Abstract
BH3-only proteins, such as Bim and Bad, contribute to tissue homeostasis by initiating apoptosis in a cell type- and stimulus-specific manner. Loss of Bim provokes lymphocyte accumulation in vivo and renders lymphocytes more resistant to diverse apoptotic stimuli and Bad has been implicated in the apoptosis of hematopoietic cells upon cytokine deprivation. To investigate whether their biological roles in apoptosis overlap, we generated mice lacking both Bim and Bad and compared their hematopoietic phenotype with that of the single-knockout and wild-type (wt) animals. Unexpectedly, bad−/− mice had excess platelets due to prolonged platelet life-span. The bim−/−bad−/− mice were anatomically normal and fertile. Their hematopoietic phenotype resembled that of bim−/− mice but lymphocytes were slightly more elevated in their lymph nodes. Although resting B and T lymphocytes from bim−/−bad−/− and bim−/− animals displayed similar resistance to diverse apoptotic stimuli, mitogen-activated bim−/−bad−/− B cells were more refractory to cytokine deprivation. Moreover, combined loss of Bim and Bad enhanced survival of thymocytes after DNA damage and accelerated development of γ-irradiation-induced thymic lymphoma. Unexpectedly, their cooperation in the thymus depended upon thymocyte-stromal interaction. Collectively, these results demonstrate that Bim and Bad can cooperate in the apoptosis of thymocytes and activated B lymphocytes and in the suppression of thymic lymphoma development.
Keywords: Bim, Bad, BH3-only protein, apoptosis, Bcl-2 family, γ-radiation, DNA damage, tumorigenesis
INTRODUCTION
Programmed cell death (apoptosis), a genetically controlled process for eliminating unwanted cells, is essential for the normal development and function of multi-cellular organisms.1 The Bcl-2 protein family plays a major role in the regulation of apoptosis in vertebrates.2 Its BH3-only members, which include Bad, Bim/Bod, Bik/Blk/Nbk, Hrk/DP5, Puma/Bbc3, Noxa, Bmf and Bid, are essential initiators of apoptosis that are activated in response to distinct developmental cues and cytotoxic signals.3,4 They selectively bind to pro-survival members of the Bcl-2 family (Bcl-2, Bcl-xL, Mcl-1, Bcl-w and A1) and trigger mitochondrial release of cytochrome c by a mechanism requiring their pro-apoptotic relatives Bax or Bak.5,6
The BH3-only protein Bad was the first pro-apoptotic Bcl-2 family member found to be regulated by extracellular survival factors.7 In cells stimulated with cytokines, such as IL-3, Bad is phosphorylated by AKT (and certain other kinases) on several serine residues, and this inhibits its pro-apoptotic activity by allowing its sequestration by 14-3-3 scaffold proteins or by directly preventing its interaction with Bcl-xL or Bcl-2.7,8 Conversely, in the absence of survival signals, Bad is de-phosphorylated, increasing its pro-apoptotic activity.8,9 Perhaps surprisingly, however, bad−/− mice were found to be largely normal. Only minor defects in apoptosis were observed upon withdrawal of IGF-1, EGF or glucose in embryonic fibroblasts or mammary epithelial cells,10,11 and some aged bad−/− mice developed lymphoma.10 Since Bad loss did not enhance the survival of cytokine-deprived myeloid progenitor cells,12 its physiological role in growth factor withdrawal-induced apoptosis remains uncertain.
In contrast, the BH3-only protein Bim, which is regulated by a range of transcriptional and post-translational mechanisms,13 plays a major role in the death of hematopoietic cells induced by cytokine deprivation and certain other apoptotic stimuli, including deregulated calcium flux and, although to a lesser extent, DNA damage.14–16 Within the animal, Bim loss provokes accumulation of hematopoietic cells, particularly lymphocytes, impairs deletion of autoreactive thymocytes17 and B cells,18 and is required for shut-down of T cell immune responses.19–21 Furthermore, in certain circumstances, Bim deficiency can provoke autoimmune disease14 and contribute to lymphoma development.22
Despite its major role in hematopoietic cell homeostasis and cytokine deprivation-induced apoptosis,14 the loss of Bim provokes less marked effects than the combined loss of Bax and Bak23,24 or Bcl-2 over-expression,15,25 suggesting that Bim has overlapping functions with other BH3-only proteins. Indeed, although mice lacking Bik are essentially normal,26 loss of both Bik and Bim arrests spermatogenesis,27 whereas combined loss of Puma and Bim renders hematopoietic cells more refractory to diverse apoptotic stimuli, including cytokine deprivation, than loss of either alone.16
Since Bad as well as Bim has been implicated in the apoptosis signalling driven by cytokine withdrawal and DNA damage, as well as in tumorigenesis, we have investigated their potential overlapping functions in cell death by generating Bim/Bad double knockout (DKO) mice. By analysis of their phenotype, and of purified lymphoid populations in culture, we show that Bim and Bad cooperate in certain apoptotic responses and in the suppression of γ-irradiation induced thymic lymphoma. Unexpectedly, some of their cooperation in vivo appears to involve lymphocyte-stromal cell interactions.
RESULTS
Lymphoid hyperplasia in Bim-deficient mice is slightly increased by concomitant loss of Bad
All progeny of bim+/−bad+/− and bim+/+bad−/− intercrosses developed in the expected Mendelian ratios, but intercrosses of bim+/−bad−/− animals yielded less than the expected number of bim−/−bad−/− double-knockout (DKO) offspring (Supplementary Table S1). Their deficit, however, was similar to that of bim−/− progeny from bim+/− intercrosses (14 and PB unpublished observations). Thus, the loss of Bad does not augment the penetrance of embryonic death evoked by Bim deficiency.
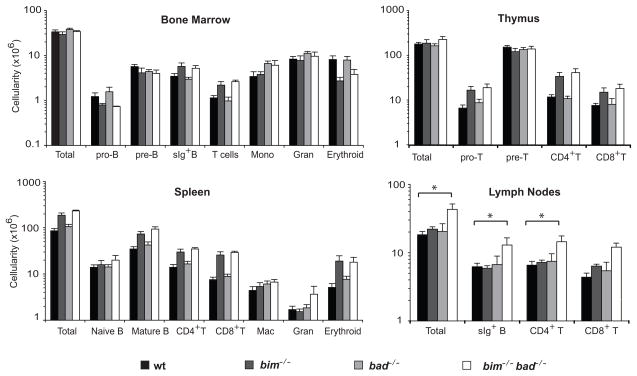
Bim/Bad doubly deficient mice were fertile and their body and organ weights (Supplementary Figure S1), appearance and behaviour were normal. Since Bim28 and Bad29 are both expressed in lymphoid and myeloid cells, we analysed the hematopoietic compartment of 6–12 week old bim−/−bad−/− and control (wt, bim−/−, bad−/−) mice to determine whether their combined loss exacerbated the leukocyte accumulation in bim−/− mice.14 Consistent with previous studies,10,14 bad−/− mice had normal numbers of lymphoid and myeloid cells, whereas Bim-deficient mice had elevated mature B and T cells (Figure 1). The bim−/−bad−/− mice displayed increases in B and T lymphocytes similar to those of bim−/− mice (Figure 1). However, the lymph node cellularity in mice lacking both Bim and Bad was slightly albeit significantly higher (p=0.05) than in bim−/− mice. There was a trend towards increased numbers of granulocytes in spleens of bim−/−bad−/− mice (Figure 1) but this did not reach statistical significance. In addition we found (significantly) increased numbers of nucleated erythroid cells in spleens of bim−/−bad−/− and to a similar extent also in bim−/− mice (Figure 1). We believe, however, that this is not due to abnormally increased survival of these cells but a consequence of a shift of erythropoiesis from the marrow to the spleen (extra-medullary erythropoiesis) due to the over-crowding of the bone marrow by supernumerary B lymphoid cells, as also seen in Eμ-bcl-2 transgenic mice.30
Figure 1.
