Summary
Nanotechnology has held great promise for revolutionizing biology. The biological behavior of nanomaterials depends primarily on how they interface to biomolecules and their surroundings. Unfortunately, interface issues like non-specific adsorption are still the biggest obstacles to the success of nanobiotechnology and nanomedicine, and have held back widespread practical use of nanotechnology in biology. Not only does the biological interface of nanoparticles needs to be understood and controlled, but nanoparticles must be treated as biological entities rather than inorganic ones. Furthermore, one can adopt an engineering perspective of the nanoparticle-biological interface, realizing that it has unique, exploitable properties.
Introduction
The combination of nanotechnology and biology has resulted in a rapidly advancing field. Since its inception about two decades ago, innovative approaches for using nanoparticles (NPs) to kill tumors, enhance drug delivery [1], assemble structures, and sense intracellular processes have been envisioned [2–5]. However, these applications are all limited by non-specific adsorption (NSA), where biomolecules stick to NPs non-covalently. Both the NP and the biomolecules it encounters are complex three-dimensional entities, thus their interface is also complex (Figure 1a). NPs are not hard spheres but crystals with facets and edges, and are coated with ligands that enable solubility and stability. Biomolecules have well-defined structures determined by numerous intramolecular and intermolecular interactions. Thus, when a NP interacts with a biomolecule, numerous non-covalent bonds can form between them [• •6], often resulting in denaturation and loss of activity.
Figure 1.
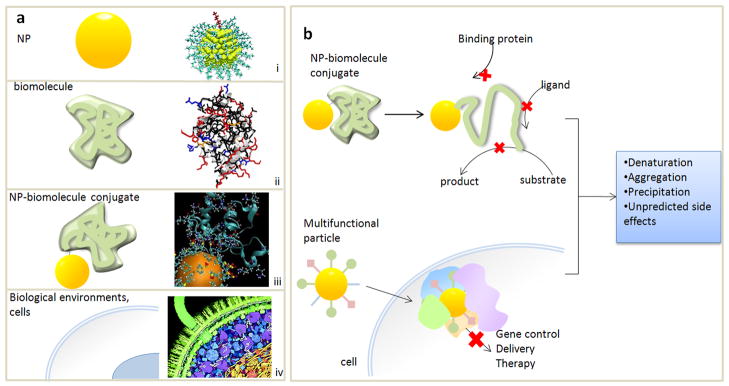
NPs and biomolecular interfaces. a) NPs, biomolecules, NP-biomolecule conjugates, and biological environments are much more complex (right) than typically depicted (left). (i) from [13] ; (ii) PDB ID 1YCC from [58], (iv) Illustration by David S. Goodsell, the Scripps Research Institute. b) Interface effects can diminish the biological function of NP-biomolecule conjugates (upper) and NPs used for therapy in cells and biological fluid, leading to undesirable and unpredictable side effects.
NSA affects not only NPs, but any interface between an inorganic surface and biology. Medical devices and implants have faced the same challenges of surface fouling. NSA can cause false positive/negative signals in sensors, compromising sensitivity. These complications dramatically intensify as the inorganic system size shrinks to the nanoscale, because surface to volume ratios increase dramatically, and nanoscale surfaces differ physically and chemically from bulk.
It is clear that the biological behavior and consequences of NPs are largely dictated by how they interface to biology (Figure 1b). Interface issues strongly influence cellular uptake, where varying NP size and shape varies uptake behavior [7,8], biodistribution [9], cytotoxicity [•10,11], and the unintended consequences of NPs such as adsorption to other species and aggregation. Clearly, NPs cannot be treated as noninteracting species, but rather as biological entities, where their interaction with the environment is mediated by the proteins that adsorb to them.
Unfortunately, the biological interface is the least understood aspect about NPs. Despite the importance of the interface, efforts to characterize it are surprisingly scarce. Biological outcomes of simple experiments such as cellular uptake and cytotoxicity of NPs are determined empirically, and we are far from being able to predict this behavior. The ligand is the most critical aspect of NPs because it is the surface presented to the biological environment. However, even fundamental studies characterizing its properties, such as surface coverage and binding strength, are few [12–15]. When biomolecules are conjugated to NPs, their behavior can be completely opposite to that of its unconjugated form. Frequently, surface issues are simply ignored, where linked proteins are assumed to be fully folded and active. To date, research on NPs in biology has focused predominantly on exploiting the size and material dependent properties of NPs, but not understanding the interface.
