Abstract
Thioredoxin-1 from Escherichia coli has frequently been used as a model substrate in protein folding studies. However, for reasons of convenience, these studies have focused largely on oxidized thioredoxin and not on reduced thioredoxin, the more physiologically relevant species. Here we describe the first extensive characterization of the refolding kinetics and conformational thermodynamics of reduced thioredoxin. We have previously described a genetic screen that yielded mutant thioredoxin proteins that fold more slowly in both the oxidized and reduced forms. In the present study, we apply our more detailed analysis of reduced thioredoxin folding to a larger number of folding mutants which includes those obtained from continuation of the genetic screen. We have identified mutant proteins that display folding defects specifically in the reduced state but not the oxidized state. Some of these substitutions represent unusual folding mutants in that they result in semi-conservative substitutions at solvent-exposed positions in the folded conformation and do not appear to affect the conformational stability of the protein. Further, the genetic selection yields mutants at only a limited number of sites, pointing to perhaps the most critical amino acids in the folding pathway and underscoring, in particular, the role of the carboxy-terminal amino acids in the folding of thioredoxin. Our results demonstrate the importance of studying the physiologically relevant folding species.
Thioredoxin-1 (TrxA) of Escherichia coli is a cytoplasmic enzyme that maintains the cysteines of substrate proteins in the reduced form (3). It does this by utilizing one of its two redox-active cysteines (cysteine-32) to attack a substrate’s disulfide bonds, thus initiating the reductive process that leads to a reduced substrate and an oxidized thioredoxin (4). The enzyme thioredoxin reductase then transfers electrons from NADPH to thioredoxin, returning its cysteines to the reduced state. Because of its abundance and relative ease of purification, thioredoxin has been the subject of numerous in vitro protein folding studies (e.g. (5–10), etc). However, most studies of thioredoxin folding have been carried out on the oxidized form of thioredoxin because it is rapidly air oxidized in the absence of a reducing agent, because oxidized thioredoxin is more stable than the reduced form, and because oxidized thioredoxin folds with simplified kinetics.
We previously described our use of a genetic screen to isolate new classes of mutants of thioredoxin that were defective for folding (11). When we examined their conformational stability and refolding kinetics in vitro, all but one of these folding mutants also showed significant defects in the refolding of the oxidized proteins. The exceptional mutant contained a relatively conservative substitution (aspartate to asparagine) at position 15, which is completely solvent exposed in structural models of thioredoxin and which had not been previously implicated in thioredoxin folding. Furthermore, the D15N mutant protein, which we presumed to cause a defect in thioredoxin folding in vivo, did not exhibit such a defect in preliminary in vitro studies on a reduced form of this mutant protein. Surprisingly, the D15N substitution had no effect on the rate of refolding of oxidized thioredoxin and even increased its conformational stability.
In the present study, we revisited our genetic screen in order to isolate more thioredoxin folding mutants. We submitted several of the purified mutant proteins to a more rigorous analysis of their conformational dynamics and refolding kinetics. In addition, we carried out the first extensive folding analysis of the reduced form of thioredoxin and applied it to some of the mutant proteins. Among the mutations that we isolated were some that caused novel substitutions at D15 as well as substitutions at D13, which is also completely solvent exposed in the thioredoxin structure. These substitutions cause defects in the refolding kinetics of the reduced, but not the oxidized, forms of these proteins. We also find that a high proportion of the folding mutations affect either the penultimate amino acid, L107, or alter the chain-terminating codon causing extension of the carboxy-terminal amino acid sequence. These findings, along with a L107A mutant constructed here, strengthen previous evidence for a key role of L107 in folding and suggest that altering the carboxy-terminus of the protein in other ways may also interfere with folding (11). Our results demonstrate the importance of studying the physiologically relevant form of a protein in vitro and suggest that genetic studies of this sort may allow a significant narrowing down of candidates for key residues in folding.
Experimental Procedures
Strain and Plasmid Construction and Growth Conditions
Strains and plasmids were constructed using standard genetic and molecular techniques (12–13). E. coli K-12 strains DRH119 (araD139 Δ(ara, leu)7697 galU galK ΔlacX74 rpsL thi ΔmalF3 Δ(phoA[PvuII]) phoR ΔdsbA::KanR ΔdsbC dsbD::mini-Tn10CamR/F′ lac-pro lacIq), DRH245 (F− ΔlacX74 galE galK thi rpsL ΔphoA(PvuII) degP41(ΔPstI)::ΩKanR ΔompT ptr-32::ΩCamR Δtsp3::ΩKanR eda51::Tn10 ΔtrxA), and JF521 (Δ(lac, pro) thi supE metE46 srl300::Tn10 trxA2(7004) recA/F′ traD36 proAB lacIQ ΔlacZ(M15)) as well as plasmids pLMD82 (bearing the phoAss-trxA3 gene under control of the phoA promoter), pCFS122 (a modified pTrc99a [Promega] bearing the sequence encoding the phoA signal sequence), and pTK10trxA were described previously (11). All restriction enzymes were obtained from New England Biolabs. The mutant trxA genes were placed under lac control by PCR amplification of a DNA fragment corresponding to the trxA gene from the mutant plasmids, into which a BspHI site was introduced at the 5′ end of the gene. The PCR fragment was subsequently digested with XbaI and BspHI and ligated with pCFS122 cut with NcoI and XbaI. Point mutations in the trxA gene were introduced into plasmid pTK10trxA by QuickChange mutagenesis (Invitrogen). Cells were generally grown at 37°C in NZ medium (14). When necessary, ampicillin was added at 200 μg/ml; chloramphenicol at 10 μg/ml; kanamycin at 40 μg/ml and tetracycline at 15 μg/ml. Induction of lac promoter constructs was accomplished by addition of IPTG to a final concentration of 10 μM.