Hematopoietic cell subset composition of bim−/−bad−/− and control mice. Cell subset composition analysis of bone marrow, thymus, spleen and lymph nodes (pooled axillary, inguinal and mesenteric) from 6–12 week old bim−/−bad−/− and control bim−/−, bad−/− or wt mice. Single cell suspensions were stained with fluorochrome-conjugated surface marker-specific monoclonal antibodies, and the percentages in each cellular compartment quantified by flow cytometric analysis. Total cell numbers were determined by trypan blue exclusion and counting in a haemocytometer. Data represent mean absolute cell numbers ± standard error of 4–7 mice of each genotype from at least 4 independent experiments. An asterisk denotes p<0.05 significance in differences between the indicated populations.
Since loss of Bim elevates serum immunoglobulin (Ig) levels,14 we measured the levels of IgG and IgM by ELISA in the sera of mice either 6–12 week or > 20 week old. Whereas loss of Bim provoked a ~5-fold increase in total serum IgG and IgM levels over wt mice, naïve bad−/− mice showed no increase, and loss of Bim plus Bad did not elevate the levels over loss of Bim alone (Supplementary Figure S2).
Bim-deficient animals (on a mixed C57BL/6x129SV background) develop an autoimmune disorder resembling human systemic lupus erythematosus,14 whereas mice lacking Bad were reported to develop diffuse large B cell lymphoma late in life.10 Hence, we monitored cohorts of bim−/−bad−/−, bim−/−, bad−/− and wt animals, all on a C57BL/6 background (8–20 generations backcrossed), until 500 days of age for signs of autoimmune disease or haematological malignancy. Only a single bim−/−bad−/− mouse succumbed to glomerulonephritis, and none of the 18 other bim−/−bad−/− animals or any of the bad−/− (n=21) or bim−/− (n=20) aged mice examined developed any disease. Hence, the autoimmune disorder that previously manifested in mice lacking Bim appears to depend upon genetic background, even when Bad is also absent, and genetic background may well also have contributed to the low-penetrance lymphoid malignancy reported for aged Bad-deficient mice.10
Bad deficiency increases platelet numbers and life-span
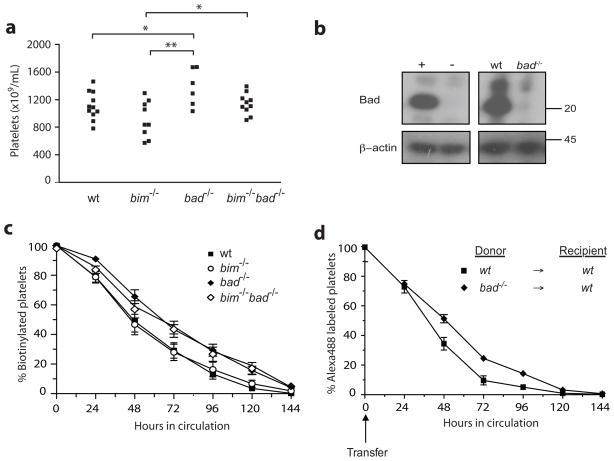
The Bcl-2 family governs the circulating life-span of platelets. These anuclear cells depend on Bcl-xL for survival, which functions to restrain pro-apoptotic Bak. Mutations in Bak extend platelet life span and cause thrombocytosis.31 We unexpectedly found that Bad-deficient mice had elevated platelet numbers in their peripheral blood (Figure 2a), revealing a previously unidentified role for Bad in platelet homeostasis. Western blotting demonstrated that platelets contain Bad protein (Figure 2b), and since the bone marrow and spleen of bad−/−mice exhibited normal numbers of megakaryocytes (data not shown), we hypothesised that platelet life-span, rather than production, was perturbed. We therefore examined platelet clearance by labelling platelets with biotin and tracking their survival in vivo. Platelet half-life - defined as the period during which 50% of labelled platelets had disappeared from the circulation - was significantly (albeit modestly) increased in bad−/− mice (Figure 2c). To examine whether this effect was cell-intrinsic, we performed adoptive transfers of labelled platelets. Upon transfer into wt recipients, the rate of disappearance of bad−/− platelets was decreased compared to wt platelets (Figure 2d). Thus, Bad regulates platelet life-span in vivo in a cell-intrinsic manner.
Figure 2.
bad−/−but not bim−/−bad−/− mice have abnormally increased numbers of platelets. (a) Peripheral blood platelet numbers were determined from 6–12 week old bim−/−bad−/−, bim−/−, bad−/− or wt mice by automated counting (Advia 2120, Bayer). Data represent absolute numbers (x109/mL) of 6–12 mice of each genotype. *P<0.05. (b) Western blot showing expression of Bad in wt platelet lysates and its absence in platelets derived from bad−/− mice. The positive control (+) for Bad expression was a lysate of wt mouse embryonic fibroblasts and the negative control (−) a lysate of bad−/− mouse embryonic fibroblasts. Results are representative of two independent experiments. (c) Platelet half-life in the circulation was determined in 6–12 week old bim−/−bad−/− (n=6), bim−/− (n=10), bad−/− (n=8) or wt (n=16) mice by injection of biotin and flow cytometric analysis of the disappearance of CD41+biotin+ platelets (see Materials and Methods). Data represent mean ± standard error. (d) The half-life of transplanted Bad-deficient platelets in the circulation of 8 week old wt recipient mice (n=8) was determined by adoptive transfer. Alexa488 labelled platelets, derived from bad−/− (n=4) or wt (n=4) donor mice, were injected into wt recipients and the disappearance of CD41+Alexa488+ platelets monitored by flow cytometry (see Materials and Methods). Data represent mean ± standard error.
The increased platelet numbers in bad−/− mice contrasts with the mild thrombocytopenia previously reported for Bim-deficient animals,14 which appears to be due to a reduction in platelet production by megakaryocytes. Remarkably, mice lacking both Bim and Bad had platelet numbers comparable to wt animals (Figure 2a), presumably because the reduced platelet production (conferred by Bim-deficiency) was counter-balanced by the enhanced platelet survival (resulting from loss of Bad).
Sensitivity of bim−/−bad−/− lymphocytes to apoptotic stimuli in vitro
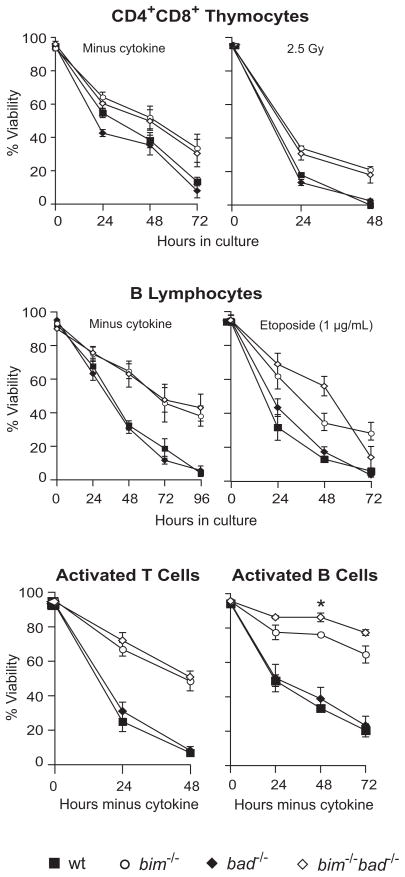
Bim is critical for the apoptosis of lymphocytes evoked by diverse cytotoxic insults, including cytokine withdrawal and deregulated calcium flux,14 and contributes modestly to DNA damage-induced apoptosis,15 even though the bim gene lacks a binding site for the tumour suppressor p53.32 To determine whether Bim and Bad have overlapping roles in apoptosis induction, we purified lymphoid sub-populations from bim−/−bad−/− and control (wt, bim−/−, bad−/−) mice and monitored their survival after exposure to a range of apoptotic stimuli in vitro. Bad-deficient lymphocytes were normally sensitive to cytokine deprivation and to DNA damage, evoked either by etoposide or γ-irradiation (Figure 3). As reported,14,15 bim−/− thymocytes were considerably more resistant than their wt counterparts to cytokine withdrawal and exhibited minor, albeit significant, resistance to γ-irradiation and etoposide (Figure 3). However, combined loss of Bim and Bad did not enhance thymocyte survival in vitro more than loss of Bim alone (Figure 3).
