Abstract
The PI3K (phosphoinositide 3-kinase) family of lipid kinases regulate cell motility in diverse organisms and cell types. In mammals, the main PI3K enzyme activated by chemokine receptor signalling is the class IB isoform, p110γ. Studies of p110γ -knockout mice have shown an essential function for this isoform in chemotaxis of neutrophils and macrophages both in vitro and in vivo. However, the roles of p110γ and other PI3K enzymes and regulatory subunits in lymphocyte motility have been more difficult to discern. Recent studies of adoptively transferred, fluorescently labelled lymphocytes have revealed complex and unexpected functions for PI3K in lymphocyte migration in vivo. In this review we highlight cell-type-specific roles for PI3K catalytic and regulatory subunits in the homing and basal motility of lymphocytes in the intact lymph node.
Keywords: chemokine, chemotaxis, leukaemia, lymphocyte, phosphoinositide 3-kinase (PI3K), two-photon microscopy
Introduction
Lymphocytes (T- and B-cells) circulate through the body in a constant search for antigens. It has become clear that chemokines play integral roles in the movement of lymphocytes across the endothelium and within lymphoid organs. Lymphocytes display both directed movement in chemokine gradients (chemotaxis) and random motility in areas of uniform chemokine concentration (chemokinesis). Two excellent reviews have discussed lymphocyte motility and the role of PI3K (phosphoinositide 3-kinase) [1,2]. Here, we expand on this topic and focus on recent studies employing TPM (two-photon microscopy) to visualize lymphocyte motility in the intact lymph node.
PI3K overview
The PI3K family consists of a group of lipid kinases that phosphorylate the 3′ hydroxy moiety of PtdIns and its derivatives [3,4]. Members of this family are grouped into four classes (IA, IB, II and III) on the basis of substrate specificity, sequence homology and regulation. Class IA and IB PI3K members mediate the acute rise of the critical second messenger PtdIns(3,4,5)P3 in response to extracellular signals and are the most extensively studied groups.
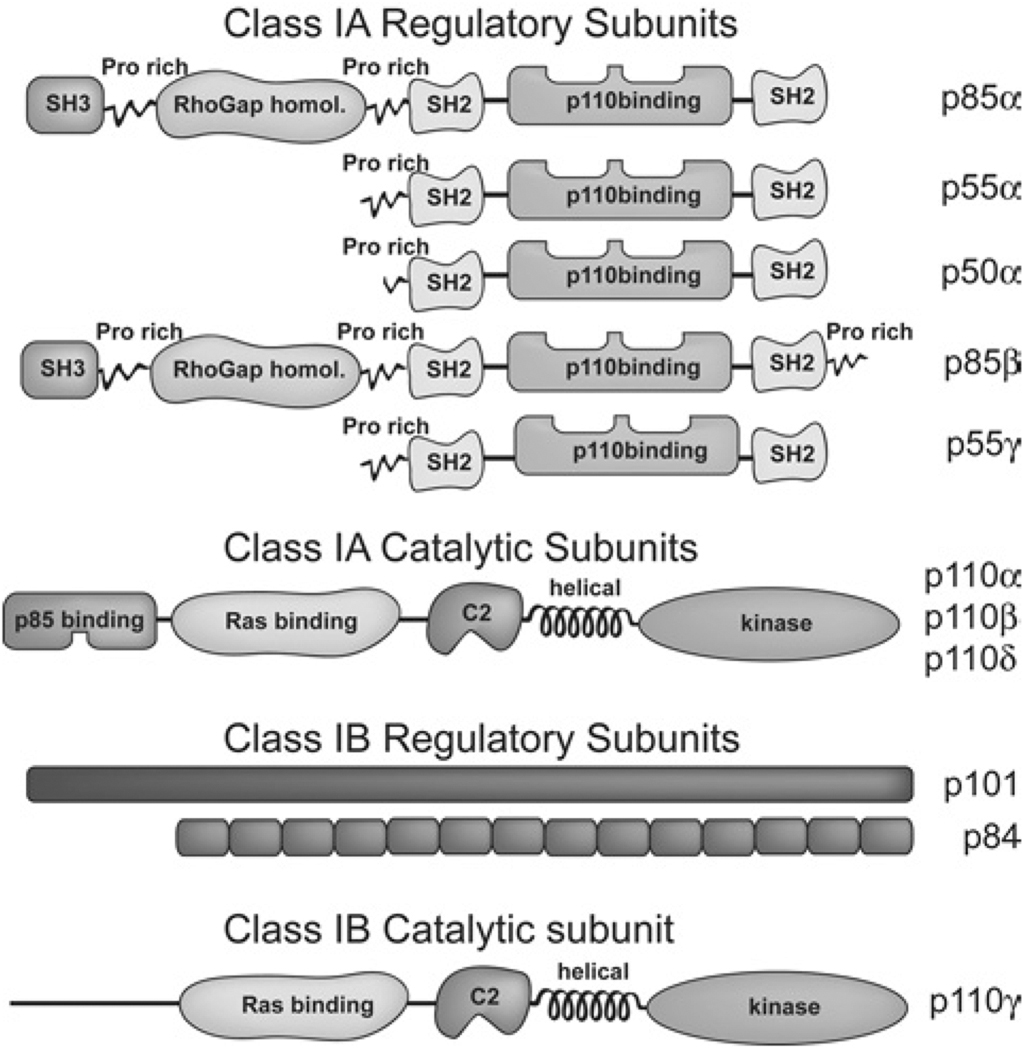