Mutagenesis
Mutagenesis of the trxA gene was carried out by mutagenic PCR using two general methods: 1) skewing the ratio of purine to pyrimidine dNTPs in the PCR reaction and 2) carrying out traditional PCR in the presence of mutagenic dNTP analogs. The first method was modified from Chen et al. (15) and is described in detail in the supplemental methods section. Alternatively, four independent PCR reactions each were carried out in the presence of either 50μM 8-oxo-dGTP (8-oxo-2′-deoxy-guanosine-5′-triphosphate) or 50μM dPTP (6H, 8H, 4-Dihydro-pyrimido[4,5-c][1,2] oxazin-7-one-8-β-D-2′-deoxy-ribofuranoside-5′-triphophate) (Jena Bioscience, Jena, Germany) using Invitrogen Platinum PCR Supermix (20 cycles of PCR) (16). To maintain the independence of any mutations that were isolated, each PCR reaction was digested and cloned separately. Each PCR reaction was individually cut with AgeI and XbaI and ligated into pLMD82 cut with the same enzymes.
Genetic screen for thioredoxin folding mutants
The genetic screen for thioredoxin folding mutants was carried out as described (11). See the supplemental methods section for a more detailed description of the genetic screen.
Subcellular Fractionation
Fractionations were performed as described previously (1).
Expression and purification of thioredoxin mutant proteins
Thioredoxin was purified as described (11). See the supplemental methods section for details.
Thermal denaturation curves and equilibrium transition curves using chemical denaturation of reduced thioredoxin
Thermal denaturation curves and equilibrium denaturation and renaturation curves in the presence of guanidinium chloride (GdmCl) of reduced thioredoxin were determined by circular dichroism spectroscopy and fitted using standard procedures derived from a two-state model. Thioredoxin was kept reduced by the inclusion of 1mM DTT in all solutions. For a more detailed description of the experimental procedures and data fitting, see the supplemental methods section.
Thioredoxin refolding kinetics
The refolding kinetics of thioredoxin were determined as previously described (11). The refolding kinetics of reduced thioredoxin were determined in the presence of 1mM DTT in order to maintain thioredoxin in the reduced state. A detailed description of the experimental procedures and data modeling can be found in the supplemental methods sections.
Results
When the post-translational signal sequence of the periplasmic protein alkaline phosphatase (PhoAss) is attached to thioredoxin-1, very little of the protein appears in the periplasm because it rapidly folds in the cytoplasm before it can be exported. However, amino acid substitutions that interfere with the folding of the PhoAss-thioredoxin fusion allow significant amounts of the protein to be exported to the periplasm (11). Although thioredoxin is normally involved in the reduction of disulfide bonds in the cytoplasm, when the mutant thioredoxins are localized to the periplasm, they can partially replace the activity of DsbA in promoting the formation of disulfide bonds (2). We previously detected such mutants by selecting for restoration of motility to an E. coli dsbA strain carrying a phoAss-trxA gene that was mutagenized using a mutD mutator strain (11). Motility is dependent on a component of the flagellum, the FlgI protein, which contains a disulfide bond essential for its activity (17).
In order to obtain new classes of mutations, we mutagenized the thioredoxin-encoding portion of the phoAss-trxA gene using mutagenic PCR. Compared to mutagenesis of the plasmid in a mutD mutator strain, PCR has the potential to generate a greater variety of types of mutations (e.g. increased frequency of transversion mutations) by altering the ratio of dNTPs present in the PCR reaction or by doping the PCR reaction with mutagenic base analogs, such as 8-oxo-dGTP (see Experimental Procedures section). After ligating the mutagenized PCR product into plasmid pLMD82 cut with AgeI and XbaI, we plated transformations of the mutagenized ligants into the dsbA screening strain (DRH119) onto the center of LB plates containing 0.35% agar and selected for increased motility by picking from the edge of the motility swarm. In order to ensure that the mutants were the result of independent mutational events, we isolated only one mutant per mutagenic PCR reaction. We purified and sequenced plasmids from isolates that 1) showed increased motility after restreaking to colonies and 2) retained the increased motility phenotype upon retransformation. A more detailed description of the screen can be found in the supplemental methods and in Huber et al. (11).
