Abstract
Tight transcriptional regulation, post-translational modifications and/or alternative splicing of BH3-only proteins fine-tune their pro-apoptotic function. Here, we characterize the gene locus of the BH3-only protein Bmf (Bcl-2 modifying factor) and describe the generation of two major isoforms from a common transcript where initiation of protein synthesis involves leucine-coding CUG. BmfCUG and the originally described isoform, Bmf short (BmfS), display comparable binding affinities to pro-survival Bcl-2 family members, localize preferentially to the outer mitochondrial membrane and induce rapid Bcl-2-blockable apoptosis. Notably, endogenous Bmf expression is induced upon forms of cell stress known to cause the repression of the CAP-dependent translation machinery such as serum-deprivation, hypoxia, inhibition of the PI3K/AKT pathway or mTOR, as well as direct pharmacological inhibition of eukaryotic translation initiation factor eIF-4E. Knock-down or deletion of Bmf reduces apoptosis under some of these conditions demonstrating that Bmf can act as a sentinel for the stress-impaired CAP-dependent protein translation machinery (150).
Keywords: apoptosis, Bcl-2 family, BH3-only proteins, Bmf
Introduction
The Bcl-2 family comprises a number of cell death- and survival-promoting proteins that are characterized by certain structural motives, called Bcl-2 homology (BH) domains. The pro-survival family members such as Bcl-2 itself, Bcl-xL, Bcl-w or Mcl-1 contain up to four homology domains (BH1-4) whereas pro-apoptotic members of the same family either possess three out of four BH-domains (BH123 proteins), e.g. Bax and Bak or only the BH3-domain, such as Bim, Bad or Bmf. According to this structural feature these proteins are referred to as BH3-only proteins and at least eight members have been described to trigger cell death in mammals (1). While their exact mode of cell killing is still subject of intense investigations genetic evidence supports the notion that these molecules act upstream of Bax and Bak in cell death signalling, since mouse embryonic fibroblasts (MEFs) deficient for both of these molecules no longer die in response to BH3-only protein overexpression (2). Biochemical evidence suggests that BH3-only proteins can interact with and antagonize partially overlapping sub-sets of Bcl-2 homologues by direct interaction, allowing for activation of Bax and/or Bak, leading to their oligomerization, subsequent destabilization of mitochondrial integrity, release of apoptogenic factors, caspase activation and cell death (3). In an alternative model, members of different groups of BH3-only proteins compete for binding to Bcl-2-like anti-apoptotic molecules. Excess of so-called “de-repressors” (e.g. Bad, Bmf, Noxa) prevents sequestration of “direct activators” (Bim, Bid and Puma) by pro-survival homologues of Bcl-2 allowing them to directly interact with and activate Bax/Bak-like molecules (4, 5). Recent genetic evidence gained from BH3-domain exchange mutants in Bim suggest that BH3-only protein-mediated killing depends on aspects of both models in vivo (6). The role of BH3-only proteins in normal physiology has been addressed primarily by analyzing mouse models lacking the individual genes (7). Loss of Puma protects neurons, primary lymphocytes, myeloid cells and mouse embryonic fibroblasts (MEF) from DNA-damage induced apoptosis but also from certain p53-independent cell death stimuli such as glucocorticoids or serum-deprivation whereas Noxa appears to play a more restricted role in mediating DNA-damage responses (8, 9). Absence of Bid prevents liver failure in response to FAS-ligation, confirming the role of Bid as the connecting element between extrinsic and intrinsic cell death pathways (10, 11). Loss of Bim causes lymphadenopathy and subsequent autoimmunity mainly due to negative selection defects and acts as a suppressor of c-myc driven lymphomagenesis (12).
Little is known about the biology of Bmf that seems to be regulated similarly to Bim. Both molecules share a highly conserved dynein light chain (DLC)-binding motif near their N-termini that targets Bmf to the actin cytoskeleton and Bim to microtubules (12). Bmf can be released from the cytoskeleton in response to UV-radiation or during detachment-induced apoptosis (anoikis), which prevents epithelial cells from colonizing elsewhere (13). Both forms of cell death, however, proceed normally in MEF and/or gastrointestinal epithelial cell from bmf−/− mice, suggesting redundancy with other BH3-only proteins (14), most likely with Bim (15). In hemopoietic cells, Bmf is widely expressed and has been implicated in cytokine withdrawal-induced apoptosis of granulocytes where the protein was reported to accumulate during in vitro culture (16). Multiple isoforms can be detected in lymphocytes and hence alternative splicing of bmf may also contribute to regulate its in vivo function (14). Two splice variants of bmf (termed bmf II and III) are expressed in normal and malignant human B cells, derived from patients with B cell lymphocytic leukemia. Surprisingly, these novel variants of Bmf lacked a functional BH3-domain and Bmf III also contains a different carboxy terminus. Overexpression of Bmf II or Bmf III in HeLa cells increased their colony forming potential, whereas Bmf, as previously reported, showed pro-apoptotic activity (17). B-CLL cells undergo rapid apoptosis in vitro and this cell death correlated with enhanced mRNA expression of the full-length form of Bmf, containing the BH3-domain necessary for apoptosis induction, but not the other two isoforms (17). Consistently, genetic ablation of all Bmf isoforms expressed in mice revealed a role in the regulation of normal B cell homeostasis and irradiation (14) as well as oncogene-driven tumorigenesis (18). Finally, a role for Bmf in necroptotic cell death, triggered by TNF in the absence of functional caspases in L929 fibroblasts was reported, but its exact role in this process remains to be fully understood (19).
Here we characterize the bmf gene locus in detail, report the molecular basis of the generation of the two major isoforms of Bmf, designated BmfS and BmfCUG, and provide evidence that Bmf can act as a sensor for stress that associates with the repression of the conventional CAP-dependent translation machinery.
Results
An additional conserved translation start site in the bmf mRNA sequence allows expression of a longer Bmf isoform, BmfCUG
Blasting the mbmf cDNA obtained in the initial library screen (20) against the NCBI-HTGS database identified RP23-266K3 as one BAC clone that contains the mouse bmf locus, annotated to chromosome 2. Further analysis of intron-exon boundaries revealed a possible alternative start codon only 4 base pairs upstream of the 5′end of the longest cDNA clone that was originally identified as putative transcription start site (TSS) (Fig.1a). Inclusion of this additional 5′sequence indicated that the mouse bmf mRNA sequence (4.8kb in size) now contains a significantly longer ORF (816bp) as the one initially published (558bp) and used for the initial characterization of Bmf protein (20). Translation of this alternative open reading frame in silicio predicted a longer version of the mouse Bmf protein, here designated Bmf-long (BmfL), with an alternative extended N-terminus, approximately 30kDa in size, but otherwise identical to the originally published protein sequence, referred to as Bmf-short (BmfS).
Figure 1. Characterization of Bmf isoforms.
(A) Schematic representation of the mouse bmf gene locus located on chromosome 2 (not to scale). ATGs that may be used for translation initiation are located in exon 1 and 3, respectively. A Kozak-flanked CTG can be found in exon 2. (B) Bmf was immunoprecipitated using rat anti-Bmf 9G10 mAb from 293T cells transiently transfected with an expression plasmid encoding BmfS cDNA or splenocytes from wild type (wt) or vav-bcl-2 transgenic (tg) mice. Immunoblotting was performed using biotinylated rat anti-Bmf specific mAb (12E10). (C) BmfL cDNA in pSKblue was used in a coupled in vitro transcription translation reaction. IVVT reactions were separated by SDS-PAGE prior immunoblotting (ns=non-specific). (D) IVVT reactions of BmfL and different mutated versions were separated by SDS-PAGE and immunoblotted using anti-Bmf antibody. Thymic cell extracts from wt and bmf−/− mice served as controls. (E) Expression of different versions of epitope tagged or untagged Bmf isoforms in transiently transfected 293T cells. (F) Amino acid sequence of mouse BmfCUG/BmfS. Amino acids specific for BmfCUG are shown in red.
