Transcriptional profiling experiments identify light- and temperature-entrained circadian transcripts in C. elegans.
Abstract
Most organisms have an endogenous circadian clock that is synchronized to environmental signals such as light and temperature. Although circadian rhythms have been described in the nematode Caenorhabditis elegans at the behavioral level, these rhythms appear to be relatively non-robust. Moreover, in contrast to other animal models, no circadian transcriptional rhythms have been identified. Thus, whether this organism contains a bona fide circadian clock remains an open question. Here we use genome-wide expression profiling experiments to identify light- and temperature-entrained oscillating transcripts in C. elegans. These transcripts exhibit rhythmic expression with temperature-compensated 24-h periods. In addition, their expression is sustained under constant conditions, suggesting that they are under circadian regulation. Light and temperature cycles strongly drive gene expression and appear to entrain largely nonoverlapping gene sets. We show that mutations in a cyclic nucleotide-gated channel required for sensory transduction abolish both light- and temperature-entrained gene expression, implying that environmental cues act cell nonautonomously to entrain circadian rhythms. Together, these findings demonstrate circadian-regulated transcriptional rhythms in C. elegans and suggest that further analyses in this organism will provide new information about the evolution and function of this biological clock.
Author Summary
Daily (circadian) rhythms in behavior and physiology allow organisms to adapt to periodic cues such as light and temperature associated with the rotation of the earth. Subsets of molecular components of the internal clock that drive these rhythms, as well as effector genes for behavioral outputs, also exhibit rhythmic expression in many organisms. While circadian rhythms in behavior have previously been described in the nematode Caenorhabditis elegans, no transcriptional rhythms or clock genes have been identified, leaving open the question of the nature of the clock in this organism. Here, we identify light- and temperature-entrained cycling genes in C. elegans via genome-wide transcriptional profiling. Transcripts showing circadian regulation (including expression with a 24-h period maintained upon removal of the entraining stimulus) and temperature compensation were identified. Light and temperature appear to entrain independent sets of genes. We also identify large sets of light- or temperature-driven genes. Mutations in a channel gene previously implicated in sensory transduction in a small set of sensory neurons abolish entrainment of gene expression by environmental signals. This work demonstrates the presence of circadian transcriptional rhythms in C. elegans, and provides the foundation for future investigations into the underlying mechanisms.
Introduction
Organisms exhibit daily (circadian) rhythms in behavior and physiology that are attuned to the earth's rotational period. The periods of these intrinsic rhythms are approximately 24 h and are synchronized by external cues or zeitgebers (time givers) such as light and temperature. The molecular composition and regulation of endogenous pacemakers that generate circadian rhythms have been extensively investigated. Both the molecules composing the core clock and the molecular mechanisms of clock function are remarkably divergent across kingdoms, as well as between eukaryotes and prokaryotes, suggesting multiple, independent evolutionary origins for the circadian clock [1]–[6]. However, genes and pathways by which circadian oscillatory rhythms are generated in animals such as Drosophila and mammals are strongly conserved [5],[7],[8], implying a single origin of the clock in the metazoan lineage [2]. Thus, it is of interest to identify and characterize the circadian clock in additional animals.
The nematode Caenorhabditis elegans is well established as an excellent model organism for the study of development and behavior [9]–[12]. In their soil habitat, worms are subjected to temperature fluctuations corresponding to solar irradiance during the day–night cycles [13],[14]. Moreover, when associated with surface-dwelling animals [15], C. elegans is likely to be exposed to daily light–dark cycles. Several studies have previously reported the presence of circadian rhythms in C. elegans. In these studies, growth-synchronized populations were entrained to light or temperature cycles, and circadian rhythms in behaviors such as locomotion or responses to osmotic stress were quantified [16]–[20]. Although these experiments demonstrated the presence of rhythms with characteristics of true circadian oscillations, these rhythms were surprisingly variable and non-robust [19]. In addition, while the C. elegans genome is predicted to encode homologs of most Drosophila and mammalian core clock components, the roles of these genes appear to be largely restricted to development [21]. Thus, whether C. elegans exhibits bona fide circadian rhythms remains unclear.
Circadian rhythmicity in behavior and physiology has been proposed to be driven by clock-regulated oscillations in transcription and translation, and/or via post-transcriptional or post-translational modifications [22]–[32]. The expression of core clock genes such as per (period) and tim (timeless) as well as many clock output genes cycles at the transcriptional level in a circadian manner in Drosophila [23],[24]. Expression profiling experiments have, therefore, provided a powerful approach to identify clock genes as well as clock output genes under circadian transcriptional control [33]–[41]. Since expression profiling is not biased by prior assumptions regarding a specific behavioral or physiological output, transcriptional rhythms may be a robust measure of the presence of a molecular circadian clock.
Here we identify light- and temperature-regulated transcriptional rhythms in C. elegans and show that subsets of these transcripts are regulated in a circadian manner. We find that light and temperature also globally drive the expression of many genes, indicating that C. elegans exhibits systemic responses to these stimuli. Light- and temperature-entrained transcripts appear to be largely nonoverlapping. We show that conserved clock gene homologs do not exhibit circadian rhythmicity at the mRNA level. We also find that the TAX-2 cyclic nucleotide-gated channel subunit, previously implicated in thermosensory signal transduction and phototransduction [42]–[44] is required to transduce both light and temperature information to the clock(s). These results demonstrate that C. elegans has circadian clock(s) that are entrained by light and temperature, and indicate that this model organism can be used to further explore the evolution and function of this critical timekeeping mechanism.
Results
Identification of Cycling Transcripts Entrained by Light and Temperature
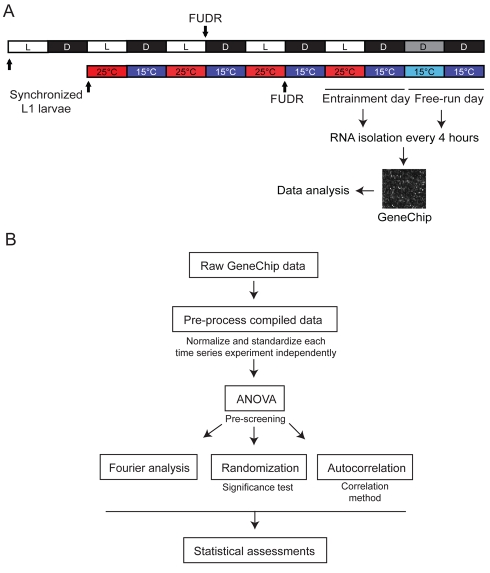
To determine whether cycling transcripts can be identified in C. elegans following entrainment to either light–dark or temperature cycles, we performed a genome-wide expression profiling analysis. Growth-synchronized populations of wild-type L1 larvae were entrained until adulthood to either 12 h∶12 h light/dark (LD) cycles at a constant temperature of 18°C, or to 25°C∶15°C (warm/cold [WC]) temperature cycles in constant darkness (DD) (Figure 1A). Following entrainment, animals were maintained under free-running conditions of DD or at a constant temperature of 15°C (constant cold [CC]) (Figure 1A). To exclude non-entrained transcripts present in the embryos of self-fertilizing C. elegans hermaphrodites, L4 larvae and adults were grown on plates containing 5-fluoro-2-deoxyuridine (FUDR), which inhibits DNA synthesis and results in embryonic lethality [45]. RNA was collected every 4 h during the last 24 h of entrainment as well as during the first 24 h of free-running conditions (Figure 1A) and hybridized to Affymetrix C. elegans Genome Arrays containing 22,500 probe sets representing more than 19,000 predicted C. elegans genes.
Figure 1. Method for RNA collection and analysis flowchart for detection of cycling transcripts.
(A) Populations of growth-synchronized wild-type L1 larvae were entrained for 5 d until adulthood to 12 h∶12 h LD cycles (500–1,000 lux) at a constant temperature of 18°C, or for 4 d to 12 h∶12 h temperature cycles (25°∶15°C [WC]) in DD. RNA was collected every 4 h during the last entrainment and the subsequent free-running days and analyzed via hybridization of Affymetrix GeneChips (see Materials and Methods). L4 larvae were transferred to FUDR-containing plates to inhibit embryonic development. (B) Normalized and standardized expression values from each independent time series experiment were prescreened with ANOVA to identify transcripts that exhibit statistically significant changes in expression over time. Appended datasets from each independent time series experiment were then examined to identify the 24-h spectral power (F 24) score of each transcript [87], and the significance was calculated via comparison with a randomly permuted dataset. The 24-h autocorrelation (AC24 score) between time points that were 24 h apart was determined by fitting the six time points collected during the entrainment day to the six time points collected during the free-running day. See Materials and Methods for additional details.