Class IA and IB PI3K have similar catalytic subunits but distinct regulatory subunits (Figure 1) and are activated by different types of transmembrane receptor [3,4]. Class IA PI3K functions downstream of receptor tyrosine kinases and exists as a stable heterodimer where one of the three catalytic isoforms p110α, p110β or p110δ associates with any of the five regulatory isoforms p85α, p55α, p50α, p85β or p55γ. Class IB PI3K consists of a p101/p87 regulatory subunit and a p110γ catalytic subunit and is activated upon GPCR (G-protein-coupled receptor) signalling.
Figure 1. Schematic diagram of class IA and IB PI3K catalytic and regulatory subunits.
In addition to the p110-binding domain, class IA regulatory subunits contain several other domains that regulate the localization and function of the p85/p110 heterodimer. The p101 or p84/p87 regulatory isoforms of class IB PI3K do not have readily identifiable structure features.
Generation of PtdIns(3,4,5)P3 by PI3K activation recruits proteins containing a PH (pleckstrin homology) domain to the cell membrane [3,4]. Several PH-domain-containing proteins mediate signalling through the PI3K pathway, including kinases [e.g. Akt, also known as PKB (protein kinase B)] and GEFs (guanine-nucleotide-exchange factors) (e.g. PREX-1). Pharmacological studies using pan-PI3K inhibitors (LY294002 or wortmannin) have shown that blocking PI3K activity leads to profound defects in lymphocyte development, activation and survival. The generation of gene-targeted mice lacking PI3K regulatory or catalytic isoforms, and the development of isoform-specific PI3K inhibitors, has uncovered unique functions of the different PI3K isoforms in lymphocyte development, function and survival (reviewed in [5–8]). Although some of these defects are likely to be attributable to defective signalling to the cell cycle and survival machinery, impaired immune function in PI3K-deficient mice may arise in part from altered homing or motility. In this review we consider the current evidence supporting the latter hypothesis.
Chemotaxis in vitro
Neutrophils and macrophages from p110γ-deficient mice show profound deficits in chemotaxis towards inflammatory chemokines [e.g. MIP-1α (macrophage inflammatory protein 1α) and IL-8 (interleukin 8)], anaphylatoxins (C5a) and bacterial products [fMLP (N-formylmethionyl-leucyl-phenylalanine)] [9–11], resulting in severely impaired recruitment to inflammatory sites. These functional deficits correlate with severe loss of PI3K signalling. Moreover, monocytes and neutrophils treated with a selective p110γ inhibitor also show decreased PI3K signalling, chemotaxis and recruitment [12].