Figure 3.
Susceptibility of bim−/−bad−/− lymphocytes to apoptotic stimuli in culture. CD4+8+ thymocytes and mature B cells (B220+sIgMlosIgDhi) were FACS sorted from thymus or lymph node cell suspensions, respectively, from 6–12 week old bim−/−bad−/−, bim−/−, bad−/− or wt mice and cultured with etoposide (1 μg/mL) or subjected to cytokine withdrawal or γ-irradiation (2.5 Gy). The activated T or B cell blasts were cultured in the absence of cytokines for the indicated times and then stained with FITC-coupled Annexin V plus propidium iodide and the percentages of viable cells quantified by FACS. Data represent mean ± standard error of cells from 3-5 mice of each genotype.
Bim-deficient mature B and T lymphocytes (CD4+8− as well as CD4−8+) were, as reported,14 significantly protected against diverse cytotoxic stimuli, including cytokine deprivation and exposure to dexamethasone, etoposide or γ-irradiation. However, the concomitant absence of Bad did not provide extra protection (Figure 3, Supplementary Figure S3 and data not shown).
Because the signalling pathways that regulate apoptosis often differ between quiescent and activated cells, we mitogenically stimulated B and T cells from bim−/−bad−/− and control mice and monitored their survival in vitro. As reported,14 ConA-activated T lymphoblasts from bim−/− mice were refractory to IL-2 deprivation and modestly protected against γ-irradiation, but the bim−/−bad−/− T lymphoblasts died at the same rate as their bim−/− counterparts (Figure 3 and data not shown). We also examined splenic B lymphocytes activated in vitro with lipopolysaccharide (LPS) plus IL-2, IL-4 and IL-5. Following cytokine deprivation, the survival of bad−/− and wt B cell blasts was similar, whereas bim−/− B lymphoblasts were highly protected (Figure 3). Interestingly, activated B cells from bim−/−bad−/− mice were modestly, but significantly (p<0.05), more refractory to cytokine deprivation than bim−/− cells (Figure 3). Thus, Bim and Bad have an overlapping role in the control of apoptosis in activated B lymphocytes.
Bim and Bad both contribute to γ-irradiation-induced thymocyte apoptosis in vivo
DNA damage potently elicits thymocyte apoptosis, mediated largely,33,34 albeit not exclusively,35 via the tumour suppressor p53. In this response, Puma, a direct p53 transcriptional target, is the rate-limiting BH3-only protein,15,36–38 but Bim also contributes14,15 and Bad might also have a role.10,39,40
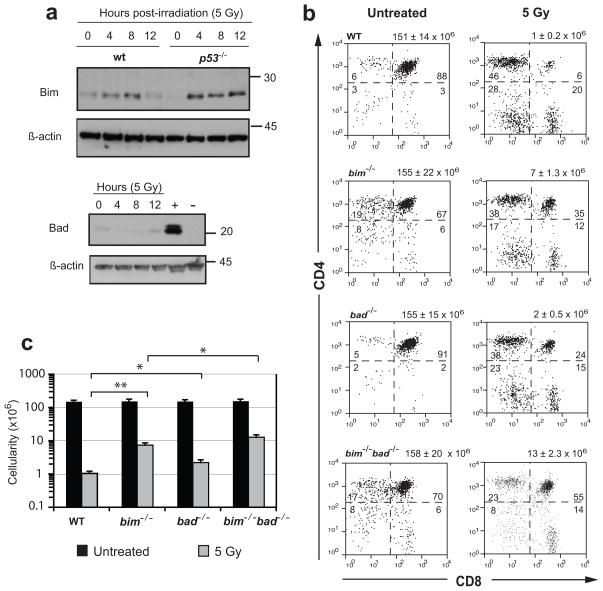
To explore the roles of Bim and Bad in the DNA damage response, we subjected wt and p53-deficient mice to γ-irradiation (5 Gy), harvested their thymi 0, 4, 8 and 12 h later, and assessed Bim and Bad expression by Western blotting. Bim levels increased in a p53-independent manner: as early as 4 h after γ-irradiation, Bim was detectable at similar levels in both wt and p53−/− thymocytes (Figure 4a). By comparison, although the available antibody readily detected Bad in wt fibroblasts (Figure 4a, lane 5), Bad levels were low in untreated wt as well as p53−/− thymocytes and did not noticeably increase after γ-irradiation (Figure 4a).
Figure 4.
Bim and Bad contribute to γ-irradiation-induced thymocyte apoptosis in vivo. (a) Western blot showing the effect of γ-irradiation on Bim and Bad expression in thymocytes, with βactin as a loading control. Wt or p53-deficient mice were left untreated or exposed to γ-irradiation (5 Gy) and lysates made of thymi harvested 0, 4, 8 and 12 h later. The positive control (+) for Bad expression was a lysate of wt mouse embryonic fibroblasts and the negative control (−) a lysate of bad−/− thymocytes. Results are representative of two independent experiments. (b) Effect of γ-irradiation on thymic cell composition. Wt, bim−/−, bad−/− or bim−/−bad−/− mice were exposed to γ-irradiation (5 Gy). Thymi were harvested 20 h later, and cell viability determined by counting single cell suspensions in a haemocytometer. Cells were stained with monoclonal antibodies to CD4 and CD8 to determine the indicated percentages of the four major thymocyte subsets. The numbers of CD4+8+ thymocytes +/- standard error before and after γ-irradiation are shown above the panels. (c) The number of CD4+8+ thymocytes following γ-irradiation. Data represent the mean ± standard error from 3 independent experiments. p<0.05 (*) and p<0.01 (**) are indicated.
Next we examined the impact of loss of Bim, Bad or both on the response of lymphoid cells to DNA damage in vivo. Animals were exposed to 5 Gy γ-irradiation and their thymus, lymph nodes and spleen harvested 20 h later. Cell numbers in each hematopoietic organ were determined and the cell subset composition quantified by immunofluorescent staining with surface marker-specific antibodies and FACS analysis. As reported,41 wt CD4+8+ thymocytes were highly sensitive to γ-irradiation (Figure 4b), their number plummeting by ~150-fold (Figure 4c). The Bad-deficient CD4−8− (pro-T) as well as mature CD4+8− and CD4−8+ thymocytes were killed as efficiently as their wt counterparts. Surprisingly, however, the percentages and total numbers of surviving CD4+8+ thymocytes were ~2-fold higher (p<0.05) in bad−/− than wt mice (Figure 4b). As reported,15 bim−/− CD4+8+ thymocytes were markedly more resistant than wt cells (p<0.0001) to γ-irradiation in vivo (Figure 4b and c). Notably, the combined loss of Bad plus Bim allowed greater survival (p<0.05) than loss of either BH3- only protein alone (Figure 4b and c). Unlike the CD4+8+ thymocytes, however, the mature CD4+ or CD8+ T cells and B cells in bad−/−bim−/− mice responded to γ-irradiation similarly to their bim−/− counterparts, and loss of Bad alone was not protective (Supplementary Figure S4). These results show that Bim and Bad have overlapping functions in γ-irradiation-induced killing of CD4+8+ thymocytes within the animal but that Bad does not contribute significantly to the death of their mature B or T cells.
Bad contributes to γ-irradiation-induced apoptosis of CD4+8+ thymocytes through a hematopoietic cell-extrinsic mechanism
The increased survival of CD4+8+ thymocytes in γ-irradiated bad−/− mice (Figure 4b and c) was surprising because the expression of Bad in thymocytes was exceedingly low and did not increase after DNA damage (Figure 4a). Thymocyte survival is influenced by its microenvironment, a network of epithelial and hematopoietic cells, including dendritic cells and macrophages.42 The observations that bad−/− CD4+8+ thymocytes exhibited enhanced survival to γ-irradiation within the animal (Figure 4b and c) but not as purified cells in culture (Figure 3) suggested that Bad might promote apoptosis of CD4+8+ thymocytes indirectly through a thymocyte-extrinsic mechanism.