We obtained a total of 20 independently isolated trxA mutants (9 single, 10 double, and 1 triple mutant) in this selection which resulted in the following amino acid substitutions: D13A, D15V, P34R, L107P, L107R, Stop109Q, Stop109Y, D9G/Stop109K, D13N/A29V, D15Y/Stop109K, L17P/Stop109K, G33C/Stop109L, P34S/Stop109Q, A46P/Stop109Q, A46T/A87V, T54P/Stop109S, I75T/Stop109K, D9N/A19V/Stop109Q (table 1). In summary, we obtained mutations that resulted in substitutions at a total of 13 amino acid positions (including mutations at the UAA stop codon which changed it to a sense codon and appended the sequence SRVDRCP to the C-terminus). Seven of these substitutions resulted in unique amino acid substitutions that were not identified in our previous screen. However, mutations at only three of these positions (changes to positions D13, I75, and Stop109) were isolated as single mutants. Mutations at the remaining five unique positions were all isolated as double or triple mutants, and only one double mutant (A46T/A87V) resulted in substitutions in which both amino acid substitutions were not otherwise isolated as single mutants. However, in this case, when separated from one another, neither single mutant caused increased motility in the dsbA screening strain. It appears that these separated mutations may not strongly affect folding unless they are combined with another similarly very weak mutation.
Table 1.
Summary of mutations obtained in genetic screen.
| Amino acid substitution* | Mutation | Independent isolations1 |
|---|---|---|
| D9N†‡ | GAC→AAC | 1(2) |
| D9G† | GAC→GGC | 1 |
| D13A | GAC→GCC | 1 |
| D13N† | GAC→AAC | 1 |
| D15V | GAT→GTT | 1(3) |
| D15N | GAT→AAT | 0(1) |
| D15Y† | GAT→TAT | 1 |
| A29V†‡ | GCA→GTA | 1(8) |
| P34S†‡ | CCG→CTG | 1(2) |
| P34R | CCG→CGG | 1 |
| I41T | ATT→ACT | 0(1) |
| I75T†‡ | ATC→ACC | 1(3) |
| S95P | TCT→CCT | 0(2) |
| L107P†‡ | CTG→CCG | 1(2) |
| L107R | CTG→CGG | 1 |
| Stop109Q | TAA→CAA | 4 |
| Stop109Y | TAA→TAT | 1 |
| Stop109S† | TAA→TCA | 1 |
| Stop109L† | TAA→TTA | 1 |
| Stop109K† | TAA→AAA | 4 |
Substitutions that were only isolated as double mutants in combination with a substitution at a position already known to cause increased PhoAss-thioredoxin export are not included.
Isolated only in combination with a second mutation in this study.
Isolated as a single mutation in Huber et al. (11)
Number in parentheses is the sum of the independent isolations in the present work and Huber et al. (11)
trxa mutants result in increased export of thioredoxin to the periplasm
The increased motility promoted by our trxA mutants could result from either increased export to the periplasm or increased activity of the small amount of thioredoxin that is translocated across the cytoplasmic membrane. If the amino acid substitutions affected protein folding, more thioredoxin should be translocated to the periplasm. We therefore placed a representative set of single mutants under the control of the inducible lactose promoter in plasmid pCFS122 and assayed for translocation by subcellular fractionation. According to a densitometric analysis of the periplasmic fractions in western blots against thioredoxin, the mutants that we tested resulted in a 7–82% increase in the steady-state amount thioredoxin in the periplasm (figure 1).
Figure 1. Mutations in thioredoxin that cause increased motility also result in increased translocation to the periplasm.
DRH245 (ΔtrxA) cells, which are deficient for multiple periplasmic proteases, were transformed with plasmids pCFS122 (PhoAss), pCFS126 (PhoAss-thioredoxin), pDRH503 (PhoAss-thioredoxin(D13A)), pDRH504 (PhoAss-thioredoxin(D15V)), pDRH506 (PhoAss-thioredoxin(L107R)), pMER209 (PhoAss-thioredoxin(L107A)) or pMER210 (PhoAss-thioredoxin(D13N)) and separated into whole cell, cytoplasm, and periplasm fractions by subcellular fractionation. (Top) Equal amounts of the whole cell or periplasmic fractions, as indicated, were analyzed by SDS-PAGE and western blotting against thioredoxin-1. (Bottom) In order to ensure that we separated the periplasmic and cytoplasmic fractions and that the fractions contained similar amounts of protein, equal amounts of the whole cell (W), cytoplasmic (C) and periplasmic (P) were resolved by SDS-PAGE and analyzed by western blotting against β-lactamase as a periplasmic control or the a subunit of RNA polymerase (RNAP-α) as a cytoplasmic control.
L107A substitution causes increased export of thioredoxin to the periplasm
Based on the folding defects in an L107P mutant of thioredoxin obtained in the previous study, we suggested that the penultimate amino acid of thioredoxin, L107, plays an important role in thioredoxin folding (11). The isolation of the L107R alteration reported here is consistent with this proposal. However, it may not be that L107 is essential for folding since both of these amino acid substitutions could have drastic effects by replacing a hydrophobic amino acid with either a strongly charged amino acid or an amino acid that can significantly alter structure. To ask whether a less dramatic change would have an effect on thioredoxin folding, we generated a mutant protein that contained an alanine substitution at L107, which reduces the length of the aliphatic side chain at position 107. The L107A substitution caused increased motility when expressed in our dsbA screening strain, and we observed an increased amount of the L107A variant in the periplasm (figure 1). We included L107A, along with several other mutants in our following studies on folding and stability.