Chemokine receptor signalling in lymphocytes appears to be more complex, with PI3K signalling acting parallel with additional pathways such as that involving the GEFDOCK2 (dedicator of cytokinesis 2) [13]. Thus T-cell migration in response to homoeostatic chemokines such as CCL21, CCL19 and CXCL12 is only partially impaired by wortmannin treatment [13,14]. As in the case with neutrophils, class IB PI3K appears to dominate chemokine-mediated PI3K signalling, as naïve p110γ −/− T-cells have a severely reduced Akt phosphorylation and partially impaired actin polymerization and cell polarization upon stimulation with CCL21 [13]. Despite these defects, however, T-cells lacking p110γ show only partially reduced chemotaxis in response to CCL21, CCL19 or CXCL12 [13,14], similar to the effects of wortmannin. These data suggest that PI3K signalling plays a minor role in T-lymphocyte chemotaxis. Class IA PI3K appears to be dispensable for T-cell chemotaxis as p110δD910A/D910A T-cells (from mice with a kinase-inactive p110δ due to D910A knock-in mutation) migrated normally upon stimulation with CCL21 and CCL19 and higher doses of CXCL12 [14].
Similarly to T-cells, B-cell migration in response to CXCL12 and CXCL13 is partially wortmannin sensitive [13]. Interestingly, however, B-cell chemotaxis is independent of class IB PI3K, as Akt phosphorylation, actin polymerization and migration in response to CXCL13 were normal in p110γ −/− B-cells [13]. Studies of p110δD910A/D910A B-cells showed that maximal response to CXCL13 requires p110δ [14], thus revealing a surprising role of class IA PI3K in signalling downstream of chemokine receptors in B-cells. Although DOCK2- and PI3K-independent Btk/PLCγ (phospholipase Cγ ) signalling play more prominent roles in B-cell migration [13,15], these results show that PI3K signalling is necessary for optimal response in vitro. The finding that class IA and IB PI3K contribute significantly to lymphocyte migration in vitro, in a cell-type-specific manner, has been confirmed and extended by in vivo experiments, as described in the next section.
Homing to lymphoid organs
Lymphocytes are programmed to circulate through lymphoid tissues where they scan APCs (antigen-presenting cells) in search of their cognate antigen. Homing refers to the process by which lymphocytes exit the circulation and enter lymphoid tissues such as the spleen, lymph nodes and Peyer’s patches where APCs reside. Several steps in lymphoid homing and localization involve signalling through chemokine and adhesion receptors. Two studies published in 2004 examined the homing and localization of PI3K-deficient lymphocytes following adoptive transfer. Nombela-Arrieta et al. [13] confirmed a role for p110γ in T-cell homing to peripheral lymphocyte organs by showing that p110γ deficiency leads to near complete abrogation of the residual migration and localization within lymphoid tissue in DOCK2−/− T-cells, albeit less severely than with wortmannin treatment (Table 1).
Table 1. Chemotaxis and motility phenotypes of PI3K-deficient or DOCK2-deficient lymphocytes.
MLN, mesenteric lymph node; N/A, not addressed; PLN, peripheral lymph node; PP, Peyer’s patches; SPL, spleen.
| Cell type | Chemotaxis | Localization | Velocity | Turning angle | Motility coefficient | Reference | |
|---|---|---|---|---|---|---|---|
| B | DOCK2 | ↓ | ↓34% | Broad | ↓93% | [13,23] | |
| DOCK2/p110γ | Same as | Same as | Same as | Same as | Same as | ||
| DOCK2−/− | DOCK2−/− | DOCK2−/− | DOCK2−/− | DOCK2−/− | [13,23] | ||
| p110γ | ↔ | ↔ | ↔ | ↔ | ↔ | [13,14,23] | |
| ↓ Homing to PP | |||||||
| p110δ | ↓ | ↓ Localization to white pulp cords |
N/A | N/A | N/A | [14] | |
| p85α | N/A | ↔ | ↓24% | N/A | ↓51% | [24] | |
| p85β | N/A | ↔ | ↓5% | N/A | ↓3% | [24] | |
| Wortmannin | ↓ | Localization to border of follicles and T-cell zones |
↓21% | N/A | [14,24] | ||
| T | DOCK2 | ↓ | ↓ Homing to PLN, MLN, PP | ↓44% | Broad | ↓93% | [13,23] |
| DOCK2/p110γ | ↓↓ | Severe homing defect to all lymphoid tissue |
Similar to DOCK2−/− |
Broad | ↓96% | [13,23] | |
| p110γ | ↓ | ↓ Homing to PLN, MLN, PP, SPL |
↔ | Slightly broader | ↓16% | [13,14,23] | |
| p110δ | ↔ | ↔ | N/A | N/A | N/A | [14] | |
| p85α | N/A | N/A | ↓12% | N/A | ↓21% | [24] | |
| p85β | N/A | N/A | ↓26% | N/A | ↓56% | [24] | |
| p85α/p55α/p50 α/p85β |
N/A | N/A | ↓37% | N/A | ↓78% | [24] | |
| Wortmannin | ↓ | ↔ | ↓26%* | N/A | ↓33% | [19,24] | |
| ↔* |
Different conclusions in cited references.