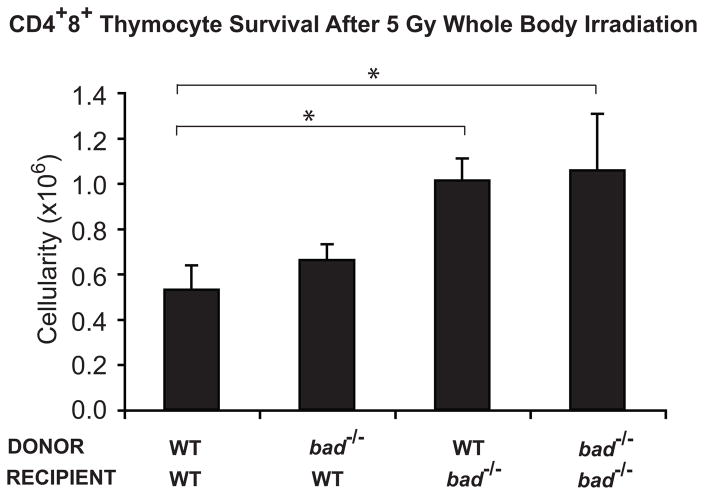
To explore this hypothesis, we generated sets of chimaeric animals by reconstituting lethally irradiated wt or bad−/− recipient mice with either a wt or bad−/− hematopoietic system. At 8–12 weeks post-reconstitution, the mice were γ-irradiated (5 Gy) and the surviving CD4+8+ thymocytes enumerated 20 h later. Both the wt as well as bad−/− thymocytes developing within a Bad-deficient stroma survived γ-irradiation significantly better (p<0.05) than wt or bad−/− thymocytes in a wt environment (Figure 5). The ~2-fold enhanced survival of wt thymocytes in bad−/− recipients was comparable to that of bad−/− thymocytes recovered from bad−/− recipients, and to that of thymocytes within un-manipulated (i.e. non-chimaeric) bad−/−animals (compare Figures 4 and 5). These results show, surprisingly, that Bad must contribute to γ-irradiation-induced apoptosis of CD4+8+ thymocytes within the animal by acting in non-hematopoietic cells, most likely thymic epithelial cells (see Discussion). Pertinently, γ-irradiation provokes both transient and persistent changes to the cellular microenvironment that can facilitate tumorigenesis.43
Figure 5.
Bad contributes to γ-irradiation-induced thymocyte apoptosis in a hematopoietic cell- extrinsic manner. Four combinations of bone marrow-derived chimaeric mice were generated by adoptively transferring bone marrow cells from wt or bad−/− mice into lethally-irradiated (2 × 5.5 Gy, 3 h apart) wt or bad−/− recipients. The reconstituted animals were exposed to 5 Gy whole body γ-irradiation 8-12 weeks post-reconstitution, and the percentages and total numbers of surviving CD4+8+ thymocytes determined 20 h later as described in Figure 4. Data represent mean ± standard error from 3 independent experiments from 4-8 mice of each chimaeric type.
Combined loss of Bim and Bad accelerates γ-irradiation-induced thymic lymphoma development
The reproducible development of thymic lymphoma by C57BL/6 mice subjected to fractionated low doses of γ-irradiation44 provides a robust experimental test for candidate tumour suppressors. Bim22,45,46 and to a lesser extent Bad10 have been implicated as tumour suppressors. Since both contribute to DNA damage-induced apoptosis of thymocytes, albeit Bim in a hematopoietic cell intrinsic (Figure 4) and Bad in an extrinsic manner (Figure 5), we investigated how their individual or combined loss affected γ-irradiation-induced thymic lymphoma development. We exposed cohorts of wt (n=10), bim−/−(n=16), bad−/− (n=22), bim−/− bad−/− (n=10) and, as a control, p53−/− (n=5) mice to fractionated low dose (4 weekly doses of 1.5 Gy) γ-irradiation44 and monitored them daily for signs of malignancy.
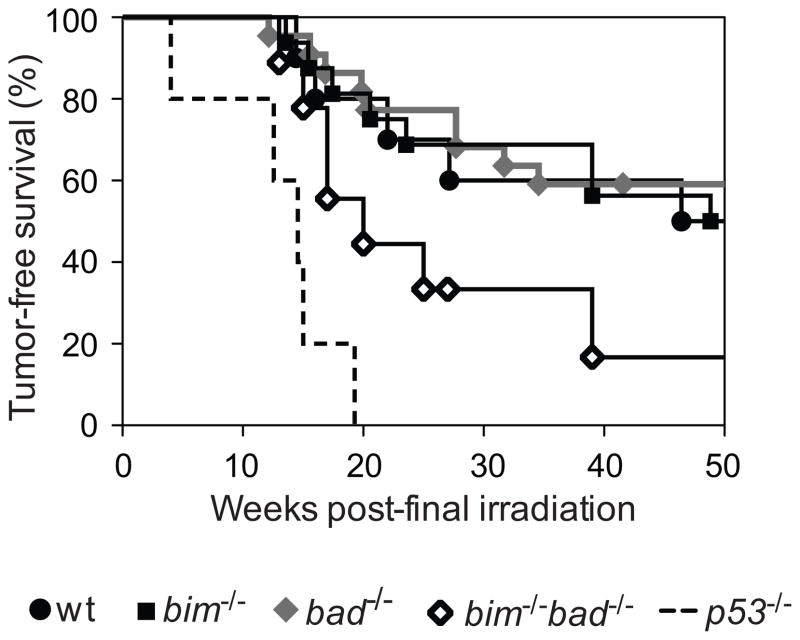
As reported,47 thymic lymphoma developed considerably faster and with higher incidence in p53−/− mice (100% mortality by 14 weeks after the final dose of γ-irradiation) than in wt animals (10% by 14 weeks and ~50% by 57 weeks; Kaplan Meier logrank p<0.001). Although some old bad−/− mice were reported to develop B lymphoma,10 their development of thymic lymphoma (~50% incidence, median latency 60 weeks) was comparable to that of wt mice (Figure 6). Although Bim deficiency (even loss of one allele) markedly accelerates B lymphoma development in Eμ-myc transgenic mice,22 bim−/− and wt mice developed γ-irradiation-induced thymic lymphoma with similar onset and incidence. Remarkably, however, the combined loss of Bim and Bad substantially accelerated (p<0.04) thymic lymphomagenesis (~80% incidence, median latency 23 weeks) over that in wt animals (~50% incidence, median latency 60 weeks; Figure 6). Histological evaluation of thymic sections from mice of all genotypes revealed a similar extent of tumour dissemination and disruption of the thymic architecture by infiltration of large blast cells (data not shown). Moreover, immuno-phenotyping revealed that most thymic lymphomas, regardless of genotype, were Thy-1+CD4+CD8+ and displayed CD43 and/or CD44 (data not shown). All were transplantable into histocompatible recipients, producing large tumours within 40 days. These results demonstrate that Bim and Bad have overlapping roles as tumour suppressors, at least in this model of γ-irradiation-induced thymic lymphomagenesis.
Figure 6.
Concomitant loss of Bim and Bad accelerates γ-irradiation-induced thymic lymphoma development. Kaplan-Meier representation of tumour latency in bim−/−bad−/− and control bim−/−, bad−/−, wt and p53−/− mice following 4 weekly doses of 1.5 Gy whole body γ-irradiation. bim−/−bad−/− mice developed γ-irradiation-induced thymic lymphoma significantly faster (p<0.04) than control mice. Mice were monitored daily for signs of tumour development (hunched posture, laboured breathing and increased spleen/lymph nodes) and sick mice were sacrificed and tissues analysed immediately.