To identify putative circadian oscillatory transcripts with high confidence, we performed a series of data analyses based on the initial assumption that these rhythms exhibit approximately 24-h periodicity (Figure 1B and Table S1; see Materials and Methods). These algorithms identify cycling transcripts based on consonances in their periods and phases while minimizing the contribution of experimental variations in oscillation amplitude [46]. Similar methods were used in a meta-analysis of circadian expression profiling data from Drosophila to identify a set of light- and temperature-entrained circadian transcripts [41],[47]. To validate the algorithms and filters applied by us, we reanalyzed a LD dataset from Drosophila (S. K. and M. R., unpublished data) and successfully identified all major core clock genes including per, clk, tim, cry, and vri, whose expression is known to cycle under these conditions (data not shown). These analyses allowed us to identify transcripts that cycle with a 24-h period in both the light- and temperature-regulated datasets, subsets of which continued to cycle under constant conditions (see below).
Transcription Rhythms Are Driven, Entrained, and Enriched for 24-h Periods
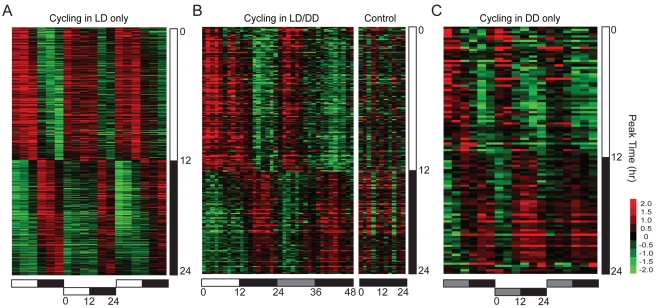
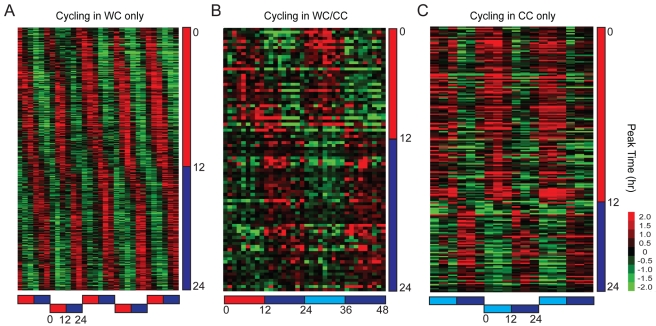
Entrainment via light or temperature cycles may drive as well as entrain transcriptional rhythms [47]. Stimulus-driven but clock-independent transcripts are expected to cycle during entrainment but not during free-running conditions, as observed in the first category of cycling transcripts (LD only or WC only) (Figures 2A and 3A). The second category included transcripts that cycled during both entrainment and free-running conditions (LD/DD or WC/CC) (Figures 2B and 3B), suggesting that expression of these genes may be under circadian control [47]. We also identified a third category of transcripts whose expression cycled during the free-running but not during the entrainment conditions (DD only or CC only) (Figures 2C and 3C). This category may include genes whose expression is also under circadian control, but whose cycling is suppressed or masked during entrainment conditions. A similar category of transcripts was previously noted in genome-wide studies of light-regulated transcriptional rhythms in Drosophila [36].
Figure 2. Light-driven and -entrained transcripts.
Phase-ordered heat maps showing cycling transcripts upon entrainment to LD cycles. (A) Transcripts that cycle in LD but not in DD. (B) Transcripts that cycle in both LD and DD. (C) Transcripts that cycle in DD but not in LD. Only transcripts that passed all applied filters and thresholds are shown (Table S1). The heat maps in (A) and (C) show appended data from three independent 1-d entrainment time series experiments and three independent 1-d free-running time series experiments, respectively. The heat map in (B) shows the average of three independent 2-d time series experiments. Data from two independent 1-d time series experiments performed in parallel in DD to control for temperature fluctuations are also shown in (B). In all panels, columns represent experimental time points, and rows represent individual transcript profiles. Rows are ordered according to the estimated peak phases of the transcript profiles across the appended or averaged datasets, indicated by the vertical bars to the right of each heat map. Expression values represented by the green-to-red color scale indicate up- or down-regulation relative to the experimental mean expression values indicated in black. Horizontal bars below the heat maps correspond to the LD entrainment protocols used in the time series experiments; white, black, and gray bars in all panels indicate the light, dark, and subjective light phases, respectively.
Figure 3. Temperature-driven and -entrained transcripts.
Phase-ordered heat maps showing cycling transcripts upon entrainment to temperature cycles (25°C∶15°C). (A) Transcripts that cycle in WC but not in CC. (B) Transcripts that cycle in both WC and CC. (C) Transcripts that cycle in CC but not in WC. Only transcripts that passed all applied filters and thresholds are shown (Table S1). The heat maps in (A) and (C) show appended data from three independent 1-d entrainment time series experiments and three independent 1-d free-running time series experiments, respectively. The heat map in (B) shows the average of three independent 2-d time series experiments. Data are presented as in Figure 2. Horizontal bars below the heat maps correspond to the WC entrainment protocols used in the time series experiments; red, blue, and light blue bars in all panels indicate the warm, cold, and subjective warm phases, respectively.
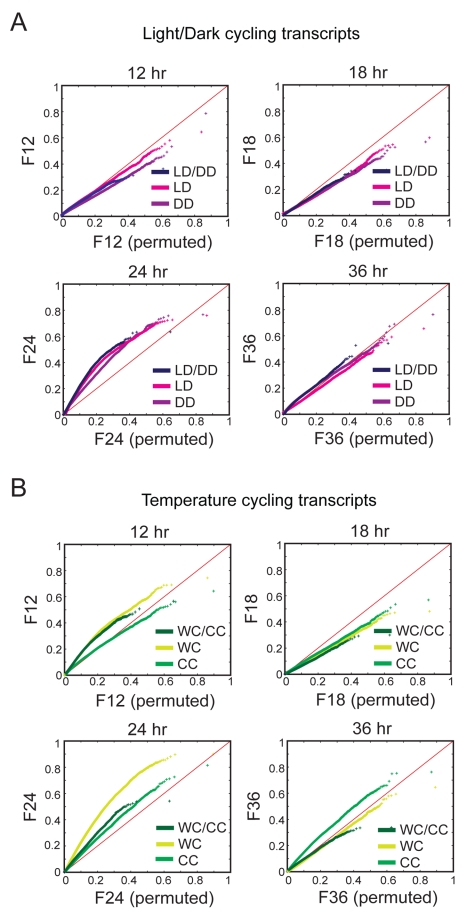
We investigated whether a 24-h circadian period is enriched in our datasets by comparing the distribution of Fourier scores with different periods with randomly permuted Fourier scores in quantile–quantile (QQ) plots. All LD datasets (LD only, LD/DD, and DD only) were significantly enriched for 24-h periods, whereas distributions of 12-h, 18-h, and 36-h period data were indistinguishable from those of randomly permuted data (Figure 4A). Although 24-h periods were also similarly enriched in the WC datasets (WC only, WC/CC, and CC only), we found lower, but significant enrichment of 12-h and 36-h periods in the WC-only and WC/CC datasets and in the CC-only datasets, respectively (Figure 4B). These results indicate robust circadian regulation of transcripts, with infradian and ultradian rhythms also observable in the temperature-regulated datasets. This pattern is similar to higher order harmonics of circadian gene expression previously identified in mouse liver [48].
Figure 4. 24-h rhythms are enriched in the light- and temperature-entrained datasets.
QQ plots of Fourier scores at four different periods (12, 18, 24, and 36 h) are shown from LD-entrained (A) and temperature-entrained (B) datasets subdivided into categories as indicated in Figures 2 and 3. The Fourier scores of each transcript (with log2-transformed expression value >10) are graphed against the randomly permuted 95th quantile Fourier scores. Enrichment of the experimental period is shown as an upward departure from the diagonal (red line), while depletion is shown as a downward departure.