Although wortmannin treatment also led to decreased B-cell homing in vivo, comparative studies of DOCK2−/− and DOCK2−/− p110γ −/− B-cells suggested that a non-class IB isoform of PI3K mediates B-cell migration. Reif et al. [14] confirmed that p110γ −/− B-cells did not display homing defects and identified p110δ as the predominant PI3K isoform in B-cell homing. Loss of p110δ led to defective homing to Peyer’s patches and decreased localization in splenic white pulp cords (Table 1). Collectively, these findings support the distinction that PI3K-dependent lymphocyte migration requires mainly p110γ in T-cells and p110δ in B-cells.
Lymphocyte motility in vivo
The use of TPM to visualize leucocyte behaviour within intact lymphoid tissues has revolutionized our understanding of the immune system [16,17]. Using this technology, many investigators have documented that resting T-cells and B-cells exhibit highly dynamic movements as they search for antigens. Both cell types exhibit an apparently ‘random walk’ pattern of movement within their respective areas of the lymph node: stopping, starting and changing directions with an average velocity of 10–12 µm/min for T-cells and 6–7 µm/min for B-cells [18]. Basal motility of T-cells requires the homoeostatic chemokines CCL19 and CCL21 (CCR7 ligands) that are abundant throughout the T-cell zone, together with adhesion ligands present on stromal cells [19–21]. Indeed, lymphocytes show random motility in vitro when plated on glass slides coated with integrin ligands in the presence of homoeostatic chemokines [22].
Is PI3K involved in regulating basal lymphocyte motility in vivo? Three groups have recently addressed this question using advanced imaging techniques. Nombela-Arrieta et al. [23] studied the motility of T- and B-cells lacking p110γ and/or DOCK2 and found that, concordant with their previous analysis of chemotaxis and homing [13], DOCK2 plays a dominant role. On the other hand, despite the established contribution of p110γ to T-cell homing and chemotaxis, they found no change in the average velocity of p110γ -deficient T-cells (Table 1). They did, however, notice that p110γ-deficient T-cells displayed increased turning angles, resulting in a small but significant decrease in motility coefficient. This could be physiologically important by decreasing the volume through which naïve T-cells scan for antigens. p110γ -deficient B-cells showed no change in velocity, turning angles or motility coefficient (Table 1). This study did not address the possible roles of other PI3K isoforms, using either knockout mice or catalytic inhibitors.
Our laboratory has used TPM to compare lymphocyte motility in wild-type lymphocytes with cells treated with the pan-PI3K inhibitor wortmannin [24]. Cells were treated with a PI3K-selective concentration of wortmannin (50 nM) for 15 min, then co-injected together with untreated cells (labelled with a different colour tracker dye) into host mice. Wortmannin-treated T- and B-cells showed mean velocities 20–25% lower than untreated controls (Table 1).Wortmannin also caused a change in B-cell localization, such that cells accumulated at the border of the follicles and the T-cell zones. Assuming that p110γ is not the relevant target, these findings suggest that other wortmannin-sensitive enzymes regulate basal lymphocyte motility and B-cell localization.
To assess the role of class IA PI3K, we measured the basal motility and localization of lymphocytes from knockout mice lacking one or more class IA regulatory isoform [24]. T-cells lacking either p85α and p85β showed reductions of velocity of 12 and 26% respectively (Table 1), whereas T-cells lacking both p85α and p85β, as well as p55α and p50α that are alternative products of the gene encoding p85α, showed a 37% decrease in velocity and a marked loss of cell polarization (Table 1). In contrast, p85α is the dominant isoform in B-cells, with knockout resulting in a 24% decrease in velocity as compared with a 5% decrease for p85β-deficient B-cells (Table 1). These experiments do not distinguish whether reduced motility results from impaired class IA PI3K signalling function or from loss of adaptor functions of the regulatory subunits independently of their role in activating the catalytic subunits. The reduced motility in wortmannin-treated cells supports at least some role for PI3K enzymatic subunits but could also be due to inhibition of other PI3K subclasses or non-PI3K targets of wortmannin. Importantly, p85α-deficient B-cells showed distinct behavioural differences relative to wortmannin-treated B-cells, exhibiting normal localization but strikingly altered morphology, extending dendritic-like projections that appeared to contact neighbouring wild-type B-cells. To directly assess the role of class IA catalytic subunits in basal motility and morphology, it will be important to analyse lymphocytes from mouse strains lacking function of one or more class IA enzyme. One might predict that p110δ contributes to basal lymphocyte motility, particularly in B-cells where altered homing and localization has already been documented and antibody responses are severely impaired.