DISCUSSION
Since both Bim48,49 and Bad7,8,50 can be activated by growth factor deprivation and certain other apoptotic stimuli, we generated Bim/Bad DKO mice to investigate potential overlaps in their apoptotic function, both within the animal and in cultured lymphoid populations. Perhaps surprisingly, the bim−/−bad−/− mice exhibited only slightly greater lymphadenopathy than the bim−/− animals (Figure 1), and their resting lymphocytes were no more resistant to cytokine deprivation than the Bim-deficient cells (Figure 3 and Supplementary Figure S3). The absence of synergy between Bim and Bad in these cells is compatible with a model in which neutralisation of all Bcl-2-like pro-survival proteins expressed within a given cell is required to initiate apoptosis efficiently.51–53 Because Bcl-2 and Mcl-1 are both highly expressed in resting mature B and T lymphocytes and critical for their survival,54,55 complete neutralisation of both these proteins is probably required for their efficient killing. Whereas Bim and Puma are particularly potent killers because they can engage all the pro-survival proteins, Bad is much less effective because it cannot bind to Mcl-1 (or A1).51,52 Thus, in any cell whose survival is sustained by Mcl-1, Bad may only marginally boost Bim’s pro-apoptotic function. In accord with that view, loss of both Puma and Bim elicits more profound and widespread effects on the lymphoid compartment16 than combined loss of Bim plus Bad.
Bim and Bad do not cooperate in the development of autoimmunity or spontaneous malignancy
Although quiescent bim−/−bad−/− and bim−/− B lymphocytes exhibited similar resistance to cytokine deprivation in vitro, mitogen-activated B cells from the doubly-deficient animals were more refractory than Bim-deficient B lymphoblasts (Figure 3). This difference may be because B lymphoblasts express higher levels than resting B cells of pro-survival proteins that Bad can neutralise, such as Bcl-xL.56 The modest increase in resistance of bim−/−bad−/− B cell blasts to cytokine deprivation did not correlate, however, with notable evidence of an abnormally activated immune system. Although many bim−/−bad−/− mice presented with larger lymph nodes than bim−/− mice, their numbers of mature B or T cells and their serum Ig levels were not significantly elevated. Moreover, although a fatal SLE-like autoimmunity was prevalent for bim−/− mice on a mixed C57BL/6x129SV genetic background,14 the bim−/− and bim−/−bad−/− mice analysed in this study, all extensively backcrossed to C57BL/6 animals, were not highly susceptible to autoimmune disease, so genetic background clearly has a critical role. Similarly, although it has been reported that aged (~18 month-old) bad−/− mice are prone to develop diffuse large B cell lymphoma,10 we observed no malignancies in cohorts of bad−/− or even bim−/−bad−/− animals monitored for a comparable period. Genetic background probably accounts for this difference, because the previous study involved mice on a mixed C57BL/6x129SV background. Pertinently, the 129SV background promotes tumorigenesis in other models of malignancy.57
Distinct roles for Bad and Bim in platelet homeostasis
We were intrigued to find that mice lacking Bad (but not those lacking Bim) have a modest elevation in peripheral blood platelet number (Figure 2a). Our data suggest that the basis for this effect is not an increase in their production, as megakaryocyte numbers were not elevated, but rather an extension of circulating platelet half-life (Figure 2c). Platelet life-span is known to be regulated by the Bcl-2 family of proteins, in particular pro-survival Bcl-xL and pro-apoptotic Bak.31 Platelets depend on Bcl-xL to restrain Bak and maintain viability in the circulation.31,58 Loss of function mutations in Bcl-xL cause dose-dependent reductions in platelet life span31 and pharmacological inhibition of Bcl-xL with the BH3-mimetic ABT- 73759 triggers platelet apoptosis and thrombocytopenia, apparently by freeing Bak and Bax from Bcl-xL. The mechanism by which this occurs at steady state in vivo is still unclear. One possibility is that BH3-only proteins regulate the entry into apoptosis. Intriguingly, a recent study has suggested that Akt-mediated inactivation of Bad facilitates the survival of human platelets in vitro.60 Our data demonstrate that in the absence of Bad, platelet life span is extended in vivo. Since ABT-737 has the same binding specificity as Bad (Bcl-2, Bcl-xL and Bcl-w),61 Bad probably contributes to the initiation of physiological platelet apoptosis by neutralising Bcl-xL.
Bim and Bad contribute in distinct ways to γ-irradiation-induced apoptosis of thymocytes in vivo
The major regulator of the cellular response to DNA damage is the p53 tumour suppressor,62 which appears to mediate apoptosis primarily by inducing Puma, with minor contributions from Noxa.15,36–38 However, p53-independent DNA damage-induced apoptotic pathways exist, as illustrated by the apoptosis following γ- or UV-irradiation observed in many tumour cells lacking functional p53.35 Indeed, Bim is up-regulated by DNA damage in γ-irradiated p53-deficient thymocytes (Figure 4a), and loss of Bim can protect (albeit only to a minor extent) certain lymphoid cell types from DNA damage-induced apoptosis both in vitro14 and in vivo.15 How genotoxic insults up-regulate Bim is uncertain, but a plausible intermediary is FOXO3A, as it is a critical inducer of bim transcription in cytokine-deprived hematopoietic cells48 and can be activated by DNA damage in a p53-independent manner.63
Surprisingly, loss of Bad had no effect on γ-irradiation-induced thymocyte killing in vitro (Figure 3), but it diminished their death in vivo (Figure 4b and c). Analysis of chimaeric mice (Figure 5) showed that this protective effect was not intrinsic to the CD4+8+ thymocyte population but rather due to loss of Bad within the thymic microenvironment. These results suggest that following γ-irradiation a nurturing stromal cell population lacking Bad survives better, presumably rendering it more able to sustain the survival of the thymic T lymphoid cells.
Bim and Bad cooperate in the suppression of γ-irradiation-induced thymic lymphoma development
Using the classic low dose γ-radiation-induced thymic lymphoma model,44 we found that combined loss of Bim and Bad, albeit not loss of either BH3-only protein alone, accelerated tumorigenesis, demonstrating functional overlap between these BH3-only proteins in tumour suppression. Loss of p53 was more potent in accelerating thymic lymphoma development than combined loss of Bim and Bad; this is consistent with the capacity of p53 to activate multiple tumour suppressive mechanisms in addition to apoptosis, including cell cycle arrest and senescence.62
Since Bim and Bad are not thought to be involved in p53-induced apoptosis, how might their combined loss accelerate γ-irradiation-induced thymic lymphoma development? It is notable that after γ-irradiation, thymocytes are considerably more numerous in bim−/− and bim−/−bad−/− animals than in wt mice (Figure 4b). Thus, protection conferred by the absence of Bim and/or Bad substantially increases the size of the putative susceptible target population. Furthermore, the elimination of more than 90% of the more mature cells by the γ-irradiation (Figure 4b) will provide a strong feedback signal to the immature cells, including nascent neoplastic clones, to proliferate and regenerate the thymus. Indeed, the repeated γ-irradiation doses both re-enforce this impetus and provide the mutagenesis contributing toward malignant progression.44 Our observation that Bad deficiency protects the thymic microenvironment from the irradiation (Figure 5) suggests that the stroma lacking Bad can more effectively sustain the emerging neoplastic clones. Accordingly, we propose that the accelerated lymphoma development in bim−/−bad−/− animals may reflect a concomitant cell-autonomous enhancement of survival in immature thymocytes, mediated largely by the absence of Bim, and a hematopoietic-extrinsic contribution by a more robust stromal support population, promoted primarily by the absence of Bad. This interpretation of the data would coincide with the view that tumorigenesis often relies on changes in the microenvironment of the emerging malignant clone.43 Alternatively, since bone marrow resident hematopoietic stem/progenitor cells that have sustained oncogenic lesions are critical for γ-radiation-induced thymic lymphoma development,44 it is also possible that combined loss of Bim and Bad accelerates tumorigenesis by extending the survival of such “lymphoma stem cells”.
Evidence is emerging that combined activation of Bim and Bad is not only important for tumour suppression but also plays a critical role in anti-cancer therapy. The killing of Bcr-Abl transformed cells by Imatinib (Gleevec) requires Bim and to a lesser extent Bad,64 which are both activated by shutdown of PI3K/AKT and ERK signalling. Consequently, our results suggest that treatment of certain types of lymphomas might be improved by agents that target these signalling pathways.