Temperature Cycles Drive the Expression of a Large Set of Genes Implicated in Metabolic Processes
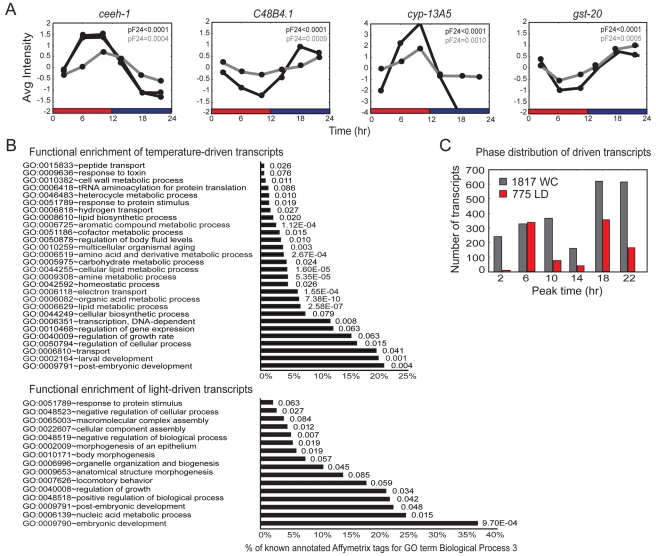
We next further analyzed the light- and temperature-driven transcripts (LD-only and WC-only datasets, respectively). The number of putative temperature-driven transcripts was significantly higher than the number of light-driven transcripts: 1,817 WC-driven compared to 775 LD-driven transcripts (Figures 2A and 3A). These comprise approximately 9% and 4% of all C. elegans genes, respectively, underscoring the critical roles of temperature and light in regulating C. elegans physiology.
We verified rhythmic expression of selected genes by performing quantitative reverse transcription PCR (qRT-PCR) analyses. Each of the 12 examined genes from the WC-only dataset exhibited significant cycling, with phases consistent with those identified in the transcriptional profiling experiments (Figures 5A and S1A). Expression was arrhythmic in the absence of imposed temperature cycles (Figure S1B). These driven genes are predicted to be implicated in diverse biological pathways (Figure 5B and Table S2). Based on the biological process Gene Ontology (GO) terms [49], categories of metabolism and electron transport were markedly enriched in the WC-only dataset (Figure 5B and Table S2). For instance, the expression of 18 genes predicted to encode cytochrome P450 enzymes was driven by WC cycles, whereas only two cytochrome P450 genes were present in the LD-driven dataset (Table S2). This suggests that temperature, but not light, modulates the expression of genes implicated in global regulation of metabolism or physiology. Genes associated with the GO term embryonic development were enriched in the LD-only-driven dataset (Figure 5B and Table S2).
Figure 5. Analyses of light- and temperature-driven transcripts.
(A) Comparison of qRT-PCR (gray) and microarray expression array (black) data for four arbitrarily selected WC-driven transcripts. Additional transcripts are shown in Figure S1. The probability of significance of circadian cycling (pF 24) as compared to a randomized dataset was calculated by appending each independent microarray or qRT-PCR time course experiment. Bars below the graphs denote the entrainment protocols, with red and blue bars indicating the warm (25°C) and cold (15°C) phases, respectively. Data shown are an average of three biologically independent replicates per time point for the microarray data, and two biologically independent replicates per time point for the qRT-PCR data. Data from two probe sets on the GeneChips for ceeh-1 are shown. (B) Functional enrichment of LD- and WC-driven transcripts. Transcripts are grouped by the Biological Process 3 GO term and analyzed for enrichment relative to all transcripts present on the GeneChip (see Materials and Methods). p-values for enrichment for each group are indicated. A complete list is provided in Table S2. (C) Histogram showing the estimated peak phases of the temperature- or light-driven transcripts. The peak phases are organized into 4-h phase groups.
We ordered the transcripts in each dataset by the average phase of peak expression (Figure 5C). We found that the peak phases were around the middle of the thermo–light or the cryo–dark phases. For LD transcripts, there were similar numbers of transcripts at these two phases, whereas more WC transcripts peaked during the cryophase than in the thermophase (Figure 5C), similar to previous observations in Drosophila [47]. Together, these results indicate that temperature and light cycles drive global changes in gene expression in C. elegans.
Putative Circadian Transcripts Represent Diverse Biological Functions
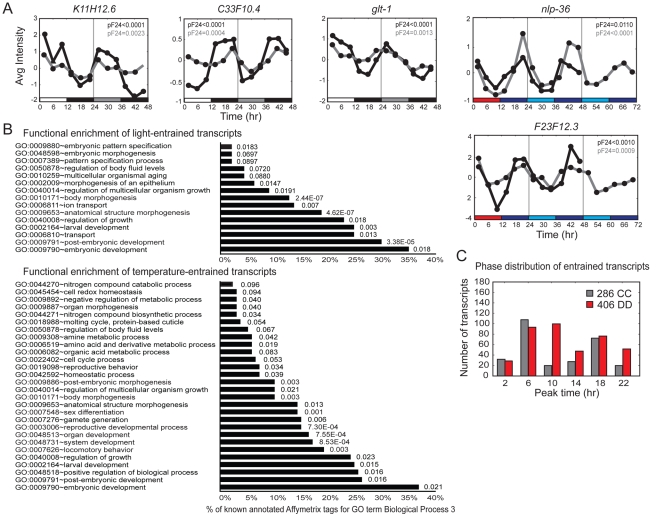
In total, 406 and 286 transcripts were found to show oscillatory expression during the DD and CC free-running conditions, respectively (see Materials and Methods). The subset of these transcripts that cycle both during the LD and WC entrainment conditions and during free-running conditions may be driven as well as entrained. We verified the cycling of selected transcripts by performing qRT-PCR (Figures 6A and S2A). Their phases were similar to those defined via expression profiling, and cycling was similarly dependent on prior entrainment (Figure S2B). Following entrainment, circadian genes sustain cycling in constant conditions. albeit with a considerably diminished amplitude in some systems over extended time periods. We therefore examined the expression of two temperature-entrained genes (nlp-36 and F23F12.3) through 2 d of free-running conditions. Both transcripts continued to exhibit rhythmic transcription during the second free-running day, with highly significant 24-h spectral power values (Figure 6A).
Figure 6. Analyses of light- and temperature-entrained circadian transcripts.
(A) Comparison of qRT-PCR (gray) and microarray expression array (black) data of randomly selected LD- and WC-entrained transcripts. Additional transcripts are shown in Figure S2. Bars below the graphs denote the entrainment protocols, with white, black, and gray bars indicating the light, dark, and subjective light phases, respectively, and with red, blue, and light blue indicating the warm (25°C), cold (15°C), and subjective warm phases, respectively. Data shown are an average of three biologically independent replicates per time point for the microarray data and two biologically independent replicates per time point for the qRT-PCR data. (B) Functional enrichment of light- and temperature-entrained circadian transcripts, as shown in Figure 5B. A complete list is provided in Table S3. (C) Histogram showing the estimated peak phases of the temperature- or light-entrained transcripts. The peak phases are organized into 4-h phase groups.
Genes associated with the GO term categories of post-embryonic morphogenesis and development were significantly enriched in both circadian datasets, in contrast to the light- and temperature-driven transcripts (Figure 6B and Table S3). We noted that genes implicated in reproductive development, and to a lesser extent locomotion, were enriched in the temperature-entrained dataset, suggesting that processes such as mating, fertilization, egg-laying, and locomotor behaviors may be under circadian control.
To determine the phase distribution of the light- and temperature-entrained transcripts, we grouped the cycling transcripts identified in the DD- and CC-entrained datasets in 4-h clusters according to their phases of peak expression. The peak phases of circadian transcripts in the light-entrained dataset occurred at around the middle of the light and dark phases, although with a relatively broad distribution (Figure 6C). The phase distribution for temperature-entrained transcripts exhibited a narrower peak in the middle of the thermophase, with another smaller peak in the middle of the cryophase. Although these distributions were not identical to those observed in the driven data, there were some similarities: they both peaked in the middle of the light (warm) and dark (cold) phases and with similar numbers of LD-entrained or -driven transcripts in the two peaks. However, more WC-driven transcripts peaked in the cryophase whereas more WC-entrained transcripts peaked in the thermophase (compare Figures 5C and 6C).