A third group has analysed the role of PI3K in lymphocyte motility using a different imaging approach employing conventional epifluorescence microscopy in lymph node slices [19]. No differences in cell velocity were observed between wild-type and wortmannin-treated T-cells (Table 1), although other findings with respect to lymphocyte velocity, trajectories and dependence on CCR7 ligands were in agreement with published results. At present it is unclear why two different teams reach different conclusions regarding the effect of wortmannin on lymphocyte motility. One clue might be provided by a recent paper showing that motility is influenced by tissue architecture: specifically, that turning angles and motility coefficients vary between the subcapsular and deep paracortical regions of the node [25]. The differing milieu of chemokines, adhesion ligands and stromal cell architecture among different regions may therefore alter the involvement of PI3K in lymphocyte motility. Future studies of pharmacological or genetic interventions should thus explicitly document motility in distinct regions of the lymph node.
Conclusions and unanswered questions
Lymphocytes must interpret and integrate signals from numerous chemokines and adhesion ligands in order to efficiently circulate, home to the proper regions of lymphoid organs and search for antigens effectively. PI3K regulates each of these processes and, although pan-PI3K inhibitors or gene knockouts generally exert only partial effects, these changes are likely to have significant impact on adaptive immunity. In the light of intensive pharmaceutical efforts to develop PI3K inhibitors for clinical use [26–28], we emphasize the importance of considering their actions not only on lymphocyte development and activation but also on motility.
We draw several general conclusions from the current literature on PI3K in lymphocyte motility. First, the class IB p110γ is the major PI3K isoform involved in T-cell chemotaxis and homing but is dispensable for these processes in B-cells. PI3K does contribute to B-cell chemotaxis and homing but the class IA isoform p110δ is the primary catalytic isoform involved. With regard to basal motility in the lymph node, p110γ is dispensable. However, class IA regulatory isoforms have unique functions required for maximum velocity of T- and B-cells during random-walk behaviour.
Changes in cell motility might contribute to defective development in some strains of PI3K-deficient mice. Strains lacking class IA PI3K catalytic (p110δ) or regulatory (p85α) subunits have marked defects in B-cell development [29–33]. Interestingly, a similar phenotype is observed in mice lacking CXCL12, a chemokine expressed by bone marrow stromal cells whose receptor (CXCR4) is expressed on B-cell progenitors. Mice lacking both p110γ and p110δ show severe defects in thymocyte development, correlating with decreased survival [34,35]; however, altered thymocyte migration could contribute to this phenotype.
Lymphocyte proliferation and differentiation is severely compromised in mice with reduced class IA function (reviewed in [5–7,36]). It is possible that altered homing or motility contributes to some of the immune defects these strains display in vivo. p110δ -deficient B-cells have defects not only in proliferation but also in homing [14]. p85α-deficient B-cells show impaired antibody responses [33] and have reduced basal motility in the lymph node and altered morphology in vivo [24]. Motility defects have not been reported in studies of p110δ-deficient T-cells. However, T-cells lacking the class IA regulatory isoforms p85α/p55α/p50α/p85β show greatly decreased basal velocity [24]. Interestingly, these mice develop an autoimmune syndrome despite impaired T-cell functional responses in vitro and in vivo [37,38]. It is conceivable that reduced T-cell motility changes the dynamics of interactions with APCs in a way that favours productive activation of self-reactive T-cells.