Conformational stability of reduced mutant thioredoxins
We proceeded to measure and compare the conformational stability of several thioredoxin folding mutant proteins obtained both here and in the previous study. To this end, we introduced mutations into the trxA gene on plasmid pTK100 resulting in L107P, L107A, D15N, D15V, and D13N substitutions by site-directed mutagenesis and purified these variants in addition to wild-type thioredoxin. Our interest in studying the substitutions at D13 and D15 comes from the surprising finding in our previous study that D15N variant of thioredoxin, which restores motility to the same extent as many of the other mutants, had an increased overall conformational stability in its oxidized state compared to wild-type thioredoxin and had no measurable effect on the kinetics of folding (11). The isolation of a mutation resulting in a D15V change reported here enhanced our interest in the role of this residue. Further, amino acid D15 and D13 are part of a local cluster of surface-exposed aspartate residues near the N-terminus of thioredoxin, which drew our attention to the D13N substitution (6). D9, which we identified in our previous study as important for thioredoxin folding, is part of the same cluster but is partially buried in the tertiary structure.
Because we hadn’t seen any effect on folding of D15N using the oxidized form of thioredoxin (11), we did most of the following experiments on the mutant thioredoxin proteins under reducing conditions (1mM DTT). Reduced thioredoxin is the form of the protein that appears from the ribosome and is almost certainly the species of the protein that folds into its final conformation.
We first probed the conformational stability of the reduced mutant thioredoxins by monitoring circular dichroism at 222nm in the presence of chemical denaturant (figure 2A) or heat (figure 2B). Although Kelley et al. (18) reported that thermal denaturation of reduced or alkylated thioredioxin is not reversible, Ladbury et al. (19) found a minimal 90 % reversibility (at a minimal protein concentration of 1.9 mg/ml) when heated to not higher than 10 °C above the TM. We therefore deduced the thermodynamic parameters from the temperature dependent ellipticity of wild type and thioredoxin variants at 0.1 mg/ml within the high temperature limit of reversible unfolding (i. e. 10 °C above the TM value). GdmCl denaturation was reversible since the folding and unfolding curves, which were acquired independently, were superimposable within the experimental accuracy. No significant difference in the ΔGH2O and half unfolding GdmCl concentration was found when the unfolding/refolding curve was established from mixing at equilibrium native and totally unfolded thioredoxin at the same concentration in the presence of GdmCl at various concentrations.
Figure 2. Chemical and thermal denaturation melting curves of reduced thioredoxin variants as determined by CD spectroscopy.
The ellipticity at 222nm (a measure of α-helical content) of wild-type thioredoxin and the indicated thioredoxin variants was measured (A) in the presence of chemical denaturant or (B) as a function of temperature in order to calculate the thermodynamic parameters for the folding of these proteins. (Black, wild-type thioredoxin; cyan, D13N thioredoxin; red, D15N thioredoxin; blue, D15V thioredoxin; pink, L107A thioredoxin; L107A thioredoxin)
As expected, both the L107P and L107A substitutions result in a substantial destabilization in the conformational stability of reduced thioredoxin (table 2). The L107P substitution causes a 16°C decrease in the TM, a 24 kCal/mol decrease in the ΔHTM according to the thermal denaturation curves, and an over 2.5 kCal/mol in the ΔGH2O according to the chemical denaturation and renaturation curves. The destabilization caused by the L107A substitution was less severe: a 10°C decrease in TM, a 15 kCal/mol decrease in ΔHTM, and a 1.5 kCal/mol decrease in the ΔGH2O.
Table 2.
Thermodynamic parameters of the reduced form of the thioredoxin variants determined from CD spectroscopy.
| Thioredoxin variant | ΔGH2O*† (kCal•mol−1) | m*† (kCal•mol−1•M−1) | [GdmCl]1/2* (M) | ΔHTm‡† (kCal•mol−1) | ΔCp‡† (kCal•mol−1•K−1) | Tm‡† °C (K) |
|---|---|---|---|---|---|---|
| wild type | 5.41 ± 0.19 | 3.39 ± 0.12 | 1.60 | 91.00 | 1.52 ± 0.12 | 78 (351) |
| ± 0.67 | ± 0.04 | |||||
| D13N | 5.59 ± 0.08 | 3.60 ± 0.05 | 1.55 | 95.07 | 2.67 ± 021 | 75 (348) |
| ± 0.95 | ± 0.04 | |||||
| D15N | 6.38 ± 0.08 | 4.09 ± 0.05 | 1.56 | 91.28 | 2.73 ± 0.04 | 80 (353) |
| ± 0.66 | ± 0.03 | |||||
| D15V | 6.69 ± 0.14 | 3.61 ± 0.08 | 1.85 | 95.81 | 2.40 ± 0.14 | 84 (357) |
| ± 2.14 | ± 0.14 | |||||
| L107P | 3.00 ± 0.13 | 4.20 ± 0.16 | 0.71 | 66.62 | 1.43 ± 0.07 | 62 (335) |
| ± 0.44 | ± 0.03 | |||||
| L107A | 4.23 ± 0.11 | 4.35 ± 0.10 | 0.97 | 75.65 | 1.55 ± 0.11 | 68 (341) |
| ± 0.59 | ± 0.04 |
Determined by chemical denaturation
Determined by thermal denaturation
Confidence intervals are given as standard error
In contrast to the L107 substitutions, the D13N, D15N, and D15V substitutions had either a minimal effect on the conformational stability or even slightly increased the stability of thioredoxin. The D13N substitution caused a small decrease (3°C) in the TM but didn’t have a significant effect on the ΔHTM or G, and the D15N substitution resulted in a moderate stabilization of reduced thioredoxin (2°C increase in TM and 0.5 kCal/mol increase in ΔG), which is consistent with the parameters reported in our previous study. We observed a larger stabilization by the D15V substitution, which caused a 6°C increase in TM and an almost 1 kCal/mol increase in ΔG, as evidenced by a shift in the transition melting curves for both chemical and thermal denaturation.
