituted with each alternate nucleotide with PCR primers containing the respective nucleotide polymorphisms (primers not shown). Mutants 1–5 used the forward primers as listed above and used the same reverse primer as wildtype ciRNA. Mutants 6–10 used the same forward primer as wildtype poliovirus ciRNA and the reverse primers as listed above. PCR generated cDNAs were transcribed by T7 polymerase (Epicentre, Madison, WI). Viral RNAs were precipitated with 0.3 M sodium acetate (pH 7.0) and two volumes of ethanol, washed with 70% ethanol, and solubilized in water. RNAs were qualitatively and quantitatively analyzed by electrophoresis in 1% agarose, ethidium bromide staining and visualization with UV light.
FRET assay of RNase L activity
A dual-labeled RNA FRET probe was used as a substrate to monitor RNase L activity.22 The substrate was derived from a 36 nucleotide intergenic sequence of respiratory syncytial virus (RSV) (UUA UCA AAU UCU UAU UUG CCC CAU UUU UUU GGU UUA) with a fluorophore (6-FAM- or 6-carboxy-fluorescein) at the 5′-terminus and a black hole quencher-1 (BHQ-1) at the 3′-terminus (Sigma-Proligo, The Woodlands, TX) (Fig. 3A). Reactions (50 μl) contained 25 mM Tris-HCl [pH 7.4], 100 mM KCl, 10 mM MgCl2, 50 μM ATP, 7 mM β-mercaptoethanol, 100 nM FRET probe, variable amounts of viral RNA (0–600 nM) CGCTG and either full-length RNase L and 2–5A (20 nM each) or Δ577 RNase L (300 nM) as indicated in figure legends. Reactions were incubated at room temperature in a black 96-well microtiter round bottom plate. Fluorescence was activated using 485 nm wavelength excitation. Five-hundred thrity-five nm wavelength emissions were detected at the indicated times in a Bio-Tek® Synergy HT Multi-Detection Microplate Reader (Winooski, VT) (Fig. 3B).
HeLa cells and poliovirus infections
HeLa cells expressing wildtype RNase L (W12 HeLa cells) and RNase L with a dominant-negative R667A mutation (M25 HeLa cells) were previously described.5 Poliovirus type 1 (Mahoney) was grown in W12 HeLa cells. An MOI of 10 PFU per cell was used for one-step growth experiments. HeLa cells (5 × 105) were seeded in 35-mm plates 18 to 22 h before infection. Cells were inoculated with virus diluted in 500 μl PBS. Following 1 h of adsorption at 37°C, the inoculum was removed and replaced with 2 ml of DMEM containing 10% fetal bovine serum, penicillin and streptomycin. G418 (250 μg/ml) was included in the medium to maintain the pcDNA3 vectors expressing wildtype and R667A RNase L within HeLa cells. After three cycles of freeze-thawing to release intracellular virus from cells the titer of infectious virus was determined by plaque assay.
Acknowledgments
We thank Eric Westhof for recognizing and alerting us to the putative loop E motif within the poliovirus RNase L ciRNA. We thank Jeffrey Kieft for critically reviewing the manuscript. We thank Chris Washenberger for help with group C enterovirus sequence alignments and Rune Hartmann (Aarhus, Denmark) for the pOAS1 clone. This work was supported by the American Cancer Society Research Scholar Grant RSG-02-063-01-MBC (DJB); by Health Service grants AI42189 (DJB), CA044059 (RHS) and T32 AI052066 (HLT); and by the Mal and Lea Bank Chair to Robert H. Silverman.