The Periods of Putative Circadian Transcripts Are Temperature-Compensated, and Their Phases Are Altered by T-Cycles
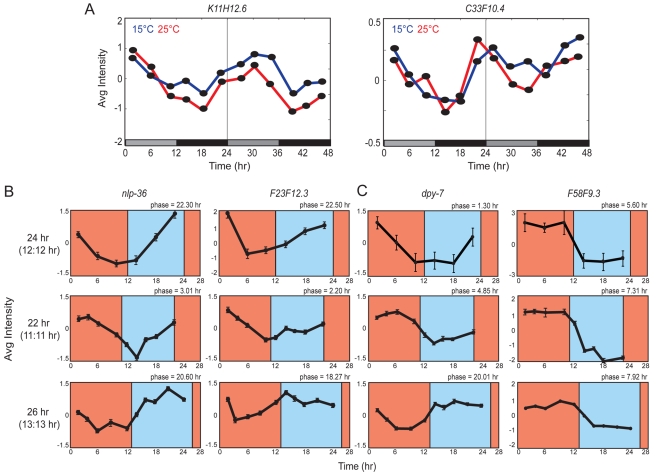
A hallmark of circadian rhythms is temperature compensation, i.e., the circadian period is almost independent of temperature [50],[51]. To compare periods of two light-entrained transcripts upon growth at 15°C and 25°C, we examined transcript levels via qRT-PCR. The calculated periods and significant periodicities for each transcript were nearly identical at both temperatures (Figure 7A and Table S4), indicating temperature compensation.
Figure 7. Temperature compensation and phase shifts of entrained transcripts.
(A) Cycling of two light-entrained transcripts during the first 2 d of free-running conditions at 15°C (blue line) or 25°C (red line) quantified via qRT-PCR. Calculated periods are as follows: K11H12.6, 23.5±0.40 h at 15°C and 23.90±0.70 at 25°C; C33F10.4, 23.35±0.25 h at 15°C and 23.45±0.25 h at 25°C. Fourier scores and associated probabilities are shown in Table S4. The bars below the graphs denote the entrainment protocol, with black and gray bars indicating the dark and subjective light phases, respectively. Data shown are from two biological replicates. (B and C) Phase shifts upon entrainment to different T-cycles. Shown is the expression of two genes each from the WC-entrained (B) and WC-driven (C) datasets as quantified from GeneChip or qRT-PCR data. RNA was collected on the fourth entrainment day. The expression of additional driven transcripts is shown in Figure S3. Data shown are the average of two technical replicates from one biological experiment.
Another characteristic of circadian rhythms is the maintenance of a phase relationship with the period (T) of the zeitgeber [52]–[55]. Typically, phases are delayed relative to those in 24-h T-cycles upon entrainment to T-cycles shorter than 24 h, and, conversely, phases are advanced when entrained to T-cycles longer than 24 h. We entrained animals to either 11 h∶11 h or 13 h∶13 h WC cycles and examined the phases of two candidate circadian transcripts by qRT-PCR. The phases of both transcripts were delayed in response to 11 h∶11 h T-cycles and advanced in response to 13 h∶13 h T-cycles (Figure 7B), further suggesting that these transcripts are under circadian regulation.
Light–Dark and Temperature Cycles Entrain Independent Gene Sets
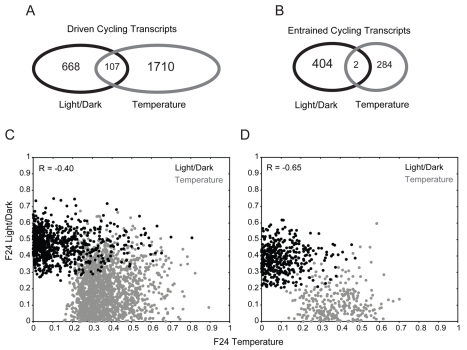
We next compared the two pairs of datasets driven and entrained by light and temperature cycles, to determine the extent to which the two zeitgebers regulate similar sets of genes. A direct comparison of the light- and temperature-driven datasets identified 107 transcripts in common, representing 106 genes (Figure 8A). In the entrained sets, there were only two genes in common (Figure 8B). One of these genes is predicted to be a pseudogene (Y102A5C.6), and the second gene (swd-3.3) is predicted to encode a homolog of the WDR5 member of the histone methyltransferase complex. Interestingly, this protein is known to associate with mammalian PER1 and regulate its functions [56].
Figure 8. Temperature and light entrain and drive the expression of distinct genes.
(A and B) Overlap between light- and temperature-driven cycling transcripts (A), and light- and temperature-entrained cycling transcripts (B). (C and D) Scatter plots of the F 24 scores of each transcript in the driven (C) or the entrained (D) datasets. F 24 scores for light–dark and temperature regulation are shown in black and gray, respectively.
Since the extent of overlap between the datasets is highly dependent on the applied filters and thresholds, we also examined the correlation between the F 24 scores for light–dark and temperature regulation for each transcript in the driven and entrained datasets. We found strong negative correlation in both cases (Figure 8C and 8D), further implying that light–dark and temperature cycles regulate independent sets of genes.
We considered the possibility that the lack of overlap between the entrained datasets may be due to the presence of additional circadian transcripts in the driven datasets. The expression of these transcripts may dampen quickly in constant conditions, precluding their inclusion in the entrained datasets. These transcripts may be distinguished from truly driven genes by being phase-shifted in response to T-cycles that are longer or shorter than 24 h. To address this hypothesis, we subjected seven arbitrarily selected transcripts from the WC-only-driven datasets to 11 h∶11 h or 13 h∶13 h T-cycles. Although the phases of six of seven of these transcripts did not exhibit the expected shifts, the phase of dpy-7 was advanced or delayed similarly to the examined entrained transcripts (Figures 7C and S3). This observation suggests that the driven datasets may include additional entrained genes, some of which may be entrained by both light and temperature.
Clock Gene Homologs in C. elegans Do Not Exhibit Rhythmic Expression in Whole Animal Profiling Experiments
Although a subset of core clock genes, such as the doubletime casein kinase gene, are expressed constitutively [57], genes such as per or tim exhibit circadian transcription in Drosophila [23]–[26]. The C. elegans genome is predicted to encode homologs of core clock genes implicated in circadian regulation in Drosophila and mammals (www.wormbase.org) [21], and animals mutant for the per homolog lin-42 exhibit an increased circadian period in locomotor behavior upon entrainment to light or temperature cycles [19]. However, upon entrainment by either light or temperature zeitgebers, no significant rhythmicity was found for the expression of the lin-42 (per), tim-1 (tim), aha-1 (clk/cyc), or atf-2 (vri) genes (Table S5). We also did not observe changes in green fluorescent protein (gfp) expression upon temperature entrainment of wild-type animals transgenic for atf-2p::gfp and aha-1p::gfp fusion genes (data not shown). It remains possible, however, that clock gene homologs cycle in a small subset of cells or tissues in C. elegans, and/or that these genes exhibit rhythmic expression at the post-transcriptional level.
Cycling Expression of the nlp-36 Neuropeptide Gene Can Be Monitored In Vivo
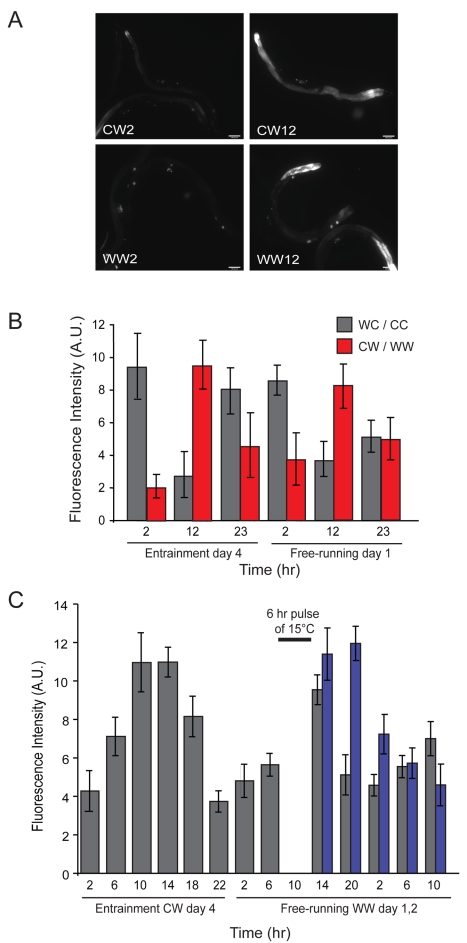
To monitor the rhythmic expression of candidate entrained genes in vivo in C. elegans, we established a real-time gfp-based reporter system. Expression of the nlp-36 neuropeptide gene exhibits strong temperature-entrained rhythmic expression (Figure 6A and Table S3). We generated strains carrying stably integrated transgenes in which the nlp-36 promoter drives expression of the gfp reporter gene fused to sequences encoding a rapidly turning over PEST domain [58]. The nlp-36p::gfp fusion gene was expressed in the intestine and additional unidentified cells in the head and tail in adult animals (Figure 9A). We subjected transgenic animals to either cold/warm (CW) or WC cycles and quantified GFP fluorescence levels in the intestine at defined time points.