Screening of the NCBI mouse EST-database confirmed that transcripts containing this upstream ATG can be found in cDNA libraries, e.g. derived from mouse E13 embryonic heart muscle tissue or E12 mouse ovary, suggesting that this longer ORF may be used in vivo to generate BmfL. Consistently, two major isoform of Bmf can be found by Western analysis in lymphocyte extracts from adult mice (14) or human leukemia cells (21) and cell lines (22). Immunoprecipitation using Bmf-specific antibodies and subsequent immunoblotting revealed that two protein products were precipitated from mouse splenocytes (Fig. 1b). The smaller product, migrating at approximately 22kDa, presumably represented the originally described mBmfS, isoform, as indicated by comparison with mBmfS transiently expressed in 293T cells (Fig. 1b). The slower migrating band was approximately 26kDa in size and we initially considered it to represent BmfL. Interestingly, two additional bands were detected in lysates and IP-complexes from Bcl-2 overexpressing lymphocytes (Fig. 1b, sFig.1a), 24kDa and 20kDa in size, that are absent or only poorly expressed in most wild type lymphocyte subsets analyzed (14). These proteins may represent either low abundance splice variants of bmf, post-translationally modified versions of the protein(s) or degradation products of the two major isoforms (see below). Phosphatase treatment of cell lysates excluded phosphorylation as a possible modification (sFig. 1a). Together, these observations suggest that (co)-transcriptional modifications such as differential splicing, alternative promoter and/or start site usage, or alternative post-translational modifications must account for the generation of the multiple Bmf isoforms.
To verify that both putative start-site ATGs contained in the mbmf mRNA can be used, we generated and subcloned a cDNA encoding the ORF for BmfL into pSK-bluescript and performed a T7-driven coupled in vitro transcription/translation reaction (IVTT) in rabbit reticulocyte lysates in the presence or absence of 35S-methionine. The reaction products were separated by SDS-PAGE, exposed to x-ray film or subjected to immunoblotting using an anti-Bmf specific mAb. Three major protein products of comparable size were detected by both methods (Fig. 1c,d, not shown). Notably, two of those products migrated with the same behaviours as those recognized in lysates from mouse thymocytes. However, the longest product generated by IVTT did not appear to have an endogenous counterpart (Fig. 1c). Mutation of the second, internal ATG2 led to the disappearance of the shortest Bmf product, indicating that only BmfS is translated from this internal ATG in exon 3 and excluding the possibility that the other two products may arise form post-translational modification of BmfS (Fig. 1d). In contrast, deletion of the first ATG1, as expected, led to the disappearance of the largest protein product but the two other products, resembling in size the endogenous proteins were still produced. Since there was no other ATG in-frame with the ORF of BmfS, we searched for the presence of alternative start sites and discovered a CTG which was flanked by a strong Kozak sequence in frame with the downstream ATG2 that gives rise to BmfS. Site-directed mutagenesis confirmed that this CTG is recognized efficiently by the translation machinery and can be used to generate a longer isoform of Bmf, designated herein as BmfCUG, since mutation of this CTG caused loss of the 26kDa product which was still produced when both ATGs in this cDNA were mutated (Fig. 1d). Overexpression of a single transcript containing this putative translation initiating CTG in the context of the surrounding Kozak sequence in 293T cells allowed efficient co-expression of BmfCUG and BmfS (lane 5), while in its absence only BmfS was translated into protein (lane 2) (Fig. 1e). Epitope tagging allowed for the efficient expression of BmfS or BmfCUG individually (lanes 3 & 4). We concluded that these two protein products most likely represent the endogenous proteins recognized by our anti-Bmf antibodies in hemopoietic cells or after IVTT (Fig. 1b,d). Sequence alignment revealed that this CTG site was conserved across all mammalian species where genomic DNA of this region was contained in the Ensemble database (Fig. 2), allowing for translation of BmfCUG, containing 24 additional amino acids at its N-terminus (Fig. 1f). In contrast, the first ATG found in exon I was either not present as in horse (equus), or, in other species such as humans and chimpanzee, this ATG1 was not in frame with the downstream ATG2, used to translate BmfS (Fig. 2).
Figure 2. The non-conventional CUG-translation initiation site of Bmf is conserved among mammals.
Alignment of Bmf genomic DNA sequence derived from various mammalian species. All but equus contain the conserved exon 1 encoding a putative translation initiation site (ATG1). All available mammalian Ensemble sequences contain exon 2 with the Kozak-flanked CTG and the originally defined ATG2 translation start site in exon 3. Arrows indicate putative transcription start sites found in ESTs from embryonic ovary and heart tissue. SA = splice acceptor, SD = splice donor site.
Taken together this indicates that at least two Bmf isoforms can be translated from a single mRNA (14), due to alternative start site usage from a common open reading frame. Importantly, all these isoforms appear to contain a functional BH3-domain, since they could be co-immunoprecipitated with Bcl-2 from splenocytes overexpressing a bcl-2 transgene (sFig. 1b)
BmfCUG and BmfS bind the same Bcl-2 family proteins and show comparable apoptotic potential as well as subcellular localization
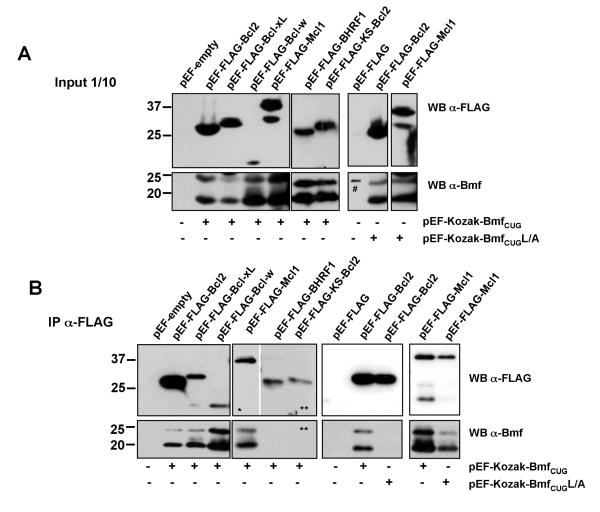
Having identified the nature of the two major isoforms expressed in lymphatic tissue we investigated if there were substantial differences in their potential to interact with known partners of the Bcl-2 family as well as in their ability to kill cells. Therefore, constructs that allow expression of either isoform alone (not shown) or both isoforms at the same time (Fig. 1e) were transiently transfected together with FLAG-tagged versions of Bcl-2, Bcl-xL, Bcl-w, Mcl-1, or the viral Bcl-2 homologues BHRF1 and KS-Bcl-2 (Fig. 3a). Co-immunoprecipitation analysis revealed that both isoforms effectively interacted with all mammalian prosurvival proteins tested, but not with their viral counterparts (Fig. 3b). As expected, introduction of a leucine to alanine mutation in the BH3-domain (L/A) abrogated interaction with Bcl-2 but we repeatedly observed residual binding of both Bmf isoforms to Mcl-1 suggesting that amino acids outside the BH3-domaine are involved in this interaction (Fig. 3b).
Figure 3. BmfS and BmfCUG show the same interaction pattern with anti-apoptotic Bcl-2 family members.