Although it is important to consider the role of motility defects in the functional phenotypes of PI3K-deficient mice, it is also crucial to gain a better understanding of the molecular pathways linking chemokine and adhesion receptors to PI3K and downstream targets. For example, how do chemokines, acting through GPCRs, connect with class IA PI3K? GPCRs have been shown to activate p110β [39], but not the p110δ isoform that is clearly important for B-cell homing and chemotaxis. It will also be important to determine which adhesion ligands are essential for basal lymphocyte motility in conjunction with chemokines and whether the cognate adhesion receptors engage PI3K. A question raised by our recent work [24] is how the adaptor functions of class IA regulatory subunits influence lymphocyte morphology and movement.
Another question is whether PI3K signalling regulates the homing and dissemination of lymphoid malignancies. It is now clear that PI3K signalling is important for proliferation and survival of many types of leukaemia and lymphoma in both humans and mice [40–43]. The homing of leukaemia cells to the bone marrow requires many of the same chemokines and adhesion molecules employed by normal lymphocytes [44]. If these signals depend on downstream activation of PI3K, one can predict that PI3K inhibitors optimized for suppressing proliferation and survival of leukaemia cells would have the added benefit of disrupting the ability of cancer cells to home to favourable bone marrow niches or extramedullary sites.
In summary, although it has taken many years to establish a consensus that PI3K is involved in regulation of lymphocyte motility, the field is now moving rapidly (and non-randomly, we hope) toward a detailed understanding of this important signalling pathway for the normal and pathophysiological functioning of lymphocytes.
Abbreviations used
- APC
antigen-presenting-cell
- DOCK2
dedicator of cytokinesis 2
- GEF
guanine-nucleotide-exchange factor
- GPCR
G-protein-coupled receptor
- PH domain
pleckstrin homology domain
- PI3K
phosphoinositide 3-kinase
- TPM
two-photon microscopy
References
- 1.Smith L, Webb A, Ward SG. Biochem. Soc. Trans. 2007;35:193–198. doi: 10.1042/BST0350193. [DOI] [PubMed] [Google Scholar]
- 2.Ward SG. Trends Immunol. 2006;27:80–87. doi: 10.1016/j.it.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Fruman DA, Meyers RE, Cantley LC. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 4.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 5.Deane JA, Fruman DA. Annu. Rev. Immunol. 2004;22:563–598. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 6.Fruman DA. Biochem. Soc. Trans. 2007;35:177–180. doi: 10.1042/BST0350177. [DOI] [PubMed] [Google Scholar]
- 7.Koyasu S. Nat. Immunol. 2003;4:313–319. doi: 10.1038/ni0403-313. [DOI] [PubMed] [Google Scholar]
- 8.Okkenhaug K, Ali K, Vanhaesebroeck B. Trends Immunol. 2007;28:80–87. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, et al. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 12.Camps M, Ruckle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Francon B, et al. Nat. Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 13.Nombela-Arrieta C, Lacalle RA, Montoya MC, Kunisaki Y, Megias D, Marques M, Carrera AC, Manes S, Fukui Y, Martinez AC, Stein JV. Immunity. 2004;21:429–441. doi: 10.1016/j.immuni.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Reif K, Okkenhaug K, Sasaki T, Penninger JM, Vanhaesebroeck B, Cyster JG. J. Immunol. 2004;173:2236–2240. doi: 10.4049/jimmunol.173.4.2236. [DOI] [PubMed] [Google Scholar]