Figure 9. Expression of an nlp-36p::gfp fusion gene is entrained by temperature cycles.
(A) gfp expression in transgenic animals carrying stably integrated copies of an nlp-36p::gfp fusion gene during temperature entrainment (CW) and free-running (WW) conditions. Adult animals were examined under 100× magnification. (B) Quantification of intestinal nlp-36p::gfp fluorescence intensity (in arbitrary units [A.U.]) at the indicated time points during entrainment or free-running days. Wild-type transgenic animals were entrained to either WC/CC (gray bars) or CW/WW (red bars) cycles. Error bars indicate the s.e.m. Data shown are from two independent experiments with 15–20 animals at each time point. (C) nlp-36p::gfp-expressing transgenic animals were subjected to CW entrainment cycles and moved to WW conditions. After 6 h at WW, a subset of animals were subjected to a pulse of 15°C temperature for 6 h and returned to WW conditions. Intestinal fluorescence intensities in animals subjected or not subjected to the cold pulse are indicated in blue and gray, respectively. Error bars indicate the standard deviation. n = 15–20 animals each at each time point.
nlp-36p::gfp expression cycled robustly both during entrainment and under free-running conditions, with the expected opposite relative phases in response to CW or WC cycles (Figure 9B). Moreover, cycling persisted for at least two subsequent days in free-running conditions, albeit with dampened amplitude (Figure 10A). Consistent with the absence of nlp-36 in the set of light-entrained transcripts, we did not observe cycling of nlp-36p::gfp with light entrainment (Figure S4).
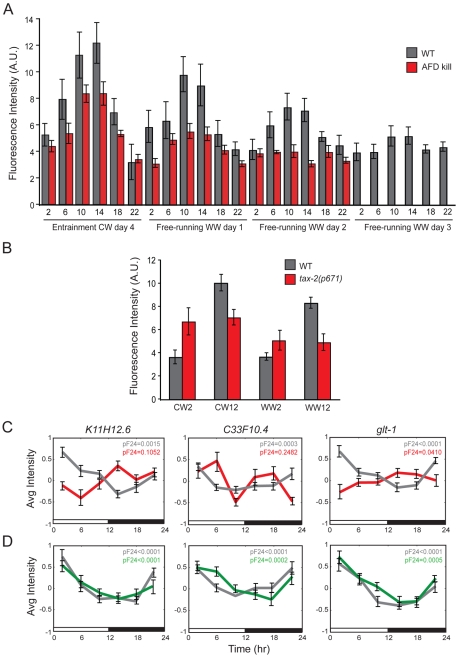
Figure 10. Rhythmic expression of nlp-36p::gfp is abolished in tax-2 mutants.
(A) Quantification of intestinal nlp-36p::gfp expression levels in wild-type (gray bars) or AFD-killed animals (red bars) at defined time points during entrainment (CW) followed by 3 d of free-running (WW) conditions. Error bars indicate the s.e.m. Data shown are from two independent experiments of 20 animals each. (B) Fluorescence intensities in wild-type animals (gray bars) or tax-2 mutant animals (red bars) carrying the nlp-36p::gfp transgene entrained to temperature cycles (CW/WW). Error bars indicate the s.e.m. Data shown are from two independent experiments. (C) Cycling of selected light-entrained transcripts in wild-type animals (gray) and in tax-2(p671) mutants (red) quantified via qRT-PCR. The probabilities of circadian cycling (pF 24) by appending each independent qPCR time course experiment are indicated. (D) Cycling of selected light-entrained transcripts in wild-type animals (gray) and in lite-1(ce314) mutants (green) quantified via qRT-PCR. The probabilities of circadian cycling (pF 24) are indicated. The bars below the graphs in (C) and (D) denote the entrainment protocol, with white and black bars indicating the light and dark phases, respectively. Data shown are from two independent biological replicates per time point. Error bars indicate the s.e.m. n = 15–20 animals each.
To determine whether the phase of cycling nlp-36p::gfp expression could be reset upon exposure to a different entraining zeitgeber schedule, we entrained nlp-36p::gfp-expressing transgenic animals to 12 h∶12 h CW cycles for 4 d. The animals were then shifted to 25°C conditions for 6 h, subjected to cold temperatures (15°C) for 6 h, and returned to 25°C. Quantification of intestinal gfp expression levels showed that administration of this temperature pulse resulted in a phase delay (Figure 9C), indicating that nlp-36 cycling is regulated by a circadian clock.
Light and Temperature Entrainment of nlp-36 Expression Requires the tax-2 Cyclic Nucleotide-Gated Channel
Molecules required to transduce light zeitgeber information to the clock have been well studied [59]–[64], although the molecular mechanisms and circuits that transduce temperature signals are less well understood. In C. elegans, the tax-2 subunit of a cyclic nucleotide-gated channel has been shown to be required for responses to temperature as well as to short-wavelength visible and ultraviolet light [42]–[44],[65],[66]. We found that temperature-entrained rhythmic expression of nlp-36p::gfp expression in the intestine was significantly affected both during entrainment and free-running conditions in tax-2 mutants (Figure 10B). Similarly, light-entrained circadian transcription of three examined genes, as measured by qRT-PCR, was also abolished in tax-2 mutants (Figures 10C), indicating that tax-2 is required for transduction of both light and temperature zeitgebers to the circadian clock(s). However, light-entrained cycling was unaffected in animals mutant for the transmembrane LITE-1 light receptor protein also shown to be required for light responses in C. elegans (Figure 10D) [42],[44],[66].
tax-2 is expressed in several sensory neuron types, including AFD thermosensory neurons, which is the primary thermosensory neuron type in C. elegans [67],[68]. We found that nlp-36p::gfp expression continued to cycle, albeit with markedly reduced amplitude, in animals in which the AFD neurons had been genetically ablated (Figure 10A; gift of Miriam Goodman, Stanford University). This result suggests that the AFD neurons are not the sole source of temperature information for the clock and that one or more tax-2-expressing sensory neurons are required to sense and transduce environmental temperature information to entrain rhythms.
Discussion
In the present study, we report the first identification, to our knowledge, of circadian-regulated transcripts in C. elegans. These transcripts are entrained by LD or WC cycles and continue to oscillate during free-running conditions with temperature-compensated periods and phases that are dependent on entraining conditions. Both light and temperature can act as zeitgebers, and they drive as well as entrain transcripts at the genome-wide level. Genome-wide circadian expression is regulated by environmental information received by a small set of sensory neuron(s), suggesting that, similar to observations in vertebrates, light and temperature information may act cell-nonautonomously to entrain the clock(s) in C. elegans. Our findings also imply that C. elegans may utilize a timekeeping mechanism that is distinct from that described previously in other animals.
In C. elegans, as well as in Drosophila, temperature cycles drive the expression of a larger set of genes than do light cycles [41],[47]. Genes implicated in metabolism appear to be enriched in the temperature-driven transcript sets, whereas genes implicated in circadian behaviors such as locomotion and reproduction are enriched in the entrained datasets in both organisms ([47] and this study). However, there is a surprising lack of overlap between the temperature- and light-entrained datasets in C. elegans. In contrast, in Drosophila, light and temperature cooperatively entrain a single transcriptional clock [47],.
What does this almost complete lack of overlap between the two C. elegans entrained datasets imply? At one extreme, light and temperature might act via different mechanisms to entrain two distinct clocks. Multiple clocks within a single organism are not unprecedented [4], although there is no evidence for a similar situation in metazoans. On the contrary, Drosophila has only a single clock that runs in multiple cells and tissues [47],[69]. Similarly, C. elegans may have a single clock that is entrained by light or temperature in different tissues. In this scenario, the different locations may have completely distinct sets of output genes, accounting for the nonoverlapping datasets. However, in this case, cycling clock mRNAs are expected to be shared in common between the two sets since in all eukaryotic circadian systems, from plants to mammals, many clock mRNAs undergo circadian oscillations at the transcriptional level [23],[24],[71],[72]. Since it is unlikely that a clock based on transcriptional feedback loops would have only one cycling mRNA (e.g., the swd-3.3 gene), C. elegans may have a novel clock mechanism. Consistent with this hypothesis, homologs of animal clock genes do not cycle in C. elegans. Recent analysis has suggested that the ascidian Ciona intestinalis also has a divergent circadian clock, further suggesting multiple evolutionary origins of the animal circadian mechanism [73]. However, it remains possible that cycling clock mRNAs were not identified in our genome-wide studies because of masking by noncycling expression in other tissue types, or that clock proteins exhibit functional rhythmicity in C. elegans via post-translational modifications [74]–[76].