(A) BmfS and BmfCUG were overexpressed simultaneously in 293T cells together with FLAG-tagged versions of Bcl-2, Bcl-xL, Bcl-w, Mcl-1, BHRF1 or KS-Bcl2. Expression of transgene-derived protein was confirmed by immunoblotting using anti-FLAG- or anti-Bmf-specific mAbs (# = marker labeling). (B) Immunoprecipitation was performed using anti-FLAG-M2 mAb. Immune complexes were separated by SDS-PAGE on 4-20% Tris-glycine gels. After electroblotting, nitrocellulose membranes were subjected to immunoblotting using the anti-Bmf mAb (17A9). Membranes were stripped and reprobed with a rat-anti-FLAG antibody. One out of three independent experiments yielding similar results is shown. (* short exposure, ** long exposure)
To assess possible differences in their proapoptotic potential we generated a series of IL3-dependent BaF3 cells that stably expressed above-mentioned pro-survival Bcl-2 family proteins alone, or together with one or the other isoform of Bmf. Upon IL3-deprivation, BaF3 cells undergo rapid cell death that can be blocked by overexpression of anti-apoptotic Bcl-2 proteins (sFig. 2a), unless these are themselves blocked by BH3-only proteins (20). In line with their ability to bind to Bcl-2, Bcl-xL or Mcl-1 in our IP-analysis, BmfCUG or BmfS antagonized the pro-survival effect of these Bcl-2 family members upon IL3-deprivation, triggering massive apoptosis in BaF3 cells (sFig. 2). Bmf overexpression failed to antagonize the anti-apoptotic effect of the two viral Bcl-2-like proteins KS-Bcl2 and BHRF1, consistent with a lack of direct interaction in our IP-analysis (Fig. 3b). Notably, BaF3 cells co-expressing Bcl-w together with BmfS or BmfCUG resisted apoptosis upon IL3-deprivation (sFig. 2), which may have been due to extremely levels of transgenic Bcl-w that, presumably, was not neutralized effectively by overexpressed Bmf (not shown).
The collective data suggested that both Bmf isoforms are equally potent in killing but since we were unable to generate BaF3 cells that overexpressed either isoform alone (not shown) it remained possible that differences may be masked by simultaneous high-level expression of a pro-survival Bcl-2 homologue. To extend our observations to a different model system, we performed lentiviral transduction of apoptosis-sensitive U2OS osteosarcoma cells with constructs encoding for BmfS or BmfCUG, as well as the BmfS L138A or BmfCUG L162A, carrying a point mutation in the BH3-domain. Parallel transduction using a GFP-encoding virus served as a control to quantify infection efficiency (usually >90%) and treatment related cell death (usually ~ 20%). While both wild type isoforms of Bmf induced comparable apoptotic cell death, assessed 24h after transduction by Annexin-V staining or sub-G1 analysis, BH3-domain point-mutant versions of Bmf failed to induce cell death of U2OS significantly, when compared to GFP-transduced control cells (Fig. 4). Seeding identical numbers of transduced U2OS cells confirmed that both isoforms of Bmf, but not its L/A-mutant versions, strongly reduced the colony formation potential of these cells, as monitored by crystal violet staining after 5 days of culture. Again, both proteins did so with comparable efficiency (Fig. 5a). Finally, we wondered whether BmfCUG and BmfS would have different preferences for Bax or Bak, in order to kill cells. Therefore, SV40-MEF expressing or lacking bak, bax or both genes were transduced with the two Bmf isoforms alone or a construct that allows simultaneous expression of both proteins. The colony formation potential of these MEF was monitored upon selection with puromycin for one week. As shown in Fig. 5b, both isoforms reduced the colony formation of wt, Bax and Bak-deficient cells suggesting that both BH123 proteins can be engaged to mediate Bmf-induced apoptosis. BmfS appeared to kill SV40 MEF somewhat more effectively and preferentially in a Bax-dependent manner. Notably, bax−/−bak−/− MEF, despite being highly resistant to the effects of etoposide treatment, also showed reduced colony formation upon transduction with Bmf when both isoforms were expressed simultaneously from a single plasmid (Fig. 5b).
Figure 4. BmfS and BmfCUG show similar potency in inducing apoptotic cell death.
U2OS cells were transduced with lentiviruses encoding wt or inactive mutant versions (L/A) of BmfS, BmfCUG or GFP, as a control. 24h later transduction efficiency was monitored by flow-cytometric analysis of the GFP-transduced cells (usually > 90% GFP+). Microscopic images were taken (A) and cell death was quantified by either APC-Annexin-V (B) or sub-G1-staining (C) followed by flow-cytometric analysis. One out of three independent experiments yielding similar results is shown.
Figure 5. BmfS and BmfCUG reduce clonal survival in U2OS cells and can engage either Bax or Bak to kill.
(A) Lentivirally transduced U2OS cells were seeded in different cell numbers and cultured for additional 5 days. Surviving colonies were stained using crystal violet. One representative experiment out of two, yielding similar results is shown. (B) SV40-MEF proficient or deficient for Bax, Bak or both were transduced with a retrovirus encoding the puromycin resistance gene alone or in combination with BmfS or BmfCUG individually, or a plasmid allowing simultaneous translation of both isoforms (BmfS/CUG). MEF were subjected to puromycin selection (2μg/ml) for 7 days and surviving colonies were stained using crystal violet. One representative experiment out of three, yielding similar results is shown. Expression of Bax or Bak in the mutant MEF lines was monitored by Western blot. MEF expressing Bax and Bak (wt) or lacking both genes were also exposed to 1.0μg of Etoposide for 72h or were left untreated to confirm their intrinsic apoptosis resistance.
To investigate if the additional 24 aa found in BmfCUG, predicted to contain an additional α-helix, may define differences in subcellular localization we transiently transfected HA-tagged versions of the individual isoforms into 293T HEK cells or NIH3T3 fibroblasts, followed by immunofluorescence analysis using an epitope tag-specific antibody. Both isoforms colocalized predominantly to mitochondria, in both cell types analyzed (Fig. 6a and data not shown). Since the anti-Bmf antibodies currently available are not suitable to distinguish between isoforms by immunofluorescence, we decided to study Bmf localization performing biochemical subcellular fractionation experiments. Analysis of cytoplasmic and mitochondrial fractions from NIH3T3 cells that were left untreated or incubated with cytochalasin D for 16h, a stimulus reported to induce translocation of Bmf from the actin-cytoskeleton to mitochondria (20), confirmed that endogenous Bmf can already be found in the mitochondrial fraction prior treatment (Fig. 6b). Alkaline extraction also indicated that Bmf is not simply loosely attached to the outer mitochondrial membrane but, similar to Bim (23), must have been actively inserted into it (Fig. 6b). Notably, the C-term end of Bmf, fused to GFP, allows for active import of this fusion protein into isolated yeast mitochondria (GH, unpublished observations). Finally, we also investigated whether both proteins may display differences in protein stability but again, at least under stady-state conditions, both isoforms were equally sensitive to CHX-treatment showing a half-life of approximately 3-4h, as assessed in WEHI231 pre-B cells (sFig. 1c), primary lymphocytes or NIH3T3 fibroblasts (not shown).
Figure 6. Bmf isoforms localize preferentially to the outer mitochondrial membrane.
(A) NIH3T3 cells were transiently transfected with HA-tagged versions of BmfS or BmfCUG. 24h after transfection cells were fixed and prepared for immunofluorescence. (B) NIH3T3 cells were left untreated or cultured in the presence of 2μM cytochalasin D for 16h. Cells were harvested, washed in PBS and lysed prior subcellular fractionation and alkaline stripping of mitochondria. Pellet and supernatant (SN) fractions were separated by SDS-PAGE, transferred onto nitrocellulose membranes and subjected to immunoblotting using anti-Bmf mAb (17A9). Membranes were reprobed with antibodies specific for the mitochondrial transmembrane protein CoxIV or cytoplasmic GAPDH.
In summary these data suggest that both isoforms show comparable half-life, identical subcellular localization, killing potential and do not differ in their ability to counteract Bcl-2 pro-survival proteins tested. Furthermore, Bmf can engage both, Bax and Bak to exert its pro-apoptotic function.