Folding kinetics of the oxidized mutant thioredoxins
In order to evaluate the impact of the D13N, D15N, D15V, L107P, and L107A substitutions on the kinetics of thioredoxin folding, we recorded kinetics after rapid dilution of unfolded protein to native conditions using a stopped-flow device under both oxidizing and reducing conditions. According to the GdmCl-induced equilibrium unfolding and refolding curves, wild-type thioredoxin and all the mutant proteins were totally unfolded at 3.5 M GdmCl and recovered an ellipticity and intrinsic fluorescence indicative of the native state at GdmCl concentrations below 0.1 M. We therefore observed the refolding by monitoring tryptophan fluorescence as a function of time after a rapid 40-fold dilution into buffer of oxidized or reduced thioredoxin denatured in 3.5M GdmHCl. Under these standard conditions, any change in the kinetic characteristics (number of phases, rate constants, amplitudes) of variants relative to wild type thioredoxin can be interpreted as a alteration in folding without having to consider the mechanism.
Oxidized WT thioredoxin folds with at least five phases: a rapid burst phase followed by four measurable folding phases (5). The initial burst phase generates a change in fluorescence with a time constant smaller than the observation dead time (2.5 ms). This phase is thought to correspond to the initial hydrophobic collapse of thioredoxin upon its return to renaturing conditions. The recorded fluorescent trace following the burst phase was best described by a curve containing three kinetic constants corresponding to three folding phases. We could not measure the slowest folding phase with stopped-flow mixing due to drift in the fluorescent signal over long periods of time. This folding phase corresponds to the slow trans to cis isomerization of P76 (5). However, none of the substitutions we identified are located immediately proximal to P76 and are thus not likely to influence the intrinsic rate of cis-trans proline isomerization at this position.
The rate constants that we derived for the first four folding phases of oxidized wild-type thioredoxin were consistent with the previously obtained kinetic constants (supplemental table S1) (5, 11). As reported in our previous study, the L107P substitution caused a substantial defect in the first and second folding phases of oxidized thioredoxin, while the D15N substitution appears to have little or no effect on the folding kinetics of oxidized thioredoxin. Of the substitutions obtained in the present study, the L107A substitution causes a defect in the rate of thioredoxin folding that is less severe than that defect caused by the L107P substitution. In addition, fitting of the L107A folding curve required the inclusion of a fourth kinetic component between the fastest and second refolding phase (k = 1.27 s−1). In contrast, the two mutants D13N and D15V did not cause any apparent defects in the folding kinetics of oxidized thioredoxin and, in the case of the D13N substitution, even appeared to speed up the first folding step following the burst phase.
Folding kinetics of the reduced mutant thioredoxins
In an effort to resolve the perplexing results with the three mutant proteins, D13N, D15N and D15V, which had no apparent folding (or stability) defects, we determined the refolding kinetics of the reduced form of these proteins, the biologically more relevant species (figure 3B). Again, we included L107P and L107A in these studies. Under a standard set of conditions in the presence of 1mM DTT, we found that reduced WT thioredoxin folds with at least six kinetic phases, including a burst phase and five observable folding phases represented by the rate constants termed k1–k5. Four of these phases, the burst phase and the three slowest observable phases (k3–k5), resulted in quenching of the tryptophan fluorescence, whereas the two remaining phases (k1 and k2) resulted in a gain in tryptophan fluorescence.
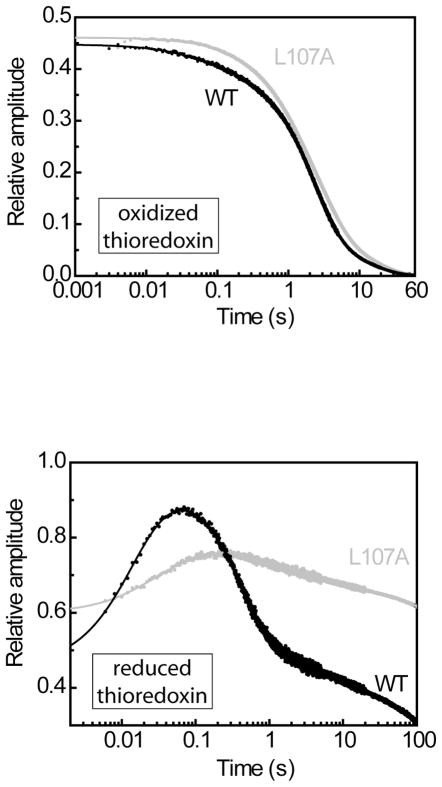
Figure 3. Example kinetic traces of tryptophan fluorescence.