An alternative possibility, consistent with the presence of a single clock, is that cycling clock mRNAs as well as common output genes are present in the two driven datasets. These datasets have a statistically significant overlap, which is expected for clock and clock-regulated mRNAs. However, these transcripts do not appear to maintain cycling in free-running conditions. Although free-running rhythms are considered to be the sine qua non for circadian genes, some bona fide circadian rhythms damp quickly in constant conditions. For example, Drosophila peripheral rhythms damp rapidly in the absence of a light–dark cycle, including transcriptional rhythms of the clock genes themselves [77],[78]. Consistent with this scenario, we identified one of seven examined transcripts in the WC-driven datasets with characteristics of an entrained rather than a strictly driven transcript. The number of entrained transcripts is, therefore, likely to be larger, and both clock mRNAs and shared output genes may be present within the set of entrained genes whose expression is severely dampened under constant conditions. Further molecular and genetic analyses will allow us to distinguish among the above possibilities.
What are the behavioral consequences of circadian rhythmicity in C. elegans? Although locomotion has been reported to be under circadian control and genes implicated in locomotor behaviors are enriched in the WC-entrained dataset, this behavioral output does not appear to be particularly robust, and exhibits marked animal-to-animal variability [16],[19]. Locomotor behaviors in C. elegans are complex and can be dissected into multiple behavioral components [79]–[83]. Reported analyses of circadian locomotor behavior relied largely on quantification of gross overall movement [16],[18], suggesting that detailed analyses of the underlying components may reveal more robust clock control. Since reproduction-related genes are also enriched in the entrained datasets, additional biologically relevant behaviors such as egg-laying or mating may also be regulated in a circadian manner in C. elegans. Identification of circadian transcriptional rhythms in C. elegans now provides the necessary reagents to characterize the molecular identity, the neuronal circuits, and the behavioral consequences of clock function in this important model organism.
Materials and Methods
C. elegans Strains and Culture Conditions
The wild-type strain used was C. elegans variety Bristol, strain N2. Animals were cultured on Escherichia coli HB101. Mutant strains used were the following: PR671 tax-2(p671) [65], KG1180 lite-1(ce314) [66], and GN112 (pgIs2) (gift of M. Goodman, Stanford University). The AFD neurons are ablated in the GN112 strain via expression of reconstituted caspases under the AFD-specific gcy-8 promoter [84],[85]. Double mutant strains were constructed using standard methods.
Entrainment Methods
Growth-synchronized populations of wild-type late L1 larvae were transferred to plates seeded with bacteria. Two populations of animals were entrained to opposing light–dark or temperature cycles in two different incubators: 12 h at 25°C followed by 12 h at 15°C (WC), or to 12 h of light followed by 12 h of dark (LD). After reaching adulthood, the two populations of animals were transferred to the same incubator and allowed to free-run at constant 15°C (CC) or in constant darkness (DD). The time point 0 h indicates lights on or the start of the warm phase, 12 h indicates lights off or the start of the cold phase. Temperature cycling experiments were conducted in DD in temperature programmable incubators (Tritech Research). Light–dark cycling experiments were conducted at 18°C in programmable incubators (Precision Scientific; cold white light [light source F20T12/CW] at about 500–1,000 lux). To inhibit progeny production during entrainment, L4 larvae were quickly transferred to plates containing bacteria and 25 µM FUDR. Adults were harvested and washed in cold 1× M9 buffer (<15 min) every 4 h during the last day of entrainment and subsequent free-running days.
Microarray Hybridization
Trizol Reagent (Invitrogen) was added to harvested worm samples and pellets were immediately frozen in liquid nitrogen. Extracted total RNA was treated with DNAase (New England Biolabs) as described [86]. cDNA synthesis was performed using T7-oligo(dT) primer and Superscript II (Affymetrix). Biotin-labeled cRNA was generated using the GeneChip IVT labeling kit (Affymetrix) and following the methods described in the Affymetrix GeneChip manual. Hybridization, washing, staining, and scanning of the cRNAs to the Affymetrix C. elegans Genome Arrays were performed as recommended by the manufacturer.
Microarray Data Analysis
Expression, normalization, and standardization
The Affymetrix software was used to scan and generate .DAT and .CEL files of microarrays. Raw data can be accessed in Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/; accession number GSE23528). The GeneChip Robust Multiarray Averaging method in the R v2.7.1 software package was used to derive log2 expression values for each probe set from Affymetrix-generated .CEL files. Expression values were normalized and standardized as described [87]. Briefly, for each time point in a time series experiment, the expression level relative to the mean (in log2 expression values) over that experiment was calculated. This normalization was performed separately for each independent time series experiment. Arrays in the independent time course experiments were standardized by setting the mean expression of each array to 0 and the variance to 1 [87]. Transcripts with low average signal intensities were excluded from further analyses.
ANOVA prescreening
Normalized and standardized expression values from each independent time series experiment were prescreened with ANOVA using a custom-written MATLAB code [87] (generously provided by K. Keegan). For each transcript on the arrays, a single-factor ANOVA was performed across all six time points during the entrainment or the free-running days, where each time point is a group and each array is an individual within the group. An ANOVA p-value of less than 0.05 was required for a transcript to pass prescreening.
Fourier analysis
Circadian oscillatory transcripts with 24-h periods were identified by Fourier transformation. Normalized and standardized data (see above) for each independent time series experiment were appended, and the 24-h spectral power (F 24) was determined for each transcript. The spectral power score and significance of this power (power p-value) generated via comparison with a randomly permuted dataset (1,000 permutations) for all analyzed periods were obtained using a custom-written MATLAB code [87] (provided by K. Keegan). We elected to use a single ordering of the days in the appended profile, since these analyses (with the exception of the AC24 score, see below) are insensitive to the order of the days in the appended time series experiments. Transcripts that exhibited pF 24-values of less than 0.02 were analyzed further.
Autocorrelation
An autocorrelation (AC24) method was used to measure the correlation between time points that are 24 h apart by fitting the six time points collected during the entrainment day to the six time points collected during the free-running day. A previously described R code [46] was used to calculate AC24 scores. Transcripts that passed this analysis were required to have an average AC24 score determined from three independent 2-d time series experiments of greater than zero.
False discovery rate analysis
To estimate the false positive discovery rate, the expression values of time series experiments for all datasets were randomly permuted (see Table S1). These randomly permuted data were used to perform ANOVA and F 24 analysis as described above. The percentage of false discovery rate in Table S1 represents the percentage of unpermuted transcripts that were identified in the randomly permuted dataset.
Fold change
Transcripts exhibiting an average fold change of greater than 1.5 (log2-transformed expression values) between their highest and lowest expression values among the six time points of a time series experiment were selected.
QQ Plots
QQ plots were generated to determine whether particular periods are enriched in the microarray datasets [41]. Briefly, 1,000 permutations of the ordering of time points for all transcripts within each appended time series experiment (LD only, DD only, LD/DD, WC only, CC only, and WC/CC datasets) were conducted, and the permuted Fourier score for the resulting time series experiments was calculated. The 1,000 permuted Fourier scores were then divided into a number of quantiles equal to the number represented in the unpermuted dataset. The distributions of Fourier scores found in unpermuted data and the permuted data quantiles were then compared and shown in QQ plots.
Gene Functional Annotation Analysis
Candidate-driven and circadian-entrained transcripts were grouped by their Biological Process 3 GO term and analyzed for enrichment relative to all transcripts present on the GeneChip using functional annotation tools in DAVID (http://david.abcc.ncifcrf.gov/).
qRT-PCR
cDNA synthesis was performed from total RNA using T7-oligo(dT) primer and Superscript III (Invitrogen). Real-time qRT-PCR analysis was performed using a Corbett Research Rotor-Gene 3000 thermal cycler. Normalization was carried out using the act-3 actin or the ard-1 short-chain alcohol dehydrogenase genes (expression levels of these genes do not appear to cycle). Primer sequences are available upon request. pF 24-values were assessed as described above.