BmfCUG and BmfS can be translated in a CAP-independent manner
Since non-ATG dependent initiation often associates with translation of proteins from intra-ribosomal entry sites (IRES) (24), we wondered whether the RNA sequence between both translation start sites possesses the capacity to enable CAP-independent translation. To test this hypothesis, we first subcloned different cDNA fragments encoding different portions of the 5′end of the bmf mRNA into a bicistronic luciferase reporter system where a minimal SV40 promoter drives expression of one mRNA endoding the Renilla (RN) luciferase, followed by a spacer and the ORF encoding for Firefly (FF) luciferase. Inserting the 258bp TSS-ATG2 fragment or the 72bp CTG-ATG2 fragment into this spacer region revealed that the longer fragment was able to promote significant expression of the FF-reporter while insertion of a non-related sequence from the GAPDH gene did not (Fig. 7a). Finally, when the same experiment was performed using modified constructs that carry an additional stem-loop structure between the SV40 promoter and the open reading frame for Renilla, hindering CAP-dependent translation, a clear shift in the ratio between RN/FF was observed in the presence of the TSS-ATG2 fragment contained in the bmf mRNA (Fig. 7a). The well-described IRES element from the cyclin dependent kinase inhibitor p27 was used as an internal control allowing comparable expression of FF-luciferase (25). To exclude residual promoter activity in the tested cDNA fragments to be responsible for the observed FF activity in our dual reporter assay, the same DNA elements were also inserted into the promoter-less pGL2luc luciferase expression vector and its activity was compared to that of a bona-fide bmf promoter fragment encoding nucleotides −600 to +70 relation to the TSS. While the latter fragment induced significant luciferase activity upon overexpression in 293T cells, all other DNA fragments did not, arguing against this possibility (Fig. 7b). To confirm this observation in a different experimental system, the same 258bp fragment (TSS-ATG2), conferring expression of FF luciferase in our dual-luciferase experiments was subcloned in between the open reading frames of two fluorescent proteins, ECFP and Venus (sFig. 3a). Fluorescence microscopy of 293T cells transfected with constructs allowing either expression of both fluorescence proteins as a single fusion protein or both ORFs separately, due to insertion of the TSS-ATG2 element preceded by a STOP-codon after ECFP, confirmed that both fluorescent proteins are translated individually in cells transfected with the latter construct (sFig. 3b). However, since we could not rule out completely that ribosomal slipping may contribute to Venus expression (or expression of the second luciferase gene in Fig. 7a), we investigated if blocking CAP-dependent translation by addition of rapamycin or the CAP-translation inhibitor 4EGI-1 favored the expression of the second fluorescent protein, Venus, over ECFP. Consistent with our idea that the 5′end of the bmf mRNA contains features of an IRES-element, we observed a clear shift towards the expression of the second fluorescence under these conditions (sFig. 3c) that should not become visible, if both proteins were synthesized in a strictly CAP-dependent manner. Finally, we transcribed bmf cDNA including a polyA tail in vitro without adding a 5′CAP and used this mRNA in a separate translation reaction. cDNA encoding GFP served as a control. The translation reactions were then separated by SDS-PAGE and immunoblotting was performed using anti-Bmf-specific and anti-GFP-specific antibodies. While the IVT of GFP mRNA did not generate any detectable protein expression, due to the lack of the 5′CAP-structure, we observed that BmfS was effectively translated from the uncapped mRNA, but, surprisingly, we also detected a weaker signal resembling BmfCUG (Fig. 7c).
Figure 7. Bmf isoforms can be translated in an IRES-dependent manner.
(A) Constructs containing a bicistronic mRNA encoding Renilla (RN) and Fire-Fly (FF) luciferase, driven by the SV40 minimal promoter and spaced by the mouse bmf cDNA elements from the predicted TSS to translation initiating CTG in exon 2 or ATG2 in exon 3, or an unrelated sequence from the human GAPDH gene were transiently transfected into 293T cells. 24h after media change cells were lysed and the different luciferase activities quantified using a Dual-Luciferase Assay kit to calculate the ratio of FF/RN activities in the absence or presence of the putative IRES element and an inhibitory stem loop structure 5′ of the first cistron, shifting the balance to IRES-mediated translation of the second cistron. Bars represent means ± SEM of three independent experiments performed in quadruplicates. Asterices* represent significant differences between RF and shRF constructs (p<0.05) (B) To exclude residual promoter activity in the putative IRES element, the indicated mouse bmf cDNA-derived sequences were subcloned into a promoter-less luciferase expression plasmid. After transient transfection into 293T cells luciferase activity was quantified as above and normalized to co-transfected Renilla luciferase. A 600bp element from the mbmf promoter served as positive control. One representative experiment performed in triplicate is shown. (C) cDNAs encoding mouse bmfL or GFP, were subcloned into the pTEX-polyA plasmid that contains a polyA tail 3′ of the multiple cloning site, as well as a 5′ SP6 promoter for in vitro transcription. Uncapped mRNA was then used in a standard in vitro translation in reticulocyte lysates. Aliquots of the reactions were separated by SDS-PAGE along with protein lysates containing Bmf isoforms or GFP as a control (ns=nonspecific). Immunoblotting analysis was performed using anti-Bmf mAb (17A9) and anti-GFP mAb (Sigma).
Together, these findings strongly suggest that the bmf mRNA contains features of an IRES element in its 5′region allowing for alternative start-site usage and, presumably, expression of both Bmf-isoforms.
Accumulation of Bmf protein and Bmf-dependent apoptosis upon inhibition of CAP-dependent translation
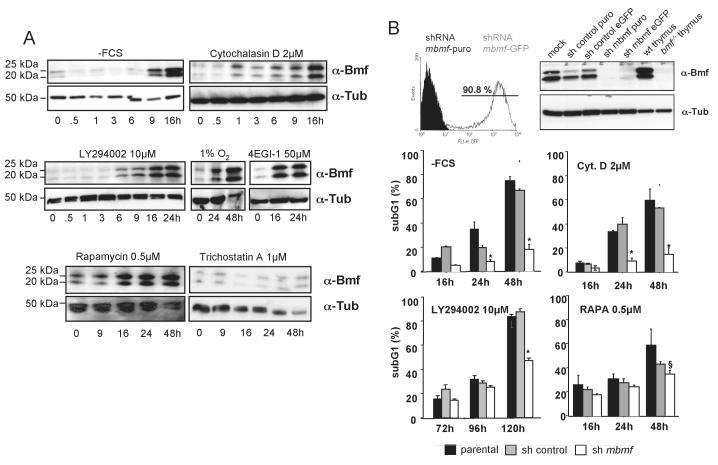
Keeping the previous findings in mind, we investigated if Bmf protein can be induced under conditions that favor CAP-independent and, hence, IRES-mediated protein translation. To test this, we deprived NIH3T3 cells and HC11 mammary epithelial cells of serum or exposed them to hypoxia, both conditions known to favor CAP-independent translation (26). Furthermore, CAP-dependent translation was compromised by inhibition of the PI3K/AKT pathway or mTOR using LY294002 or rapamycin, respectively (27). Finally, we directly interfered with conventional translation using 4EGI-1, an inhibitor of the eukaryotic initiation factor (eIF) 4E - eIF4G interaction and hence, CAP-dependent translation (28). Notably, all these stimuli caused accumulation of both Bmf isoforms, while treatment of cells with the ER-stressor tunicamycin, the DNA-damaging drug etoposide or the pan-kinase inhibitor staurosporine had no such effect (Fig. 8a; not shown). Notably, treatment of both cell types with the actin-depolymerizing agent cytochalasin D also lead to an increase in Bmf protein levels (Fig. 8a; sFig. 1d).
Figure 8. Bmf accumulates prior cell death in NIH3T3 cells.
(A) NIH-3T3 fibroblasts deprived of serum or cultured in the presence of the indicated stimuli and concentrations over time. Cell extracts were separated by SDS-PAGE and immunoblotting was performed using the Bmf-specific mAb (17A9). Membranes were reprobed with an anti-tubulin antibody to compare protein loading. (B) NIH3T3 were lentivirally transduced with shRNA targeting human (sh control) or mouse bmf. Transduction and knock-down efficiency was confirmed by flow-cytometric analysis and Western blotting. Stable knock-down clones were selected using puromycin and used for survival analysis. Cells were deprived of serum or exposed to the indicated drugs. Apoptosis was quantified by sub-G1 staining of cells followed by flow-cytometric analysis. Bars represent means ± SE of three independent experiments performed in duplicates using two independent sh_clones. [*] indicates statistically significant differences between parental, sh_control and sh_bmf expressing cells and [§] indicates significant differences between parental and sh_bmf expressing cells (p< 0.037 for –FCS; p< 0.002 for Cyt. D; p< 0.0019 for LY294002; p< 0.05 for rapamycin).