MATERIALS AND METHODS
Mice
All animal work followed the guidelines of the Melbourne Directorate Animal Ethics Committee. The generation and genotyping of bim−/− mice,14 bad−/− mice,10 and p53−/− mice65 has been described previously. These strains of mice were all originally generated on a mixed C57BL/6x129SV background, using 129SV-derived ES cells, but had been backcrossed with C57BL/6 mice for 8–15 generations prior to use in the studies described here. The bim−/−bad−/−mice were generated by intercrossing bim−/− and bad−/− mice. Animals were analysed at 6–12 weeks of age unless otherwise specified.
Immunofluorescence staining, flow cytometric analysis and cell sorting
Single cell suspensions were prepared from bone marrow, spleen, lymph nodes, peripheral blood or thymus. Cell subset composition was determined by immunofluorescent staining with the following rat or hamster monoclonal antibodies: RA3-6B2 (anti-B220), S7 (anti-CD43), 333.12 or 5.1 (anti-IgM), 11-26C (anti-IgD), H129 or YTA321 (anti-CD4), YTS169 (anti-CD8), MI/70 (anti-Mac-1), RB6-8C5 (anti-Gr-1), T3.24.1 (anti-Thy-1), IM781 (anti- CD44) and Ter119 (anti-erythroid marker), followed by flow cytometric analysis in a FACScan (BD Biosciences). Staining was performed in the presence of anti-Fcγ receptor II (FcγRII) antibody (2.4G2) plus 2% normal rat serum to prevent non-specific binding of antibodies. Antibodies were produced in our laboratory and conjugated to biotin (Molecular Probes), fluorescein isothiocyanate (FITC, Molecular Probes), cyanine 5 (Cy5, Amersham), R-phycoerythrin (R-PE, Prozyme), or allophycocyanin (APC, Prozyme) according to the manufacturers’ instructions. Biotinylated antibodies were visualised by secondary staining using FITC-, PE- or Tricolor-streptavidin conjugates (Caltag). Dead cells were excluded by staining with propidium iodide (2 μg/mL). Cell sorting was performed using a MoFlo (Cytomation) or DiVa (BD Biosciences) high-speed flow cytometer.
Enzyme-linked immunosorbent assay (ELISA)
Immunoglobulin levels were measured using ELISA as described.66 ELISA plates were coated with goat anti-total mouse immunoglobulin antibodies (Southern Biotechnology Associates) in carbonate buffer. For measurement of total IgG levels, biotinylated goat antibodies specific to mouse IgG1, IgG2a, IgG2b and IgG3 (Southern Biotechnology Associates) were combined for detection, followed by incubation with avidin-HRP and enzymatic detection. For determination of IgM levels, a biotinylated goat anti-IgM antibody was used as the secondary reagent.
Platelet clearance analysis
Mice were injected intravenously (i.v.) with 600 μg N-hydroxysuccinimidobiotin (NHS- biotin; Sigma) in buffer containing 140 mM NaCl and 10% DMSO. At various time points, whole blood was isolated from the tail vein and mixed with Resuspension Buffer Mix, which comprised 25% (v/v) Aster Jandl Anticoagulant (85 mM sodium citrate dihydrate, 69 mM citric acid anhydrans, 10 mM glucose, pH 4.6) and 75% (v/v) Resuspension buffer (10 mM HEPES, 140 mM NaCl, 3 mM KCl, 0.5 mM MgCl2 hexahydrate, 0.5 mM NaHCO3, 10 mM glucose, pH 7.4). The buffy coat, containing the platelet fraction, was separated by centrifugation at 125 g for 5 min and stained with FITC-conjugated rat anti-CD41 monoclonal antibody (Becton Dickinson) and PE-conjugated streptavidin for 40 min at room temperature. Samples were washed in Wash buffer (140 mM NaCl, 5 mM KCl, 12 mM sodium citrate, 10 mM glucose, 12.5 mM sucrose, pH 6.0) by centrifugation at 860 g for 5 min. The platelet pellet was resuspended in resuspension buffer prior to flow cytometric analysis on a FACSCalibur (Becton Dickinson).
Adoptive transfer of platelets
Donor mice (wt or bad−/−; ~12 week-old) were injected with 0.1 μg/kg Alexa488-conjugated antibody specific for GPIβ subunit of GPIβ-V-IX for in vivo platelet labelling (Emfret). Labelling efficiency was assessed by flow cytometry, and only donor mice with >90% of the platelet population Alexa488+ were used for experiments. Platelets were purified from these donors and injected intravenously into (untreated; ~8 week-old) C57BL/6 recipients. At various time points, whole blood was isolated from the tail vein of recipients as described (see above under ‘Platelet clearance analysis’) and the disappearance of Alexa488+ platelets analysed by flow cytometry.
Immunoblotting
Protein lysates were fractionated using 12% SDS-PAGE gels (Invitrogen), and electro-blotted onto nitrocellulose membranes (Hybond C-extra, Amersham). Membranes were probed with rabbit polyclonal anti-Bad (Cell Signalling) or rat monoclonal anti-Bim (clone 3C5, Alexis) antibodies. Membranes were probed with mouse monoclonal anti-β-actin (clone AC-74, Sigma) antibodies to demonstrate equal loading. Horseradish peroxidase (HRP)-conjugated sheep-anti-rabbit immunoglobulin (Ig), goat anti-rat Ig (Southern Biotech) or sheep anti-mouse Ig antibodies (Chemicon) were used as secondary reagents and visualised using enhanced chemiluminescence (ECL) reagent (Amersham Biosciences). The molecular weight of proteins was measured using the rainbow molecular weight marker cocktail (RPN 756 Amersham Biosciences).
Cell culture and cell death assays
Cells were cultured in the high-glucose version of Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 250 μM L-asparagine, 50 μM 2-mercaptoethanol and 10% heat-inactivated foetal calf serum (JRH Biosciences). Activated B cells were generated by culturing splenocytes for 3 days in medium supplemented with 20 μg/mL lipopolysaccharide (Difco) plus IL-2, IL-4 and IL-5 (each at 100 U/mL). Activated B cells were FACS sorted by staining with a cocktail of FITC-conjugated antibodies to CD4, CD8, Gr-1, Mac-1 and Ter119 plus PI and collecting the unstained (FITC−PI−) fraction. Sorted activated B cells were typically >98% pure as assessed by staining for the B lymphoid specific marker B220. Activated T cells were generated by culturing spleen cells for 3 days in medium containing Concanavalin A (Con A, 2 μg/mL) plus IL-2 (100 U/mL), followed by 2 days of culture in medium containing IL-2 alone (100 U/mL). Viable T cell blasts were FACS sorted by staining with a cocktail of FITC-conjugated antibodies to B220, CD19, Gr-1, Mac-1 and Ter119 plus PI and collecting the unstained (FITC−PI−) fraction. Sorted T cell blasts were typically >98% pure as assessed by staining with antibodies to CD4 and CD8. Viability of cultured cells was assessed by staining with FITC-coupled Annexin V plus propidium iodide (2μg/mL) and analysis using a FACScan (Becton Dickinson).
Generation of bone marrow chimaeric mice
Wild-type C57BL/6-Ly5.1, C57BL/6-Ly5.2 or bad−/− (C57BL/6-Ly5.2) mice were subjected to lethal γ-irradiation (2 × 5.5 Gy - 3 h interval) from a 60 Co source at 3.6 Gy/min. Bone marrow cells (5 × 106) from C57BL/6-Ly5.2 or bad−/− (C57BL/6-Ly5.2) mice were used to reconstitute lethally irradiated C57BL/6-Ly5.1 mice. In addition, we reconstituted lethally irradiated bad−/− (C57BL/6-Ly5.2) mice with bone marrow from C57BL/6-Ly5.1 or bad−/−(C57BL-6-Ly5.2) mice to serve as controls. Animals were maintained on neomycin supplemented drinking water (1.6 g/L) for 14 days post-irradiation to prevent infection, and analysed 8–12 weeks post-reconstitution.