Wild-type (black) and L107A (gray) thioredoxin, denatured in 3.5M GdmCl, were rapidly diluted 40-fold to refolding conditions by stopped-flow mixing. The refolding kinetics were recorded on 3 adjacent regions of time with the 3 following sampling periods: 0.001 s from 0 to 5 s, 0.01 s from 5 to 20 s, 0.2 s from 20 s to the 600s. The refolding was monitored by the intrinsic fluorescence of tryptophans 28 and 31, and the kinetics were normalized to the total amplitude between the native and the unfolded states. (A) Refolding under oxidizing conditions; (B) Refolding in the presence of 1mM DTT. The solid lines correspond to the best fit from non linear least square analysis using a multiexponential equation.
In contrast to our studies with the oxidized form of thioredoxin mutant proteins, we found that all of the reduced mutant thioredoxins examined were defective in at least one of the five observable folding phases (table 3). As expected from our previous study (11), the L107P substitution was the most severe and resulted in defects in four of the five observable folding phases (k1–k4). The L107A substitution appeared to cause less severe, but significant, defects in two of the folding phases (k3 and k4). This is consistent with our in vivo results which suggest that the folding of L107A thioredoxin is intermediate between WT and L107P thioredoxin in the cell.
Table 3.
Folding rate constants of the reduced thioredoxin variants determined from tryptophan fluorescence.
| Variant | Burst (%)* | k1 (s−1) † | k2 (s−1) † | k3 (s−1) † | k4 (s−1) † | k5 (s−1)† |
|---|---|---|---|---|---|---|
| WT | 57 | 75.6 | 20.4 | 2.52 | 0.248 | 8.7 10−4 |
| ± 7.5† | ± 2.7 | ± 0.05 | ± 0.037 | ± 2.9 10−4 | ||
| D13N | 43 | 104 | 45.6 | 2.19 | 0.090 | 9.0 10−4 |
| --- | ± 21 | ± 17.5 | ± 0.23 | ± 0.020 | ± 1.8 10−4 | |
| D15N | 54 | 100 | 14.3 | 2.19 | 0.109 | 0.00110 |
| --- | ± 28 | ± 1.3 | ± 0.11 | ± 0.013 | ± 2.6 10−4 | |
| D15V | 43 | 34.0 | 4.61 | 0.734 | 0.087 | 7.4 10−4 |
| --- | ± 7.9 | ± 0.27 | ± 0.017 | ± 0.026 | ± 2.9 10−4 | |
| L107P | 57 | 46.3 | 6.76 | 0.933 | 0.070 | 9.5 10−4 |
| --- | ± 10.7 | ± 1.48 | ± 0.172 | ± 0.013 | ± 1.4 10−4 | |
| L107A | 40 | 75.1 | 20.6 | 0.751 | 0.082 | 6.5 10−4 |
| --- | ± 8.8 | ± 4.5 | ± 0.099 | ± 0.029 | ± 1.0 10−4 |
The amplitude of burst phase is given as the percentage of the total fluorescence change.
The confidence intervals (standard deviation) were calculated by fitting at least five individual kinetics.
The D15V mutant protein also had severe effects, causing defects in the first four folding phases of reduced thioredoxin. The defects caused by the D13N and D15N substitutions were more subtle. Both of these substitutions caused a moderate apparent increase (~25%) in k1, and the D13N substitution caused a substantial increase (~2-fold) in k2. However, both substitutions caused a greater than 2-fold defect in the fourth rate constant (k4). Since k4 is nearly two orders of magnitude slower than k1 or k2, this defect could make up for the moderate increase in the rate of folding caused by the D13N and D15N substitutions. The defects observed for the reduced form of each of these three proteins is in marked contrast to the lack of any effects when the oxidized form of the proteins was studied.
Discussion
We describe here the isolation of 20 catalytically active mutants of thioredoxin, which when fused to a post-translational signal sequence, display more activity in the periplasm than the wild-type protein, presumably because they are better exported to the periplasm. When combined with those obtained in a previous study (11), we have now identified a total of 39 independently isolated mutants that result in increased activity of thioredoxin in the periplasm. In our previous publication, we mention several other attributes of thioredoxin that these mutations could affect (11). However, the combined observations that (i) all of the mutants that we examined display increased export of thioredoxin to the periplasm in vivo and that (ii) all of the mutant proteins that we examined in vitro are affected for folding suggest that majority of the mutations that we identified cause folding defects. In some cases, the effect of these substitutions on thioredoxin folding seemed small. However, these small differences may be sufficient to allow more productive interaction with a component of posttranslational translocation machinery, such as the dedicated secretion chaperone SecB.
Although we used different mutagenic techniques with different mutational biases, many of the amino acid positions in the protein sequence were identified multiple times, sometimes with mutations that resulted in substitutions of different amino acids (often with different properties) in the same position. Some of these sites could represent mutational hotspots, although different mutagenic techniques usually give different hotspots (20–23). However, it seems likely that further screening using the present method would not result in the identification of novel amino acid positions that are important for folding, which may be due to the limitations discussed in our initial paper (11).