- 15.de Gorter DJ, Beuling EA, Kersseboom R, Middendorp S, van Gils JM, Hendriks RW, Pals ST, Spaargaren M. Immunity. 2007;26:93–104. doi: 10.1016/j.immuni.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Cahalan MD, Parker I. Curr. Opin. Immunol. 2006;18:476–482. doi: 10.1016/j.coi.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahalan MD, Parker I, Wei SH, Miller MJ. Curr. Opin. Immunol. 2003;15:372–377. doi: 10.1016/s0952-7915(03)00079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller MJ, Wei SH, Parker I, Cahalan MD. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 19.Asperti-Boursin F, Real E, Bismuth G, Trautmann A, Donnadieu E. J. Exp. Med. 2007;204:1167–1179. doi: 10.1084/jem.20062079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bajenoff M, Egen JG, Koo LY, Laugier JP, Brau F, Glaichenhaus N, Germain RN. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worbs T, Mempel TR, Bolter J, von Andrian UH, Forster R. J. Exp. Med. 2007;204:489–495. doi: 10.1084/jem.20061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stachowiak AN, Wang Y, Huang YC, Irvine DJ. J. Immunol. 2006;177:2340–2348. doi: 10.4049/jimmunol.177.4.2340. [DOI] [PubMed] [Google Scholar]
- 23.Nombela-Arrieta C, Mempel TR, Soriano SF, Mazo I, Wymann MP, Hirsch E, Martinez AC, Fukui Y, von Andrian UH, Stein JV. J. Exp. Med. 2007;204:497–510. doi: 10.1084/jem.20061780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matheu MP, Deane JA, Parker I, Fruman DA, Cahalan MD. J. Immunol. 2007;179:2261–2269. doi: 10.4049/jimmunol.179.4.2261. [DOI] [PubMed] [Google Scholar]
- 25.Huang JH, Cardenas-Navia LI, Caldwell CC, Plumb TJ, Radu CG, Rocha PN, Wilder T, Bromberg JS, Cronstein BN, Sitkovsky M, et al. J. Immunol. 2007;178:7747–7755. doi: 10.4049/jimmunol.178.12.7747. [DOI] [PubMed] [Google Scholar]
- 26.Ruckle T, Schwarz MK, Rommel C. Nat. Rev. Drug Discov. 2006;5:903–918. doi: 10.1038/nrd2145. [DOI] [PubMed] [Google Scholar]
- 27.Workman P, Clarke PA, Guillard S, Raynaud FI. Nat. Biotechnol. 2006;24:794–796. doi: 10.1038/nbt0706-794. [DOI] [PubMed] [Google Scholar]
- 28.Wymann MP, Zvelebil M, Laffargue M. Trends Pharmacol. Sci. 2003;24:366–376. doi: 10.1016/S0165-6147(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 29.Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, Gray A, Humphries LA, Rawlings D, Reynolds H, Vigorito E, Turner M. J. Exp. Med. 2002;196:753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fruman DA, Snapper SB, Yballe CM, Davidson L, Yu JY, Alt FW, Cantley LC. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 31.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Wang D, Ihle JN. Mol. Cell. Biol. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, Koyasu S. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 34.Swat W, Montgrain V, Doggett TA, Douangpanya J, Puri K, Vermi W, Diacovo TG. Blood. 2006;107:2415–2422. doi: 10.1182/blood-2005-08-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb LM, Vigorito E, Wymann MP, Hirsch E, Turner M. J. Immunol. 2005;175:2783–2787. doi: 10.4049/jimmunol.175.5.2783. [DOI] [PubMed] [Google Scholar]
- 36.Fruman DA. Biochem. Soc. Trans. 2004;32:315–319. doi: 10.1042/bst0320315. [DOI] [PubMed] [Google Scholar]
- 37.Deane JA, Kharas MG, Oak JS, Stiles LN, Luo J, Moore TI, Ji H, Rommel C, Cantley LC, Lane TE, Fruman DA. Blood. 2007;109:2894–2902. doi: 10.1182/blood-2006-07-038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oak JS, Deane JA, Kharas MG, Luo J, Lane TE, Cantley LC, Fruman DA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16882–16887. doi: 10.1073/pnas.0607984103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yart A, Chap H, Raynal P. Biochim. Biophys. Acta. 2002;1582:107–111. doi: 10.1016/s1388-1981(02)00144-0. [DOI] [PubMed] [Google Scholar]
- 40.Billottet C, Grandage VL, Gale RE, Quattropani A, Rommel C, Vanhaesebroeck B, Khwaja A. Oncogene. 2006;25:6648–6659. doi: 10.1038/sj.onc.1209670. [DOI] [PubMed] [Google Scholar]
- 41.Kharas MG, Fruman DA. Cancer Res. 2005;65:2047–2053. doi: 10.1158/0008-5472.CAN-04-3888. [DOI] [PubMed] [Google Scholar]
- 42.Sujobert P, Bardet V, Cornillet-Lefebvre P, Hayflick JS, Prie N, Verdier F, Vanhaesebroeck B, Muller O, Pesce F, Ifrah N, et al. Blood. 2005;106:1063–1066. doi: 10.1182/blood-2004-08-3225. [DOI] [PubMed] [Google Scholar]
- 43.Vivanco I, Sawyers CL. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 44.Dick JE, Lapidot T. Int. J. Hematol. 2005;82:389–396. doi: 10.1532/IJH97.05144. [DOI] [PubMed] [Google Scholar]