Whole body γ-irradiation and thymic lymphoma induction experiments
Mice were subjected to 2.5 or 5 Gy γ-irradiation. Bone marrow, spleen, thymus and lymph nodes were harvested 20 h post-irradiation. Single cell suspensions were prepared and total cell numbers determined by trypan blue staining and counting in a haemocytometer. Absolute numbers of cell subsets were calculated by multiplying the percentage of a cell type with the total organ cellularity. For studies on γ-irradiation induced thymic lymphoma development, cohorts of wt, bim−/−, bad−/−, bim−/−bad−/− and p53−/− mice aged between 25–35 days were subjected to 1.5 Gy γ-irradiation at 7 day intervals over 4 consecutive weeks as described.44 Mice were monitored daily for signs of malignancy and sick animals sacrificed and subjected to post mortem analysis.
Statistical analysis
Results are expressed as mean ± standard error. Statistical analysis was performed using the Students t test. P values less than 0.05 were considered to be statistically significant differences. Kaplan-Meier tumour-free survival curves were constructed using GraphPad Prism software (Version 5), and statistical analysis was performed using the logrank (Mantel- Cox) test.
Supplementary Material
Progeny frequencies during breeding of bim−/−bad−/− mice. (a) bim+/−bad+/− mice were mated with bad−/− mice in order to fix the bad knockout allele. (b) Intercrosses of the resulting bim+/−bad−/− progeny generated bim−/−bad−/− offspring. Values in brackets indicate expected yields. The frequency of bim−/−bad−/− offspring (68%) was significantly different (p<0.0003) from the predicted Mendelian ratio but the deficit was not greater than that of bim−/− progeny from bim+/− parents; thus, the loss of Bad did not exacerbate the extent of embryonic lethality produced by Bim loss.
Mice deficient for both Bim and Bad develop normally. Whole body and organ weights (g) were determined from 6–12 week old (a) male and (b) female bim−/−bad−/−, bim−/−, bad−/− and wt mice. Data represent mean weight ± SEM from 4–6 mice of each genotype.
Serum immunoglobulin levels in bim−/−bad−/− and control mice. (a) Serum IgM and (b) total IgG levels from naïve bim−/−bad−/− and control bim−/−, bad−/− and wt mice (aged 6–12 weeks or >20 weeks of age) were quantified by ELISA.
Susceptibility of bim−/−bad−/− CD4+ T and CD8+ T lymphocytes to apoptotic stimuli. Mature CD4+ T (CD4+8−) and mature CD8+ T (CD4−8+) cells were purified from the lymph nodes of 6–12 week old mice by FACS sorting and cultured with either etoposide (1 μg/mL) or subjected to cytokine withdrawal. Cells were stained with FITC-conjugated Annexin V plus propidium iodide and the percentage of viable cells (Annexin V−PI−) quantified by FACS analysis. Data points represent mean ± standard error of 3–5 mice of each genotype.
Bim and Bad do not have overlapping roles in γ-irradiation-induced apoptosis of mature lymphocytes in vivo. Mice (bim−/−bad−/− and control bim−/−, bad−/− and wt) were subjected to γirradiation (5 Gy) or were left untreated (control). Lymph nodes were harvested 20 h following γ-irradiation and single cell suspensions stained with monoclonal antibodies to CD4, CD8 and B220. Absolute cell numbers were determined by trypan blue staining and counting in a haemocytometer. Data are expressed as mean ± SEM of 4–6 mice of each genotype from 3 independent experiments. p<0.05 (*) and p<0.01 (**) are indicated.
Acknowledgments
We thank Drs S Cory, the late AW Harris, LA O’Reilly and L Lee for gifts of mice, reagents and advice; the late Prof S Korsmeyer and Dr N Danial for bad−/− mice, K Vella, G Siciliano, A Naughton, N Iannarella, K Pioch, J Merryfull and M James for expert animal care, B Herbert, C Young, L Tai and M Robati for genotyping, Dr F Battye, V Milovac, C Tarlinton, C Young and J Garbe for cell sorting, J Corbin for automated blood analysis, Dr S Mihajlovic, E Tsui and A Hasanein for histology, D Quilici, T Nikolaou, G Thomas, S Kwok and D Baum for γ-irradiation. This work was supported by fellowships and grants from the Cancer Council of Victoria, the Australian National Health and Medical Research Council (Program Grant 257502, Project Grant 516725), the Leukaemia Foundation of Australia, the Sylvia and Charles Viertel Charitable Foundation, the Leukemia and Lymphoma Society (SCOR grant 7015) and the US National Cancer Institute (CA 43540).
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
CONFLICT OF INTEREST/DISCLOSURE: The authors declare no competing financial interests.
References
- 1.Strasser A, O’Connor L, Dixit VM. Apoptosis signaling. Ann Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007 Feb 26;26(9):1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang DCS, Strasser A. BH3-only proteins – essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 4.Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol. 2005;5:189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- 5.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292(5517):727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 2001;15(12):1481–1486. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14–3-3 not Bcl-xL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 8.del Peso L, González-Garcia M, Page C, Herrera R, Nuñez G. Interleukin-3–induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 9.Zha JP, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer SJ. BH3 domain of BAD is required for heterodimerization with BCL–XL and pro-apoptotic activity. Journal of Biological Chemistry. 1997;272(39):24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 10.Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100(16):9324–9329. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424(6951):952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 12.Ekert PG, Jabbour AM, Manoharan A, Heraud JE, Yu J, Pakusch M, et al. Cell death provoked by loss of Interleukin-3 signalling is independent of Bad, Bim, and PI3 Kinase, but depends in part on Puma. Blood. 2006 May 16;108(5):1461–1468. doi: 10.1182/blood-2006-03-014209. [DOI] [PubMed] [Google Scholar]
- 13.Puthalakath H, Strasser A. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 2002;9(5):505–512. doi: 10.1038/sj.cdd.4400998. [DOI] [PubMed] [Google Scholar]
- 14.Bouillet P, Metcalf D, Huang DCS, Tarlinton DM, Kay TWH, Köntgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 15.Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for {gamma}-radiation and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005 Aug 23;106(3):4131–4138. doi: 10.1182/blood-2005-04-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erlacher M, Laabi V, Manzl C, Bock G, Tzankov A, Haecker G, et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. Journal of Experimental Medicine. 2006;203(13):2939–2951. doi: 10.1084/jem.20061552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouillet P, Purton JF, Godfrey DI, Zhang L-C, Coultas L, Puthalakath H, et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- 18.Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreative B cells. J Exp Med. 2003;198:1119–1126. doi: 10.1084/jem.20030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hildeman DA, Zhu Y, Mitchell TC, Bouillet P, Strasser A, Kappler J, et al. Activated T cell death in vivo mediated by pro-apoptotic Bcl-2 family member, Bim. Immunity. 2002;16:759–767. doi: 10.1016/s1074-7613(02)00322-9. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini M, Belz G, Bouillet P, Strasser A. Shut down of an acute T cell immune response to viral infection is mediated by the pro-apoptotic Bcl-2 homology 3-only protein Bim. Proc Natl Acad Sci U S A. 2003;100(24):14175–14180. doi: 10.1073/pnas.2336198100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes PD, Belz GT, Fortner K, Budd RC, Strasser A, Bouillet P. Apoptosis regulators Fas and Bim cooperate in shutdown of chronic immune responses and prevention of autoimmunity. Immunity. 2008;28(2):197–205. doi: 10.1016/j.immuni.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004 Apr 20;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsten T, Ross AJ, King A, Zong W, Rathmell JC, Shiels HA, et al. The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol Cell. 2000;6(6):1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rathmell JC, Lindsten T, Zong W-X, Cinalli RM, Thompson CB. Deficiency in Bak and Bax perturbs thymic selection and lymphoid homeostasis. Nature Immunology. 2002;3(10):932–939. doi: 10.1038/ni834. [DOI] [PubMed] [Google Scholar]