In Vivo Monitoring of gfp-pest Expression
About 3.5-kb upstream sequences of the nlp-36 predicted neuropeptide gene were fused to gfp-encoding sequences that included a PEST domain [58]. Multiple copies of the nlp-36p::gfp-pest transgene were stably integrated into the genome of wild-type animals together with the unc-122p::dsRed marker to generate the PY7644 (oyIs80) strain. To obtain a growth-synchronized population of L1 larvae, ten adult animals were transferred to seeded plates and were allowed to lay approximately 50 eggs. Wild-type animals, tax-2 mutants, and AFD-killed L1 larvae carrying the oyIs80 transgene were entrained for 4 d to temperature cycles (12 h∶12 h) under well-fed conditions, and intestinal nlp-36p::gfp-pest fluorescence intensity was determined at specific time points during entrainment and free-running days. No FUDR was added during the entrainment protocol. Images of 15–20 adult animals were acquired at 100× magnification using a compound microscope (Zeiss) equipped with epifluorescence and a CCD camera (Hamamatsu Photonics) at each time point. Levels of intestinal expression were quantified in arbitrary units using Image J (National Institutes of Health).
Supporting Information
Analysis of temperature-driven transcripts. (A) Comparison of qRT-PCR (gray) and microarray expression array (black) data of randomly selected WC-driven transcripts. The probability of significance of circadian cycling (pF 24) as compared to a randomized dataset was calculated by appending each independent dataset. There are two probe sets each for acs-2 and cyp-35C1 on the GeneChips. Bars below the graphs denote the entrainment protocols, with red and blue bars indicating the warm (25°C) and cold (15°C) phases, respectively. (B) qRT-PCR data of temperature-driven transcripts under constant conditions (15°C). Data shown are an average of three biologically independent replicates per time point for the microarray data and two biologically independent replicates per time point for the qRT-PCR data.
(0.70 MB TIF)
Analysis of light- and temperature-entrained transcripts. (A) Comparison of qRT-PCR (gray) and microarray expression array (black) data of arbitrarily selected light- and temperature-entrained transcripts. The probability of significance of circadian cycling (pF 24) as compared to a randomized dataset was calculated by appending each independent microarray or qRT-PCR time course experiment. Bars below the graphs denote the entrainment protocols, with white, black, and gray bars indicating the light, dark, and subjective light phases, respectively, and with red, blue, and light blue bars indicating the warm (25°C), cold (15°C), and subjective warm phases, respectively. Data shown are an average of three biologically independent replicates per time point for the microarray data, and two biologically independent replicates per time point for the qRT-PCR data. (B) qRT-PCR data of light- and temperature-entrained transcripts under constant conditions. Data shown are an average of two biologically independent replicates per time point for the microarray data, and two biologically independent replicates per time point for the qRT-PCR data, with the exception of K11H12.6 and C33F10.4, for which data from one experiment were analyzed.
(0.80 MB TIF)
Phases of transcripts from the WC-driven datasets. Animals were entrained to the indicated T-cycles for 3 d and RNA was collected on the fourth day. qRT-PCR data for each time point are the average of two technical replicates from one biological experiment.
(0.84 MB TIF)
nlp-36 p ::gfp expression does not cycle upon light entrainment. Fluorescence intensities in wild-type animals (gray bars) or tax-2 mutant animals (red bars) carrying the nlp-36p::gfp transgene entrained to light cycles (LD/DD). Error bars indicate the standard error of the mean (s.e.m). Data shown are from two independent experiments.
(0.18 MB TIF)
False discovery rate analysis of datasets.
(0.11 MB DOC)
GO categories and lists of all light- and temperature-driven transcripts.
(1.18 MB XLS)
GO categories and lists of all light- and temperature-entrained transcripts.
(0.47 MB XLS)
Fourier analyses of periods at different temperatures.
(0.06 MB DOC)
Transcripts of C. elegans clock genes do not oscillate in a circadian manner.
(0.06 MB DOC)
Acknowledgments
We are grateful to Felix Naef, Herman Wijnen, and Kevin Keegan for help with data analyses, Rinho Kim for technical assistance, and Miriam Goodman for reagents.
Abbreviations
- CC
constant cold
- CW
cold/warm
- DD
constant darkness
- FUDR
5-fluoro-2-deoxyuridine
- gfp
green fluorescent protein
- GO
Gene Ontology
- LD
light/dark
quantile–quantile
- qRT-PCR
quantitative reverse transcription PCR
- s.e.m.
standard error of the mean
- WC
warm/cold
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the National Institutes of Health (RO1 GM081639 to PS; P30 NS045713, PO1 044232 to MR; F31 NS060584 to SW), a National Science Foundation IGERT training grant (DGE-0549390 to MB and JPR), a Human Frontiers Science Program Long-term Fellowship and Career Development Award (SK), and the Howard Hughes Medical Institute (MR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Young M. W, Kay S. A. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 2.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo T. A cyanobacterial circadian clock based on the Kai oscillator. Cold Spring Harb Symp Quant Biol. 2007;72:47–55. doi: 10.1101/sqb.2007.72.029. [DOI] [PubMed] [Google Scholar]
- 4.Bell-Pedersen D, Cassone V. M, Earnest D. J, Golden S. S, Hardin P. E, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu W, Hardin P. E. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 6.Johnson C. H, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allada R, Emery P, Takahashi J. S, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- 8.Allada R, Chung B. Y. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bono M, Maricq A. V. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- 10.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:e129. doi: 10.1371/journal.pgen.0030129. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conradt B. Genetic control of programmed cell death during animal development. Annu Rev Genet. 2009;43:493–523. doi: 10.1146/annurev.genet.42.110807.091533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramot D, MacInnis B. L, Lee H. C, Goodman M. B. Thermotaxis is a robust mechanism for thermoregulation in Caenorhabditis elegans nematodes. J Neurosci. 2008;28:12546–12557. doi: 10.1523/JNEUROSCI.2857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson A. F. Movement of five nematode species through sand subjected to natural temperature gradient fluctuations. J Nematol. 1994;26:46–58. [PMC free article] [PubMed] [Google Scholar]
- 15.Kiontke K, Sudhaus W. Ecology of Caenorhabditis species. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saigusa T, Ishizaki S, Watabiki S, Ishii N, Tanakadate A, et al. Circadian behavioural rhythm in Caenorhabditis elegans. Curr Biol. 2002;12:R46–R47. doi: 10.1016/s0960-9822(01)00669-8. [DOI] [PubMed] [Google Scholar]
- 17.Kippert F, Saunders D. S, Blaxter M. L. Caenorhabditis elegans has a circadian clock. Curr Biol. 2002;12:R47–R49. doi: 10.1016/s0960-9822(01)00670-4. [DOI] [PubMed] [Google Scholar]
- 18.Simonetta S. H, Golombek D. A. An automated tracking system for Caenorhabditis elegans locomotor behavior and circadian studies application. J Neurosci Methods. 2007;161:273–280. doi: 10.1016/j.jneumeth.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Simonetta S. H, Migliori M. L, Romanowski A, Golombek D. A. Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS ONE. 2009;4:e7571. doi: 10.1371/journal.pone.0007571. doi: 10.1371/journal.pone.0007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonetta S. H, Romanowski A, Minniti A. N, Inestrosa N. C, Golombek D. A. Circadian stress tolerance in adult Caenorhabditis elegans. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194:821–828. doi: 10.1007/s00359-008-0353-z. [DOI] [PubMed] [Google Scholar]
- 21.Jeon M, Gardner H. F, Miller E. A, Deshler J, Rougvie A. E. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 22.McDonald M. J, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardin P. E, Hall J. C, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci U S A. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers M. P, et al. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 25.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 26.Shearman L. P, Zylka M. J, Weaver D. R, Kolakowski L. F, Jr, Reppert S. M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 27.Gallego M, Virshup D. M. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 28.Mehra A, Baker C. L, Loros J. J, Dunlap J. C. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Garceau N. Y, Loros J. J, Dunlap J. C. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 30.Garceau N. Y, Liu Y, Loros J. J, Dunlap J. C. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 32.Kadener S, Menet J. S, Sugino K, Horwich M. D, Weissbein U, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 34.Ceriani M. F, Hogenesch J. B, Yanovsky M, Panda S, Straume M, et al. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald M. J, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 36.Ueda H. R, Matsumoto A, Kawamura M, Iino M, Tanimura T, et al. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 37.Akhtar R. A, Reddy A. B, Maywood E. S, Clayton J. D, King V. M, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 38.Correa A, Lewis Z. A, Greene A. V, March I. J, Gomer R. H, et al. Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci U S A. 2003;100:13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duffield G. E, Best J. D, Meurers B. H, Bittner A, Loros J. J, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 40.Nowrousian M, Duffield G. E, Loros J. J, Dunlap J. C. The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young M. W. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward A, Liu J, Feng Z, Xu X. Z. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat Neurosci. 2008;11:916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hedgecock E. M, Russell R. L. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1975;72:4061–4065. doi: 10.1073/pnas.72.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Ward A, Gao J, Dong Y, Nishio N, et al. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat Neurosci. 2010;13:715–722. doi: 10.1038/nn.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell D. H, Stiles J. W, Santelli J, Sanadi D. R. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- 46.Wijnen H, Naef F, Young M. W. Molecular and statistical tools for circadian transcript profiling. Methods Enzymol. 2005;393:341–365. doi: 10.1016/S0076-6879(05)93015-2. [DOI] [PubMed] [Google Scholar]