Abbreviations
- RNase L
ribonuclease L
- ciRNA
competitive inhibitor RNA
- ORF
open reading frame
- FRET
fluorescence resonance energy transfer
- 6FAM
6-carboxyfluorescein
- BHQ-1
black hole quencher-1
- MOI
multiplicity of infection
- PFU
plaque forming units
- PV
poliovirus
- OPV
oral poliovirus vaccine
References
- 1.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–40. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 2.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005;33:605–10. doi: 10.1093/nar/gki447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woese CR, Pace NR. Probing RNA structure, function and history by comparative analysis. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 5.Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J Virol. 2007;81:5561–72. doi: 10.1128/JVI.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA. 2008;14:1026–36. doi: 10.1261/rna.958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol. 1998;283:571–83. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 8.Owens RA, Baumstark T. Structural differences within the loop E motif imply alternative mechanisms of viroid processing. RNA. 2007;13:824–34. doi: 10.1261/rna.452307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zhong X, Itaya A, Ding B. Evidence for the existence of the loop E motif of Potato spindle tuber viroid in vivo. J Virol. 2007;81:2074–7. doi: 10.1128/JVI.01781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J Mol Biol. 1998;279:773–93. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 11.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 12.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2008 doi: 10.1016/j.virusres.2008.06.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–8. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 14.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–10. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 15.Bailey JM, Tapprich WE. Structure of the 5′ nontranslated region of the coxsackievirus B3 genome: Chemical modification and comparative sequence analysis. J Virol. 2007;81:650–68. doi: 10.1128/JVI.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szewczak AA, Moore PB. The sarcin/ricin loop, a modular RNA. J Mol Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 18.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Bakkers JM, Galama JM, Bruins Slot HJ, Pilipenko EV, Agol VI, et al. Structural requirements of the higher order RNA kissing element in the enteroviral 3′UTR. Nucleic Acids Res. 1999;27:485–90. doi: 10.1093/nar/27.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melchers WJ, Hoenderop JG, Bruins Slot HJ, Pleij CW, Pilipenko EV, Agol VI, et al. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–96. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown B, Oberste MS, Maher K, Pallansch MA. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J Virol. 2003;77:8973–84. doi: 10.1128/JVI.77.16.8973-8984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thakur CS, Xu Z, Wang Z, Novince Z, Silverman RH. A convenient and sensitive fluorescence resonance energy transfer assay for RNase L and 2′,5′ oligoadenylates. Methods Mol Med. 2005;116:103–13. doi: 10.1385/1-59259-939-7:103. [DOI] [PubMed] [Google Scholar]
- 23.Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46:1051–5. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wreschner DH, James TC, Silverman RH, Kerr IM. Ribosomal RNA cleavage, nuclease activation and 2–5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–81. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odoom JK, Yunus Z, Dunn G, Minor PD, Martin J. Changes in population dynamics during long-term evolution of Sabin type 1 poliovirus in an immunodeficient patient. J Virol. 2008;82:9179–90. doi: 10.1128/JVI.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans DM, Dunn G, Minor PD, Schild GC, Cann AJ, Stanway G, et al. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature. 1985;314:548–50. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- 27.Bouchard MJ, Lam DH, Racaniello VR. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J Virol. 1995;69:4972–8. doi: 10.1128/jvi.69.8.4972-4978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westrop GD, Wareham KA, Evans DM, Dunn G, Minor PD, Magrath DI, et al. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J Virol. 1989;63:1338–44. doi: 10.1128/jvi.63.3.1338-1344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, et al. outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science. 2002;296:356–9. doi: 10.1126/science.1068284. [DOI] [PubMed] [Google Scholar]
- 30.Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- 31.Player MR, Torrence PF. The 2–5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman RH. Viral encounters with OAS and RNase L during the IFN antiviral response. J Virol. 2007;81:12720–9. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′–5′) oligoadenylate—dependent endonuclease. Science. 1981;212:1030–2. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 34.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–7. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 35.Han JQ, Wroblewski G, Xu Z, Silverman RH, Barton DJ. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage NY: Cold Spring sites. J Interferon Cytokine Res. 2004;24:664–76. doi: 10.1089/jir.2004.24.664. [DOI] [PubMed] [Google Scholar]
- 36.Washenberger CL, Han JQ, Kechris KJ, Jha BK, Silverman RH, Barton DJ. Hepatitis C virus RNA: Dinucleotide frequencies and cleavage by RNase L. Virus Res. 2007;130:85–95. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ida-Hosonuma M, Iwasaki T, Yoshikawa T, Nagata N, Sato Y, Sata T, et al. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J Virol. 2005;79:4460–9. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshikawa T, Iwasaki T, Ida-Hosonuma M, Yoneyama M, Fujita T, Horie H, et al. Role of the alpha/beta interferon response in the acquisition of susceptibility to poliovirus by kidney cells in culture. J Virol. 2006;80:4313–25. doi: 10.1128/JVI.80.9.4313-4325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou A, Nie H, Silverman RH. Analysis and origins of the human and mouse RNase L genes: mediators of interferon action. Mamm Genome. 2000;11:989–92. doi: 10.1007/s003350010194. [DOI] [PubMed] [Google Scholar]
- 40.Dong B, Niwa M, Walter P, Silverman RH. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA. 2001;7:361–73. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman RH, Dong B, Maitra RK, Player MR, Torrence PF. Selective RNA cleavage by isolated RNase L activated with 2–5A antisense chimeric oligonucleotides. Methods Enzymol. 2000;313:522–33. doi: 10.1016/s0076-6879(00)13033-2. [DOI] [PubMed] [Google Scholar]
- 42.Rusch L, Dong B, Silverman RH. Monitoring activation of ribonuclease L by 2′,5′-oligoad-enylates using purified recombinant enzyme and intact malignant glioma cells. Methods Enzymol. 2001;342:10–20. doi: 10.1016/s0076-6879(01)42531-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2–5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–65. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]