The isolation of an L107R substitution, along with the previously obtained L107P substitution (11), suggest that position L107 is critical for stable folding of thioredoxin in vivo, and our finding that an L107A substitution also altered protein folding appears to support this notion. Structural modeling studies suggest that the shortening of the side chain at position 107 as in the L107A and L107P variants could reduce the number of van der Waals contacts with L53, L80 and L102 and could open the hydrophobic cluster formed by these residues to access by the solvent. In addition, these studies suggest that the arginine residue in the L107R variant does not fit in the hydrophobic pocket and would protrude into the solvent, which would open the pocket to access by the solvent. Moreover, loss of the L107 hydrophobic anchor might slow down the zipping rate of the amphiphilic terminal helix. Santos et al. (7) have recently reported studies on the folding of a sub-domain of thioredoxin that also suggests L107 plays an important role. Our results with L107A thioredoxin suggest that alanine scanning in combination with our genetic screen could be a useful approach for identifying additional thioredoxin folding mutants.
The isolation of multiple, independently isolated mutations that alter residues near the C-terminus of thioredoxin (e.g. mutations that cause substitutions at S95, L107, and the termination codon) seem to suggest the general importance of the C-terminal region of thioredoxin for its folding. It is unknown by what mechanism the C-terminal extension, which resulted from a mutations at the termination codon, could cause increased motility in vivo. However, this C-terminal extension could also slow protein folding, perhaps by a mechanism similar to that of signal sequences (24–26). Regardless of the mechanism, the isolation of these substitutions could provide insight into the mechanism of protein folding in vivo. For example, the sequestration of residues critical for thioredoxin folding (e.g. L107) in the polypeptide exit channel of the ribosome ensures that thioredoxin cannot stably fold until it is fully synthesized. If this phenomenon is generalizable, other proteins, or domains of proteins, could contain residues critical to their folding near their C-termini. Alternatively, extending the open reading frame by 8 amino acids in the termination codon mutants could allow more time for the cotranslational association of the translocation machinery with the nascent chain (1). Other studies on the export of PhoAss-thioredoxin have reported that extending the reading frame at the N-terminus between the signal sequence- and thioredoxin-encoding portions of the phoA-trxA gene results in a similar increase in export (2).
Because the D13N, D15N, and D15V substitutions did not cause altered folding of oxidized thioredoxin, our in vitro characterization of these variants led us to a more extensive analysis of folding of the reduced form of thioredoxin. This characterization revealed that the kinetics of folding of these three mutant proteins is affected only in the reduced form. In contrast, the three mutant proteins have either an enhanced or unaltered conformational stability compared to the wild-type thioredoxin. Taken together with our kinetic analysis, these results suggest that it is the rate of folding, rather than its stability, that is the more important factor for causing increased export of thioredoxin in vivo.
D13 and D15, along with D9 and D10, are part of series of negatively charged surface-exposed residues in the first helix of thioredoxin and, with the exception of D9 (6), do not appear to contribute substantially to the thermostability of the protein. Thus, the finding that substitutions at these residues could affect folding of thioredoxin was unexpected. Indeed, in our previous study, we did not observe an effect on folding of the D15N substitution (11). However, an effect on the folding of reduced thioredoxin became apparent in the present study with more measurements and at longer refolding times. One explanation for the observed effect on folding for these substitutions is that these residues could be involved in interactions required for the stabilization of an important folding step, interactions which no longer exist in the native structure. For example, according to our structural analysis D13 forms a fully hydrated salt-bridge with K18 in the same helix. The elimination of this interaction by the introduction of an asparagine residue does not appear to affect the thermostability of thioredoxin significantly but could slow the formation of this helix. Similarly, D15 could be involved in a transient interaction required for some step in thioredoxin folding, which is no longer present in the fully folded conformation. In addition, D15 is located above a hydrophobic pocket in the thioredoxin structure, and substitution with valine could result in the burying of the amino acid side-chain. This artificial interaction could stabilize the fully folded conformation but may interfere with folding of thioredoxin. This possibility is supported by the small, but significant, increase in the ΔHTM, which suggests an increase in the degree of order of the protein that should result in a negative difference in the TΔS terms between the D15V mutant and the WT proteins. Indeed, the Δ(TΔS) calculated from the experimental values of ΔHTM and ΔCp deduced from the fits of the thermal transitions (see Table 2) is negative for all temperatures up to 64 °C (unpublished results).
Supplementary Material
Acknowledgments
We thank T. Rose for his help in structural prediction and energy calculations, and we thank members of the Beckwith lab for many helpful conversations.
Footnotes
This work was supported by grant GM041883 from the National Institute of General Medical Sciences and by the Institut Pasteur. JB is an American Cancer Society Professor.