In order to evaluate whether Bmf is also important for cell death induction under these conditions, apoptosis was quantified in NIH3T3 cells stably expressing an shRNA targeting bmf mRNA for degradation by RNAi (Fig. 8b). Consistent with a rate-limiting pro-apoptotic role of Bmf under these conditions, its knockdown rendered NIH3T3 cells refractory to cell death induced by stimuli that interfere with CAP-dependent translation including serum deprivation or inhibition of PI3K/AKT/mTOR network. Knock-down of Bmf also delayed cell death induced by cytochalasin D treatment, which does not associate with CAP-independent translation, but apoptosis induced by UV-radiation, anoikis, the proteasome inhibitor Velcade or treatment with the histone deacetylase inhibitor Trichostatin A occurred normal (Fig. 8b and sFig. 4). Notably, cell death induced by rapamycin, inhibiting mTOR complex 1 activity, or the PI3K-inhibitor LY294002 was both only partially blocked. This may have been due to incomplete knockdown of Bmf expression or involvement of other pro-apoptotic factors. Therefore, we wanted to confirm our observations in a clean genetic back-ground and investigated whether Bmf could be induced in SV40 immortalized MEF, which usually do not express significant levels Bmf protein (AV, unpublished), and whether absence of the protein in bmf−/− MEF could confer resistance to apoptosis induction by these stimuli. Serum-deprived wt MEF, as well as cells treated with inhibitors of the PI3K/AKT/mTOR network all induced Bmf protein. When wt MEF were first serum-deprived for 24h, triggering Bmf accumulation, subsequent exposure to cycloheximide prevented further protein accumulation more potently than inhibition of transcription with actinomycin D (Fig. 9a). Similar observations were made in MEF treated with rapamycin or L294002 (Fig. 9b). Together these observations again supported the idea that inhibition of CAP-dependent translation favors Bmf protein accumulation under these conditions, at least in part by IRES-mediated translation. Consistently, inhibition of the PI3K/AKT/mTOR network by LY294002 or rapamycin triggered cell death in bmf−/− MEF less effectively. Notably, these cells still succumbed to apoptosis equally fast, or even faster upon treatment with staurosporine (Fig. 9c). Serum deprivation yielded inconsistent results in the different SV40 MEF clones tested and the observed differences did not yield statistically significant differences (Fig. 9c) suggesting that some of the observed effects are cell type specific or depend of the type of immortalization.
Figure 9. Bmf-deficiency delays apoptosis by cell stress associated with CAP-independent translation in primary thymocytes and SV40 MEF.
(A) SV40 MEF were deprived of serum in the absence or presence of actinomycin D or cycloheximide. Protein lysates were generated at the indicated time-points and Bmf expression was analyzed by immunoblotting. (B) Wt or bmf−/− SV40 MEF were exposed to rapamycin or LY294002 up to 48h (upper panel). Alternatively, actinomycin D or cycloheximide was added at t=0h or after 24h of incubation with rapamycin or LY294002 (lower panel). (C) Cells lacking or expressing Bmf were treated with the indicated compounds over time. Thymocyte apoptosis was quantified by AnnexinV/PI staining (n=4/genotype). Apoptosis of MEF was assessed by sub-G1 staining and flow cytometric analysis. Bars represent means ± SE of three independent experiments. Experiments using three independent MEF clones of each genotype yielded comparable results and were therefore pooled (STS=staurosporine). Asterices * indicate statistically significant differences between wt and bmf−/− MEF (p< 0.006 for LY294002; p< 0.001 for rapamycin). (D) Wt and Bmf-deficient mice were injected i.p. with a single dose of polyIC (100μg/mouse), or saline. After 20h, mice were sacrificed and the percentage of naïve CD8+CD44− T cells quantified by flow cytometric analysis. Bars represent means of ± SEM of three animals per genotype and group. * indicates statistically significant differences between wt and bmf−/− (p<0.003).
Since viral infection often associates with the inhibition of CAP-dependent translation, we wondered if loss of Bmf prevents attrition of T cells observed to occur early in such anti-viral immune responses (29). This situation can be mimicked to a certain degree by injection of polyIC that triggers the typ I IFN-dependent apoptotic loss of CD8+ T cells early in an immune response (29). Therefore, wt and bmf−/− mice were injected with a single dose of polyIC and the percentage of CD8+ T cells was quantified 20 h later. CD8+ T cell numbers declined but naïve CD8+CD44− T cells were most affected by polyIC treatment in wt animals. In contrast, in mice lacking Bmf that presented with an overall reduced percentage of naïve T cells, polyIC treatment had no such effect (Fig. 9d). Together our results support the hypothesis that Bmf can act as a sentinel for cell stress that associates with the inhibition of CAP-dependent translation in a cell type and stimulus dependent manner.
Discussion
BH3-only proteins are essential regulators of cell death that sense countless endogenous and exogenous stress signals translating them into the activation of the apoptotic cell death machinery. In order to provide a cell with the biggest possible flexibility to respond to highly diverse forms of stress, multiple modes of regulation are usually required to control the activation and function of individual BH3-only proteins. For example, DNA-damage can trigger the transcription of Puma and Noxa (30-32), while phosphorylation/dephosphorylation events upon growth factor receptor-engagement can modulate the activity of Bad by cytoplasmic sequestration (33, 34) or abundance of Bim via the proteasome (35). Furthermore, N-myristylation of Bid secures its proper mitochondrial targeting and caspase-cleavage its pro-apoptotic activity (36). Protein-protein interaction, e.g. with motor complex associated proteins such as dynein-light chain molecules can lead to distinct subcellular localization of Bim or Bmf, at least in some cell types that depend on integrin signals and/or contact to the ECM for survival (12). Alternative splicing as a means of regulation of BH3-only protein activity has been described, e.g. for Puma (30), Bim (37) but also Bmf (17). However, none of the major isoforms of Bmf described here is generated by differential splicing but by different start site usage and, therefore, do not represent equivalents of human BMF II or BMF III, found in CLL cells (17). BMF II and BMF III as well as the γ- and δ-isoforms of Puma, do lack a functional BH3-domain (30) and their physiological functions remains unclear. In contrast, all Bmf isoforms detected in our study could be co-immunoprecipitated together with Bcl-2 from lymphocytes, indicating that they all must contain a functional BH3-domain (sFig. 1b). Whether the additional 24 aa encoded in the N-terminus of BmfCUG can serve as an interface for specific protein-protein interactions outside the Bcl-2 family is subject of our current investigations.
Functional analysis failed to reveal any differences in the binding pattern of Bmf isoforms to pro-survival Bcl-2 proteins, similar to observations made using alternatively spliced isoforms of Bim (37). BmfCUG bound to Bcl-2, Bcl-x, Bcl-w and Mcl-1, as initially also reported for BmfS (20), but neither isoform bound effectively to the viral Bcl-2-like proteins BHRF1 or KS-Bcl2 in overexpression studies (Fig. 3). Although Biacore analysis defined a low affinity of the Bmf BH3-domain for Mcl-1 (38), this interaction can be readily detected under conditions of overexpression (Fig. 3 and (20)). Recently, we also observed interaction of endogenous Bmf with endogenous Mcl-1 in mouse B lymphoma cells (AV, unpublished), suggesting that the use of BH3-domain peptides cannot recapitulate entirely the interaction behaviour of the full-length protein.
Similar to the major Bim isoforms, shorter BmfS appeared somewhat more potent in reducing the colony forming potential of MEF, however this effect was not observed in U2OS cells. Both isoforms can engage Bax as well as Bak to promote cell killing but BmfS seems to have a preference for Bax over Bak (Fig. 5). To our surprise, we observed that MEF lacking Bax and Bak also showed reduced colony formation when both isoforms were overexpressed from a single plasmid and at the moment, we can only speculate that these cells may either activate Bok under these conditions or die by necroptosis (19). However, a possible impact of Bmf isoform overexpression on the proliferation of these MEF has not been formally excluded. Hence, the physiological functions of both isoforms remain largely elusive and will require the generation of knock-in mutant mouse strains expressing only one or the other isoform of Bmf.