- 47.Boothroyd C. E, Wijnen H, Naef F, Saez L, Young M. W. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007;3:e54. doi: 10.1371/journal.pgen.0030054. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes M. E, DiTacchio L, Hayes K. R, Vollmers C, Pulivarthy S, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner M, Ball C. A, Blake J. A, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardin P. E. Essential and expendable features of the circadian timekeeping mechanism. Curr Opin Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Hastings J. W, Sweeney B. M. On the mechanism of temperature independence in a biological clock. Proc Natl Acad Sci U S A. 1957;43:804–811. doi: 10.1073/pnas.43.9.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roenneberg T, Dragovic Z, Merrow M. Demasking biological oscillators: properties and principles of entrainment exemplified by the Neurospora circadian clock. Proc Natl Acad Sci U S A. 2005;102:7742–7747. doi: 10.1073/pnas.0501884102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merrow M, Brunner M, Roenneberg T. Assignment of circadian function for the Neurospora clock gene frequency. Nature. 1999;399:584–586. doi: 10.1038/21190. [DOI] [PubMed] [Google Scholar]
- 54.Aschoff J, Pohl H. Phase relations between a circadian rhythm and its zeitgeber within the range of entrainment. Naturwissenschaften. 1978;65:80–84. doi: 10.1007/BF00440545. [DOI] [PubMed] [Google Scholar]
- 55.Roenneberg T, Daan S, Merrow M. The art of entrainment. J Biol Rhythms. 2003;18:183–194. doi: 10.1177/0748730403018003001. [DOI] [PubMed] [Google Scholar]
- 56.Brown S. A, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 57.Kloss B, Price J. L, Saez L, Blau J, Rothenfluh A, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 58.Frand A. R, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berson D. M, Dunn F. A, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- 60.Hattar S, Lucas R. J, Mrosovsky N, Thompson S, Douglas R. H, et al. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature. 2003;424:76–81. doi: 10.1038/nature01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 62.Emery P, So W. V, Kaneko M, Hall J. C, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 63.van der Horst G. T, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 64.Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, et al. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- 65.Coburn C, Bargmann C. I. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 66.Edwards S. L, Charlie N. K, Milfort M. C, Brown B. S, Gravlin C. N, et al. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 2008;6:e198. doi: 10.1371/journal.pbio.0060198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prahlad V, Cornelius T, Morimoto R. I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- 69.Yoshii T, Vanin S, Costa R, Helfrich-Forster C. Synergic entrainment of Drosophila's circadian clock by light and temperature. J Biol Rhythms. 2009;24:452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- 70.Matsumoto A, Matsumoto N, Harui Y, Sakamoto M, Tomioka K. Light and temperature cooperate to regulate the circadian locomotor rhythm of wild type and period mutants of Drosophila melanogaster. J Insect Physiol. 1998;44:587–596. doi: 10.1016/s0022-1910(98)00046-8. [DOI] [PubMed] [Google Scholar]
- 71.Gekakis N, Staknis D, Nguyen H. B, Davis F. C, Wilsbacher L. D, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 72.Alabadi D, Oyama T, Yanovsky M. J, Harmon F. G, Mas P, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 73.Minamoto T, Hanai S, Kadota K, Oishi K, Matsumae H, et al. Circadian clock in Ciona intestinalis revealed by microarray analysis and oxygen consumption. J Biochem. 2009;147:175–184. doi: 10.1093/jb/mvp160. [DOI] [PubMed] [Google Scholar]
- 74.Cha J, Huang G, Guo J, Liu Y. Posttranslational control of the Neurospora circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:185–191. doi: 10.1101/sqb.2007.72.010. [DOI] [PubMed] [Google Scholar]
- 75.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harms E, Kivimae S, Young M. W, Saez L. Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- 77.Hardin P. E. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plautz J. D, Kaneko M, Hall J. C, Kay S. A. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 79.Chalfie M, Sulston J. E, White J. G, Southgate E, Thomson J. N, et al. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Croll N. A. Components and patterns in the behavior of the nematode Caenorhabditis elegans. J Zool (1987) 1975;176:159–176. [Google Scholar]
- 81.Gray J. M, Hill J. J, Bargmann C. I. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pierce-Shimomura J. T, Morse T. M, Lockery S. R. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Srivastava N, Clark D. A, Samuel A. D. Temporal analysis of stochastic turning behavior of swimming C. elegans. J Neurophysiol. 2009;102:1172–1179. doi: 10.1152/jn.90952.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu S, Avery L, Baude E, Garbers D. A. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chelur D. S, Chalfie M. Targeted cell killing by reconstituted caspases. Proc Natl Acad Sci U S A. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Portman D. S The C. elegans Research Community, editor. Profiling C. elegans gene expression with DNA microarrays. WormBook. 2006. doi/10.1895/wormbook.1.104.1. [DOI] [PMC free article] [PubMed]
- 87.Keegan K. P, Pradhan S, Wang J. P, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of temperature-driven transcripts. (A) Comparison of qRT-PCR (gray) and microarray expression array (black) data of randomly selected WC-driven transcripts. The probability of significance of circadian cycling (pF 24) as compared to a randomized dataset was calculated by appending each independent dataset. There are two probe sets each for acs-2 and cyp-35C1 on the GeneChips. Bars below the graphs denote the entrainment protocols, with red and blue bars indicating the warm (25°C) and cold (15°C) phases, respectively. (B) qRT-PCR data of temperature-driven transcripts under constant conditions (15°C). Data shown are an average of three biologically independent replicates per time point for the microarray data and two biologically independent replicates per time point for the qRT-PCR data.
(0.70 MB TIF)
Analysis of light- and temperature-entrained transcripts. (A) Comparison of qRT-PCR (gray) and microarray expression array (black) data of arbitrarily selected light- and temperature-entrained transcripts. The probability of significance of circadian cycling (pF 24) as compared to a randomized dataset was calculated by appending each independent microarray or qRT-PCR time course experiment. Bars below the graphs denote the entrainment protocols, with white, black, and gray bars indicating the light, dark, and subjective light phases, respectively, and with red, blue, and light blue bars indicating the warm (25°C), cold (15°C), and subjective warm phases, respectively. Data shown are an average of three biologically independent replicates per time point for the microarray data, and two biologically independent replicates per time point for the qRT-PCR data. (B) qRT-PCR data of light- and temperature-entrained transcripts under constant conditions. Data shown are an average of two biologically independent replicates per time point for the microarray data, and two biologically independent replicates per time point for the qRT-PCR data, with the exception of K11H12.6 and C33F10.4, for which data from one experiment were analyzed.
(0.80 MB TIF)
Phases of transcripts from the WC-driven datasets. Animals were entrained to the indicated T-cycles for 3 d and RNA was collected on the fourth day. qRT-PCR data for each time point are the average of two technical replicates from one biological experiment.
(0.84 MB TIF)
nlp-36 p ::gfp expression does not cycle upon light entrainment. Fluorescence intensities in wild-type animals (gray bars) or tax-2 mutant animals (red bars) carrying the nlp-36p::gfp transgene entrained to light cycles (LD/DD). Error bars indicate the standard error of the mean (s.e.m). Data shown are from two independent experiments.
(0.18 MB TIF)
False discovery rate analysis of datasets.
(0.11 MB DOC)
GO categories and lists of all light- and temperature-driven transcripts.
(1.18 MB XLS)
GO categories and lists of all light- and temperature-entrained transcripts.
(0.47 MB XLS)
Fourier analyses of periods at different temperatures.
(0.06 MB DOC)
Transcripts of C. elegans clock genes do not oscillate in a circadian manner.
(0.06 MB DOC)