Abbreviations used: WT, wild-type; PhoA, alkaline phosphatase; PhoAss, alkaline phosphatase signal sequence; IPTG, isopropyl-β-D-thio-galactopyranoside; 8-oxo-dGTP, 8-oxo-2′-deoxy-guanosine-5′-triphosphate; dPTP, 6H, 8H, 4-Dihydro-pyrimido[4,5-c][1,2] oxazin-7-one-8-β-D-2′-deoxy-ribofuranoside-5′-triphophate; DTT, dithiothreitol; GdmCl, guanidinium chloride
Supporting information available
The supporting information includes a more detailed description of the materials and methods. This material is available free of charge via the Internet at http://pub.acs.org.
References
- 1.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol. 2003;185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debarbieux L, Beckwith J. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc Natl Acad Sci U S A. 1998;95:10751–10756. doi: 10.1073/pnas.95.18.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritz D, Beckwith J. Roles of thiol-redox pathways in bacteria. Annu Rev Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 4.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 5.Georgescu RE, Li JH, Goldberg ME, Tasayco ML, Chaffotte AF. Proline isomerization-independent accumulation of an early intermediate and heterogeneity of the folding pathways of a mixed alpha/beta protein, Escherichia coli thioredoxin. Biochemistry. 1998;37:10286–10297. doi: 10.1021/bi9805083. [DOI] [PubMed] [Google Scholar]
- 6.Mancusso R, Cruz E, Cataldi M, Mendoza C, Fuchs J, Wang H, Yang X, Tasayco ML. Reversal of negative charges on the surface of Escherichia coli thioredoxin: pockets versus protrusions. Biochemistry. 2004;43:3835–3843. doi: 10.1021/bi0354684. [DOI] [PubMed] [Google Scholar]
- 7.Santos J, Sica MP, Buslje CM, Garrote AM, Ermacora MR, Delfino JM. Structural selection of a native fold by peptide recognition. Insights into the thioredoxin folding mechanism. Biochemistry. 2009;48:595–607. doi: 10.1021/bi801969w. [DOI] [PubMed] [Google Scholar]
- 8.de Lamotte-Guery F, Pruvost C, Minard P, Delsuc MA, Miginiac-Maslow M, Schmitter JM, Stein M, Decottignies P. Structural and functional roles of a conserved proline residue in the alpha2 helix of Escherichia coli thioredoxin. Protein Eng. 1997;10:1425–1432. doi: 10.1093/protein/10.12.1425. [DOI] [PubMed] [Google Scholar]
- 9.Jeng MF, Campbell AP, Begley T, Holmgren A, Case DA, Wright PE, Dyson HJ. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2:853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- 10.Gleason FK. Mutation of conserved residues in Escherichia coli thioredoxin: effects on stability and function. Protein Sci. 1992;1:609–616. doi: 10.1002/pro.5560010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber D, Cha MI, Debarbieux L, Planson AG, Cruz N, Lopez G, Tasayco ML, Chaffotte A, Beckwith J. A selection for mutants that interfere with folding of Escherichia coli thioredoxin-1 in vivo. Proc Natl Acad Sci U S A. 2005;102:18872–18877. doi: 10.1073/pnas.0509583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- 13.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 14.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JC, Minev M, Beckwith J. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J Bacteriol. 2002;184:695–705. doi: 10.1128/JB.184.3.695-705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 17.Dailey FE, Berg HC. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley RF, Shalongo W, Jagannadham MV, Stellwagen E. Equilibrium and kinetic measurements of the conformational transition of reduced thioredoxin. Biochemistry. 1987;26:1406–1411. doi: 10.1021/bi00379a029. [DOI] [PubMed] [Google Scholar]
- 19.Ladbury JE, Kishore N, Hellinga HW, Wynn R, Sturtevant JM. Thermodynamic effects of reduction of the active-site disulfide of Escherichia coli thioredoxin explored by differential scanning calorimetry. Biochemistry. 1994;33:3688–3692. doi: 10.1021/bi00178a027. [DOI] [PubMed] [Google Scholar]
- 20.Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JH, Low KB. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell. 1984;37:675–682. doi: 10.1016/0092-8674(84)90400-8. [DOI] [PubMed] [Google Scholar]
- 22.Miller JH. Mutagenic specificity of ultraviolet light. J Mol Biol. 1985;182:45–65. doi: 10.1016/0022-2836(85)90026-9. [DOI] [PubMed] [Google Scholar]
- 23.Miller JH. Mutational specificity in bacteria. Annu Rev Genet. 1983;17:215–238. doi: 10.1146/annurev.ge.17.120183.001243. [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Topping TB, Randall LL. Physiological role during export for the retardation of folding by the leader peptide of maltose-binding protein. Proc Natl Acad Sci U S A. 1989;86:9213–9217. doi: 10.1073/pnas.86.23.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S, Liu G, Topping TB, Cover WH, Randall LL. Modulation of folding pathways of exported proteins by the leader sequence. Science. 1988;239:1033–1035. doi: 10.1126/science.3278378. [DOI] [PubMed] [Google Scholar]
- 26.Beena K, Udgaonkar JB, Varadarajan R. Effect of signal peptide on the stability and folding kinetics of maltose binding protein. Biochemistry. 2004;43:3608–3619. doi: 10.1021/bi0360509. [DOI] [PubMed] [Google Scholar]
- 27.Lodish HF. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J Mol Biol. 1970;50:689–702. doi: 10.1016/0022-2836(70)90093-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.