Our observation that the generation of BmfCUG involves translation initiation from a CUG, encoded in exon 2 suggested that IRES-mediated translation might serve as a mechanism to initiate translation. This possible feature was overlooked initially when the short isoform of Bmf that starts from an ATG encoded at the beginning of exon 3 was first described (20). Our follow up analysis suggests that actually both major isoforms appear to be translated in an IRES-dependent manner from alternative start-sites contained in the bmf mRNA (Figs. 2,7). Alternative start site usage has also been reported for Bad, leading to the expression of two main isoforms, but in this case ATG serves for initiation of translation in both cases (39). During apoptosis, eukaryotic initiation factors are modified early on, e.g. by dephosphorylation of 4E-BP1 and subsequent caspase-cleavage of eukaryotic eIF4G, both enforcing CAP-independent translation (26). Consistently, a number of apoptosis regulators including p53, Bcl-2, Apaf-1 and XIAP were reported to be generated from IRES-elements under conditions of stress, thought to maintain a functional apoptotic machinery when CAP-dependent translation is compromised (40). However, some of these proposed IRES-elements were subsequently proven to be false and the activity observed in dual reporter systems due to cryptic splicing events generating a 3′cistron of the second reporter (41-43). To strengthen our claim that Bmf can be translated in an IRES-mediated manner, we have (i) excluded residual promoter activity in the DNA sequences tested, (ii) demonstrated that insertion of a hair-pin structure favours expression of the second cistron in dual reporter assays and, most importantly, performed (iii) sequential transcription translation reaction using uncapped mRNA that confirmed translation of Bmf protein in the absence of a 5′CAP structure (Fig. 7). Consistently, endogenous Bmf expression was clearly induced by different stimuli that associate with the repression of CAP-dependent translation (Figs. 8,9). The latter observation proposes a role for Bmf isoforms as sentinels that become activated in response to stress that associates with reprogramming of the translation machinery from CAP-dependent to CAP-independent translation such as growth factor deprivation, hypoxia or inhibition of the PI3K/AKT/mTOR signaling axis (Fig. 8,9; sFig.1). Consistent with a role for Bmf as a sensor of this type of stress, knock-down or absence of Bmf interfered but ultimately failed to prevent apoptosis in response to some of the tested stimuli suggesting that it must collaborate with other cell death regulators, presumably other BH3-only proteins, such as Bim (15) in a cell type and stimulus dependent manner. Whether Bmf plays also a rate-limiting role for apoptosis induced upon physiological triggers of CAP-independent translation in vivo, e.g. during viral infection or hypoxia-induced tumor cell death will be subject of our future investigations.
Material & Methods
Cell culture, primary cells and reagents
All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. Mouse embryonic fibroblasts were isolated from E14.5 embryos after removal of internal organs, brain and fetal liver by trypsin digestion of the remaining carcass. All experiments were performed using MEFs that were immortalized with the SV40 large T antigen. U2OS cells, MEFs and NIH3T3 murine fibroblasts were cultured in the DME medium supplemented with 10% FCS (PAA), 250μM L-glutamine (Invitrogen) and penicillin/streptomycin (1U/ml, Sigma-Aldrich). IL3-dependent BaF3 pro-B lymphoid cells cells were cultured in RPMI 1640 medium, supplemented with 250μM L-glutamine, 50μM 2-mercaptoethanol, penicillin/streptomycin (1U/ml) and 10% FCS, supplemented with 15% of WEHI-3B cell supernatant. HC11 mouse mammary epithelial cells were cultured in RPMI 1640 medium, supplemented with 5μg/ml bovine insulin (Sigma), 10ng/ml murine EGF (Peprotec), 250μM L-glutamine, 50μM 2-mercaptoethanol, penicillin/streptomycin (1U/ml) and 10% FCS. The generation of vav-bcl-2 transgenic (44) and bmf−/− mice has been described (14).
Apoptosis-induction, sub-G1 and AnnexinV/propidium iodide staining
For the induction of cell death the following reagents were used: 2μM Cytochalasin D (Sigma), 10μM LY-294002 (InvivoGen), 500nM rapamycin (Alexis), 50μM 4EGI-1 (Alexis), 50nM Staurosporine (Sigma), 1μg/ml Tunicamycine (Sigma). Cells cultured under hypoxic conditions were cultured in a humidified atmosphere containing 1% O2.
For sub-G1 analysis, cells were collected, along with their supernatant, centrifuged for 5 minutes at 1500rpm, 4°C. Then the pellet was washed once with PBS and centrifuged again. After supernatant removal, pellet was resuspended in 500μl 70% Methanol (in PBS) and fixed at −20°C. Cells were then washed with PBS and centrifuged for 5 minutes at 3000rpm, 4°C. The pellet was resuspended in 475μl PBS with RNAseA (Sigma; final concentration 10μg/ml) and incubated at 37°C for 20 minutes. After that, 25μl of propidium iodide (final concentration 2μg/ml) were added to the cell suspension, which was incubated for 10 minutes at 4°C before analysis in flow cytometer, FACScan (Becton Dickinson). Alternatively, the percentage of viable cells in culture was determined by staining them with 2μg/mL propidium iodide plus APC- or FITC-coupled Annexin-V in Annexin binding buffer (Becton Dickinson) and subsequent flow cytometric analysis.
Individual in vitro transcription and in vitro translation
cDNA encoding mbmfL or GFP cDNA was subcloned in the pTEX plasmid containing a polyA sequence downstream of the multiple cloning site. DNA was linearized with NotI, run on a 1.5% agarose TAE gel and then purified using a gel extraction kit (Qiagen). In vitro transcription using 25% volume of eluted DNA was performed using the SP6 MEGAscript Kit (Ambion). RNA was recovered by LiCl precipitation (final concentration 4.5M). RNA concentration was quantified in a spectrophotometer and 1μg of in vitro transcribed RNA was subsequently translated using the Retic Lysate IVT kit (Ambion) according to the manufacturers recommendation, in 25mM or 50mM potassium-acetate containing reaction buffer. Protein was precipitated using 12% TCA and resuspended in 2x Laemmli buffer and separated by SDS-PAGE prior immunoblotting.
Coupled in vitro transcription/translation reaction
Usually, 1μl plasmid DNA (100ng) was mixed with 10μl translation mix containing reticulocyte extract and, optional, 1μl 35S methionine (Amersham). Samples were incubated for 90 minutes at 30°C and then frozen until SDS-PAGE was performed.
SDS-PAGE: 3μl of each sample were diluted in ONYX lysis buffer, mixed 1:3 with laemmli loading buffer and applied on a 14-20% precast gradient polyacrylamide gel. On one lane a protein size marker was loaded. Electrophoresis was performed at 70 volt in Tris-Glycine buffer. After electrophoresis the gel was vacuum-dried and exposed to an X-ray film for 2 days. Results were obtained after developing the film using AGFA Curix 60 photo imager. Alternatively, non-radioactive reactions were electroblotted onto nitrocellulose membranes and subjected to immunoblotting using anti-Bmf mAb 17A9.