- 25.Ogilvy S, Metcalf D, Print CG, Bath ML, Harris AW, Adams JM. Constitutive bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci U S A. 1999;96(26):14943–14948. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coultas L, Bouillet P, Stanley EG, Brodnicki TC, Adams JM, Strasser A. Proapoptotic BH3-only Bcl-2 family member Bik/Blk/Nbk is expressed in hemopoietic and endothelial cells but is redundant for their programmed death. Mol Cell Biol. 2004 Feb;24(4):1570–1581. doi: 10.1128/MCB.24.4.1570-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coultas L, Bouillet P, Loveland KL, Meachem S, Perlman H, Adams JM, et al. Concomitant loss of proapoptotic BH3-only Bcl-2 antagonists Bik and Bim arrests spermatogenesis. Embo J. 2005 Nov 16;24(22):3963–3973. doi: 10.1038/sj.emboj.7600857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Reilly LA, Cullen L, Visvader J, Lindeman G, Print C, Bath ML, et al. The pro-apoptotic BH3-only protein Bim is expressed in hemopoietic, epithelial, neuronal and germ cells. American Journal of Pathology. 2000;157:449–461. doi: 10.1016/S0002-9440(10)64557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mok C-LGG-G, Williams O, Coles M, Taga S, Tolaini M, et al. Bad can act as a key regulator of T cell apoptosis and T cell development. Journal of Experimental Medicine. 1999;189(3):575–586. doi: 10.1084/jem.189.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci U S A. 1991;88:8661–8665. doi: 10.1073/pnas.88.19.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, Ellis S, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007 Mar 23;128(6):1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, et al. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mammalian Genome. 2001;12(2):163–168. doi: 10.1007/s003350010242. [DOI] [PubMed] [Google Scholar]
- 33.Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 34.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 35.Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 36.Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 37.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 38.Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types the tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008 Jun;15(6):1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta SR, Ranger AM, Lin MZ, Sturgill JF, Ma YC, Cowan CW, et al. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Developmental Cell. 2002;3(5):631–643. doi: 10.1016/s1534-5807(02)00326-x. [DOI] [PubMed] [Google Scholar]
- 40.Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosispoptosis. Mol Cell Biol. 2006 Dec;26(23):9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strasser A, Harris AW, Cory S. Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 42.van Ewijk W, Wang B, Holländer G, Kawamoto H, Spanopoulou E, Itoi M, et al. Thymic microenvironments, 3-D versus 2-D? Seminars in Immunology. 1999;11(1):57–64. doi: 10.1006/smim.1998.0158. [DOI] [PubMed] [Google Scholar]
- 43.Barcellos-Hoff MH, Park C, Wright EG. Radiation and the microenvironment -tumorigenesis and therapy. Nat Rev Cancer. 2005 Nov;5(11):867–875. doi: 10.1038/nrc1735. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan HS, Brown MB. A quantitative dose-response study of lymphoid-tumor development in irradiated C57 black mice. Journal of the National Cancer Institute. 1952;13:185–208. [PubMed] [Google Scholar]
- 45.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005 Feb 17;24(8):1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 46.Sturm I, Stephan C, Gillissen B, Siebert R, Janz M, Radetzki S, et al. Loss of the tissue-specific proapoptotic BH3-only protein Nbk/Bik is a unifying feature of renal cell carcinoma. Cell Death Differ. 2005 Dec;2:1–9. doi: 10.1038/sj.cdd.4401782. [DOI] [PubMed] [Google Scholar]
- 47.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nature Genetics. 1994;8:66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 48.Dijkers PF, Medema RH, Lammers JJ, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Current Biology. 2000;10(19):1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 49.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005 Aug;12(8):1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 50.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005 Feb 4;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 52.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 Domains of BH3-Only Proteins Differentially Regulate Bax-Mediated Mitochondrial Membrane Permeabilization Both Directly and Indirectly. Mol Cell. 2005 Feb 18;17(4):525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Pro-apoptotic Bak is sequestered by Mc1–1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005 Jun 1;19(11):1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75:229–240. doi: 10.1016/0092-8674(93)80065-m. [DOI] [PubMed] [Google Scholar]
- 55.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL- 1. Nature. 2003 Dec 11;426(6967):671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 56.Grillot DAM, Merino R, Pena JC, Fanslow WC, Finkelman FD, Thompson CB, et al. bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. Journal of Experimental Medicine. 1996;183:381–391. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donehower LA, Harvey M, Vogel H, McArthur MJ, Montgomery CA, Jr, Park SH, et al. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog. 1995 Sep;14(1):16–22. doi: 10.1002/mc.2940140105. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, Nimmer PM, Tahir SK, Chen J, Fryer RM, Hahn KR, et al. Bcl-2 family proteins are essential for platelet survival. Cell Death and Differentiation. 2007;14:943–951. doi: 10.1038/sj.cdd.4402081. [DOI] [PubMed] [Google Scholar]
- 59.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005 Jun 2;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 60.Catani MV, Gasperi V, Evangelista D, Finazzi Agro A, Avigliano L, Maccarrone M. Anandamide extends platelets survival through CB(1)-dependent Akt signaling. Cell Mol Life Sci. 2010 Feb;67(4):601–610. doi: 10.1007/s00018-009-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vousden KH, Vande Woude GF. The ins and outs of p53. Nature Cell Biology. 2000;2(10):E178–E180. doi: 10.1038/35036427. [DOI] [PubMed] [Google Scholar]
- 63.Yang JY, Xia W, Hu MC. Ionizing radiation activates expression of FOXO3a, Fas ligand, and Bim, and induces cell apoptosis. Int J Oncol. 2006 Sep;29(3):643–648. [PMC free article] [PubMed] [Google Scholar]
- 64.Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14907–14912. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 66.Fischer SF, Bouillet P, O’Donnell K, Light A, Tarlinton DM, Strasser A. Pro-apoptotic BH3-only protein Bim is essential for developmentally programmed death of germinal center-derived memory B cells and antibody forming cells. Blood. 2007 Dec 1;110(12):3978–3984. doi: 10.1182/blood-2007-05-091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Progeny frequencies during breeding of bim−/−bad−/− mice. (a) bim+/−bad+/− mice were mated with bad−/− mice in order to fix the bad knockout allele. (b) Intercrosses of the resulting bim+/−bad−/− progeny generated bim−/−bad−/− offspring. Values in brackets indicate expected yields. The frequency of bim−/−bad−/− offspring (68%) was significantly different (p<0.0003) from the predicted Mendelian ratio but the deficit was not greater than that of bim−/− progeny from bim+/− parents; thus, the loss of Bad did not exacerbate the extent of embryonic lethality produced by Bim loss.
Mice deficient for both Bim and Bad develop normally. Whole body and organ weights (g) were determined from 6–12 week old (a) male and (b) female bim−/−bad−/−, bim−/−, bad−/− and wt mice. Data represent mean weight ± SEM from 4–6 mice of each genotype.
Serum immunoglobulin levels in bim−/−bad−/− and control mice. (a) Serum IgM and (b) total IgG levels from naïve bim−/−bad−/− and control bim−/−, bad−/− and wt mice (aged 6–12 weeks or >20 weeks of age) were quantified by ELISA.
Susceptibility of bim−/−bad−/− CD4+ T and CD8+ T lymphocytes to apoptotic stimuli. Mature CD4+ T (CD4+8−) and mature CD8+ T (CD4−8+) cells were purified from the lymph nodes of 6–12 week old mice by FACS sorting and cultured with either etoposide (1 μg/mL) or subjected to cytokine withdrawal. Cells were stained with FITC-conjugated Annexin V plus propidium iodide and the percentage of viable cells (Annexin V−PI−) quantified by FACS analysis. Data points represent mean ± standard error of 3–5 mice of each genotype.
Bim and Bad do not have overlapping roles in γ-irradiation-induced apoptosis of mature lymphocytes in vivo. Mice (bim−/−bad−/− and control bim−/−, bad−/− and wt) were subjected to γirradiation (5 Gy) or were left untreated (control). Lymph nodes were harvested 20 h following γ-irradiation and single cell suspensions stained with monoclonal antibodies to CD4, CD8 and B220. Absolute cell numbers were determined by trypan blue staining and counting in a haemocytometer. Data are expressed as mean ± SEM of 4–6 mice of each genotype from 3 independent experiments. p<0.05 (*) and p<0.01 (**) are indicated.