Transient transfections, immunoblotting, immunoprecipitation and phosphatase treatment
293T human embryonic kidney cells or NIH3T3 fibroblasts were transiently transfected in 6-well plates using 3μl of Lipofectamine 2000 reagent (Invitrogen) and a total of 1μg of plasmid DNA. Cells were incubated in Optimem (Gibco) with the transfection mix for 6h before replacing the medium. After 48h cells were harvested and lysed in ONYX buffer (20mM TrisHCl pH 7.5, 135mM NaCl, 1.5 mM MgCL2, 1mM EGTA, 1mM EDTA, 1% TritonX-100, 10% glycerol) for 30 min on ice. Insoluble debris was cleared by centrifugaiton and protein concentration was quantified using the Bradford assay. Usually, 75-100μg of protein separated by SDS-PAGE using 4-20% PAGE Novex-ready gels (Invitrogen). After transfer, membranes were probed with rat monoclonal antibodies specific for Bmf (clone 9G10, 17A9 or 12E10) or specific for the FLAG epitope (mouse anti-FLAG M2, Sigma). Equal loading of proteins was confirmed by probing filters with antibodies specific for α–tubulin (DM1A, Santa Cruz Biotechnologies), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 71.1, Sigma-Aldrich), or rabbit anti CoxIV polyclonal (Abcam) for mitochondrial fractions. Horseradish peroxidase (HRP)-conjugated sheep-anti rat Ig antibodies (Silenus) served as secondary reagents and the enhanced chemiluminescence (ECL) system was used for detection. Immunoprecipitation was performed using Protein G-sepharose (GE Healthcare). For dephosphorylation, single cell suspensions were prepared from thymus or spleen from wt mice. Resuspended cells were washed twice in PBS then lysed 30 min at 4°C in Chaps buffer (20 mM Tris-HCl, 5 mM MgCl2, 137 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.5% Chaps pH 7.5) supplemented with (Complete™, Roche). Cellular debris were removed by centrifugation at 10.000xg for 20 min at 4°C. Protein extract was dephosphorylated with lambda protein phosphatase (7u/μg of protein extract) (BioLabs) and 2 mM MnCl2 2h at 4 or 30°C. The reaction was stopped by addition of SDS-PAGE loading buffer.
Luciferase assays
Lipofectamine was used to transiently transfect 293T cells with different dual-luciferase constructs for 6h. After additional 24h, cells were harvested and Renilla and Firefly-luciferase activity was quantified using the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturers recommendation.
Immunofluorescence stainings
Cells were cultured on glass cover slips and 24h or 48h after lipofectamine transfection cultures were briefly washed with PBS and incubated for 30min at 37°C with 100nM MitoTracker Red CMXRos (Molecular Probes, Invitrogen, Austria) in culture medium to stain the mitochondria. Then cultures were briefly washed with PBS, medium was removed and cells were fixed for 15min at room temperature with 4% PFA (in PBS). Cells were then washed 3 times with PBS and further incubated for 1h with PBS containing 0.1% Triton X-100, 1% bovine serum albumin and 10% fetal calf serum for permeabilization and blocking. Incubation with primary antibody (mouse anti-HA clone: 12CA5, Sigma, 1:100 in blocking solution) was performed overnight at 4°C. After 3 further washes with PBS, cells were incubated with secondary antibody (Alexa-fluor-488-labelled anti-mouse, Molecular Probes, Invitrogen, Austria; 1:100 in blocking solution) for 1h at room temperature, then washed again 3 times and incubated with DAPI (1μg/ml, in PBS) for nuclear staining for 10min at room temperature. Finally, following another washing step, cells were fixed with Vectashield antifade mounting medium (Vector Laboratories Burlingarne, CA) and the glass cover slips were mounted on microscope slides. Cell images were collected with a confocal laser scanning microscope (Zeiss LSM 510 Axiovert 100M, 63x/1.4 oil immersion lens; Carl Zeiss MicroImaging, Inc.), with the resolution set to 1024 ×1024 pixels, and pinholes set to acquire images of below 1 μm thickness. Further image processing, i.e. background correction, adjustment of brightness and contrast and export to tif-format, were made using Image J software.
For the fluorescence detection of IRES-mediated expression of YFP 293T HEK cells cultured on glass cover slips were transfected with plasmid constructs containing cDNA encoding a ECFP-Venus fusion protein, or a sequence where both open reading frames were separated by the putative Bmf-IRES, and were then exposed to 50 μM 4EGI-1, 0.5 μM rapamycin for 24h or were left untreated. Next, cells were fixed with 4% PFA in PBS at room temperature, washed with PBS for 3 times and mounted on microscope slides with Vectashield antifade mounting medium. Images were then acquired using a Leica SP5 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) with a 63xglycerol immersion objective, applying the LAS AF acquisition software Version 2.1.0. Cells were alternately excited either with the 458nm or the 514 nm laser line of an Argon laser and emission filter sliders set to 460-495nm or to 520-590nm to detect CFP and YFP, respectively. Image resolution was set of 512 × 512 pixels and pinholes to 1 Airy unit. Relative emission intensities of acquired images were finally quantified with Image J software.
Mitochondrial subfractionation
After 16h of incubation with 2μM Cytochalasin D or solvent control, NIH3T3 cells were harvested in 300μl of isotonic mitochondrial buffer (MB) (70mM sucrose, 210mM mannitol, 1mM EDTA, 10mM Hepes pH7.5) supplemented with proteases inhibitor cocktail (Complete, Roche). Cells were left for 15min on ice and then they were broken by 7 passages repeated for 3 times through a 25G 0.5- by 25mm needle fitted on a 1ml syringe. Cells suspension were then centrifuged at 2000g at 4°C for 5 min to remove unbroken cells. Lysates were spun further for 10min at 13000g at 4°C to pellet mitochondria, ER and nuclei. The cytosolic supernatant fraction was subsequently centrifuged for 1h at 4°C at 39000rpm in an ultracentrifuge. Supernatant was then precipitated with 12%TCA and resuspended in 45μl of 2x Laemmli buffer prior boiling (5min, 95°C). The 13K pellet = heavy membrane fraction was resuspended in 65μl MB-EGTA buffer (containing 0,5mM EGTA instead of EDTA) and further centrifuged at 500g for 3min at 4°C to separate the nuclear fraction. The supernatant containing mitochondria was subdivided into 3 aliquotes and centrifuged at 10000g, for 10min at 4°C. The pellet resulting from one aliquot was resuspended in 15μl of MB-EGTA buffer containing 1% Triton and used for protein quantification by Bradford. The two other mitochondrial pellets were subjected to alkali extraction. Pellets were resuspended in 125μl of H2O and 125μl of NaCl (2M) or Na2CO3 (2M). Samples were incubated on ice for 30 minutes mixed by vortexing every 10 minutes followed by centrifuged at 100000g for 30 minutes at 4°C. Alkaline-sensitive fractions (supernatant) were precipitated adding 12% TCA (final concentration) and then resuspended in 15μl of 2x Laemmli buffer prior boiling. The alkaline-resistant pellet fraction was boiled in the same volume. Finally 15μl of each fraction were loaded on a 4-20% Tris-HCl gel (Biorad) and separated by SDS gel electrophoresis.
Viral Transduction & Colony Formation Assays
U2OS cells and NIH3T3 cells were transduced with lentiviruses encoding different isoforms of Bmf, MEF lacking Bax or Bak were transduced retrovirally.
For overexpression of the various Bmf isoforms, the corresponding cDNAs were PCR amplified and recombined into pDONR-207 (Invitrogen) using Invitrogen’s B/P recombination kit. Sequence verified clones were used for L/R recombination with pHR-tetCMV-dest (45) thereby generating the lentiviral plasmids pHR-tetCMV-BmfS, pHR-tetCMV-BmfCUG, pHR-tetCMV-BmfSL/A, pHR-tetCMV-BmfCUGL/A and pHR-tetCMV-eGFP.
Generation of lentiviral plasmid for constitutive knock-down of human BMF were described before (45). For the knock-down of mouse Bmf, 5′GATCCCCtcagtcgactccagctcttTTCAAGAGAaagagctggagtcgactgaTTTTTGGAAA3′ (sense) and 5′AGCTTTTCCAAAAAtcagtcgactccagctcttTCTCTTGAAaagagctggagtcgactgaGGG3′ (antisense) oligonucleotides were phosphorylated, annealed and cloned into the BglII-HindIII site of pENTR-THT (45). Thereafter, sequence verified clones were used for L/R recombination into the final lentiviral destination vectors pHR-dest-SFFV-eGFP and pHR-dest-SFFV-Puro, thereby generating pHR-THT-mBmf shRNA-SFFV-eGFP and pHR-THT-mBmf shRNA-SFFV-Puro.
For lentiviral transfection, human HEK 293T cells were transiently transfected with lentiviral plasmids containing cDNAs coding for mouse BmfS, BmfCUG, BmfS L/A mutant, BmfCUG L/A mutant or eGFP as control, along with the packaging plasmids pSPAX and pVSV-G. After 48h and 72h lentiviral supernatant was collected and sterile filtered (0.2μM), supplemented with Polybrene to a final concentration of 4μg/ml and added to the target cells overnight.
For retroviral transduction of MEF, Lipofectamine (Invitrogen) was used to transiently transfect Phoenix packaging cells with constructs encoding for Bmfs, BmfCUG, both isoforms, or empty control subcloned in the retroviral pBABE-puro backbone. After 48h, supernatant was collected and 0.20μm filter-sterilized. Polybrene was then added to reach a final concentration of 4μg/ml. The viral supernatant was added to MEF lacking Bax or Bak by spin infection. The following morning, fresh medium was added to the infected cells. After 48h, medium was replaced by medium containing 2μg/ml puromycin. Selection was performed for one week where medium was changed twice.
For assessment of colony formation, cells were washed once with PBS and incubated for 10 minutes in 0.5ml of crystal violet staining solution (0.2% crystal violet in 50% methanol). Plates were then rinsed 3 times with bi-distilled water, air-dried and stored in the dark until photographic documentation.
Statistical Analysis
Statistical analysis was performed using the unpaired Student t test and a Stat-view 4.1 software program. P-values of <0.05 were considered to indicate statistically significant differences.
Supplementary Material
Acknowledgments
We thank Profs A Strasser, C Borner, M Schuler and W Doppler for cells and reagents. S Kiessling and F Müllauer for first experiments initiating this work, W Chmelewskij for help with MEF experiments, B Tomaselli for help with hypoxia experiments, Irene Gaggl for expert technical assistance, as well as all our colleagues in the lab for insightful discussions. This work was supported by grants from the AICR, St. Andrews, UK (#06-0440), the Graduate School for Molecular Biology and Oncology (MCBO) and SFB021, both funded by the Austrian Science Fund (FWF).
Abbreviation List
- (Bmf)
Bcl-2-modifying factor
- (BH)
Bcl-2 homology domain
- (IVT)
in vitro transcription
- (IVTT)
in vitro transcription and translation
- (TSS)
transcription start site
- (IRES)
intra ribosomal entry site
- (FF)
Fire-fly
- (RN)
Renilla
- (ECFP)
enhanced cyan fluorescent protein
- (SV)
simian virus
- (MEF)
mouse embryonic fibroblasts
- (sh)
short hairpin
- (PFA)
para-formaldehyde
References
- 1.Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008 Dec;27(Suppl 1):S128–136. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 2.Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 2001;15(12):1481–1486. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science (New York, NY. 2007 Feb 9;315(5813):856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 4.Letai A, Bassik M, Walensky L, Sorcinelli M, Weiler S, Korsmeyer S. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2(3):183. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 5.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005 Feb 18;17(4):525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Merino D, Giam M, Hughes PD, Siggs OM, Heger K, O’Reilly LA, et al. The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J Cell Biol. 2009 Aug 10;186(3):355–362. doi: 10.1083/jcb.200905153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008 Jan;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 8.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003 Oct;4(4):321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 9.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science (New York, NY. 2003 Nov 7;302(5647):1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 10.Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DC, et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature. 2009 Aug 20;460(7258):1035–1039. doi: 10.1038/nature08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin X-M, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400(6747):886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 12.Pinon JD, Labi V, Egle A, Villunger A. Bim and Bmf in tissue homeostasis and malignant disease. Oncogene. 2008 Dec;27(Suppl 1):S41–52. doi: 10.1038/onc.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puthalakath H, Strasser A, Huang DCS. Rapid selection against truncation mutants in yeast reverse two-hybrid screens. BioTechniques. 2001;30(5):984–988. doi: 10.2144/01305st03. [DOI] [PubMed] [Google Scholar]
- 14.Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O’Reilly L, et al. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J Exp Med. 2008 Mar 17;205(3):641–655. doi: 10.1084/jem.20071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubner A, Cavanagh-Kyros J, Rincon M, Flavell RA, Davis RJ. Functional cooperation of the proapoptotic Bcl2 family proteins Bmf and Bim in vivo. Mol Cell Biol. 2010 Jan;30(1):98–105. doi: 10.1128/MCB.01155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villunger A, Scott C, Bouillet P, Strasser A. Essential role for the BH3-only protein Bim but redundant roles for Bax, Bcl-2, and Bcl-w in the control of granulocyte survival. Blood. 2003 Mar 15;101(6):2393–2400. doi: 10.1182/blood-2002-07-2132. [DOI] [PubMed] [Google Scholar]
- 17.Morales AA, Olsson A, Celsing F, Osterborg A, Jondal M, Osorio LM. Expression and transcriptional regulation of functionally distinct Bmf isoforms in B-chronic lymphocytic leukemia cells. Leukemia. 2004 Jan;18(1):41–47. doi: 10.1038/sj.leu.2403183. [DOI] [PubMed] [Google Scholar]
- 18.Frenzel A, Labi V, Chmelewskij W, Ploner C, Geley S, Fiegl H, et al. Suppression of B-cell lymphomagenesis by the BH3-only proteins Bmf and Bad. Blood. 2010 Feb 4;115(5):995–1005. doi: 10.1182/blood-2009-03-212670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008 Dec 26;135(7):1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puthalakath H, Villunger A, O’Reilly LA, Beaumont JG, Coultas L, Cheney RE, et al. Bmf: a pro-apoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science (New York, NY. 2001;293:1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- 21.Mackus WJ, Kater AP, Grummels A, Evers LM, Hooijbrink B, Kramer MH, et al. Chronic lymphocytic leukemia cells display p53-dependent drug-induced Puma upregulation. Leukemia. 2005 Mar;19(3):427–434. doi: 10.1038/sj.leu.2403623. [DOI] [PubMed] [Google Scholar]
- 22.Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, et al. The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia. 2007 Nov 29; doi: 10.1038/sj.leu.2405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber A, Paschen SA, Heger K, Wilfling F, Frankenberg T, Bauerschmitt H, et al. BimS-induced apoptosis requires mitochondrial localization but not interaction with anti-apoptotic Bcl-2 proteins. J Cell Biol. 2007 May 21;177(4):625–636. doi: 10.1083/jcb.200610148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, et al. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003 May-Jun;95(3-4):169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 25.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002 Dec 1;16(23):3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008 Jan;100(1):27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006 Oct;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 28.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007 Jan 26;128(2):257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 29.Bahl K, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, et al. IFN-induced attrition of CD8 T cells in the presence or absence of cognate antigen during the early stages of viral infections. J Immunol. 2006 Apr 1;176(7):4284–4295. doi: 10.4049/jimmunol.176.7.4284. [DOI] [PubMed] [Google Scholar]
- 30.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 32.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the bcl-2 family and candidate mediator of p53-induced apoptosis. Science (New York, NY. 2000;288(5468):1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 33.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 34.del Peso L, González-Garcia M, Page C, Herrera R, Nuñez G. Interleukin-3–induced phosphorylation of BAD through the protein kinase Akt. Science (New York, NY. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 35.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Diff. 2005 Aug;12(8):1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 36.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science (New York, NY. 2000;290(5497):1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 37.O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17(2):384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005 Feb 4;17(3):393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 39.Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2003 Aug 5;100(16):9324–9329. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol Biosyst. 2007;3(12):825–834. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 41.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33(20):6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, Logg CR. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proceedings of the National Academy of Sciences of the United States of America. 2008 Mar 25;105(12):4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saffran HA, Smiley JR. The XIAP IRES activates 3′ cistron expression by inducing production of monocistronic mRNA in the {beta}gal/CAT bicistronic reporter system. RNA. 2009 Aug 27; doi: 10.1261/rna.1557809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogilvy S, Metcalf D, Print CG, Bath ML, Harris AW, Adams JM. Constitutive bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):14943–14948. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, et al. The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia. 2008 Feb;22(2):370–377. doi: 10.1038/sj.leu.2405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.