Consequently, challenges remain for not only characterizing the biological interface but also controlling it. Surface treatments are highly variable and difficult to reproduce. There is enormous diversity in ligand types (small molecules, branched species, polymers), and in how they bind (covalent or non-covalent, monolayers or multilayers [16–18]). Also, ligand can come on and off the particle, and free ligand can influence biological behavior [•19]. Therefore, the biological interface of NPs is a significant challenge that needs to be addressed for their development and application.
Different classes of NSA
It is helpful to categorize NSA and interface effects. NSA can occur by adsorption of 1) the linked biomolecule or 2) other species present in the environment.
Self-adsorption
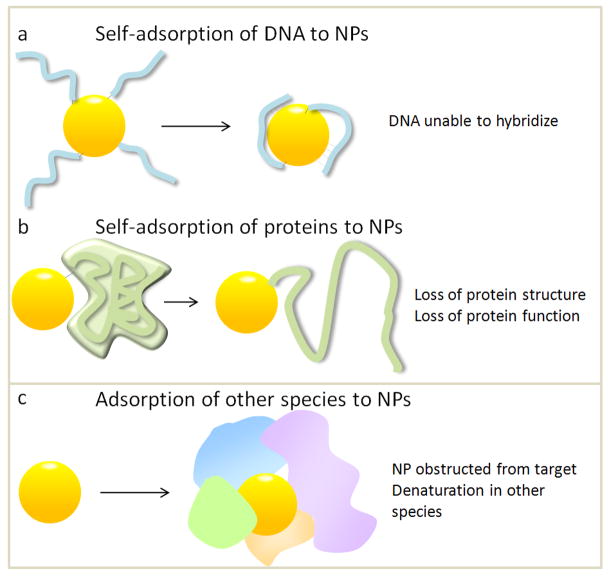
DNA in NP-DNA conjugates adsorb to NPs via the nucleotides (Figure 2a), as reported by Gearheart and Murphy et al. [20] and Zanchet and Alivisatos et al. [21]. DNA self-adsorption inhibits its ability to bind to a target [22], compromising its use for assembly or sensing. Self-adsorption depends on coverage, where lower coverage increases self-adsorption because of larger exposed NP surface areas. Increasing coverage decreases NSA, but can also reduce hybridization ability due to steric hindrance. This varies with NP size, as higher curvature allows for a higher density of oligonucleotides, while limiting steric hindrance. Self-adsorption also depends on DNA sequence [23], since each nucleotide has a different affinity for gold surfaces [24]. Self-adsorption can be alleviated by spacers or chemical modification with thiols for gold NPs, which block DNA adsorption and its ability to hybridize [25].
Figure 2.
Different classes of NSA. Self adsorption of a covalently linked a) DNA molecule or b) protein; c) adsorption of other species to the NP.
Proteins also adsorb onto NPs and denature, damaging protein function (Figure 2b). This is further complicated by the fact that the NP sterically hinders substrates from accessing the active site, so activity loss can be due to both effects. Because proteins are more complex than DNA, probing protein-NP interfaces is challenging. However, there has been progress in determining “design rules” for self-adsorption onto NPs [26–30]. Cytochrome c unfolds on gold NPs with charged ligands but not for neutral ligands. However, for CoFe2O4 NPs, the dominant interaction is between the COO− on residues and Co or Fe surface atoms. In this case, PEG was not effective as molecules containing COO− for reducing NSA. Experiments changing the labeling site on cytochrome c [29] have elucidated that denaturation is partial and varies with labeling position, as different protein motifs have different stabilities and roles in folding.
Adsorption of other species
Another type of NSA is where other species adsorb to the NP (Figure 2c). This type of NSA is impossible to avoid because biological environments are innately crowded [• •6]. Biological fluids such as blood are highly concentrated [31,32], and intracellular protein concentration is >300 mg/mL, significantly higher than the dilute solutions used for NP conjugation and biophysical characterization. Consequently, when NPs are introduced to these environments, proteins adsorb to the NP, shrouding it in a “protein corona,” which can follow the migrating particle [••33]. Unfortunately, adsorption of other species is complex and difficult to prevent. [34] It is also challenging to predict, where NPs coated with ligands thought to be inert, such as PEG, still encounter NSA even in dilute solutions. [30,35] Furthermore, NSA in cells or biological fluids results in NP aggregation or precipitation and deleterious side effects. [36] There have been advances in surface chemistry to render NPs inert to adsorption [32,37,38], such as cloaking particles with polymers [39]. Allen and Bawendi et al. [40] have coated quantum dots with polymeric imidazole ligands which resist adsorption, and thus have yielded unprecedented images of tumor vasculature. However, this level of control over surface chemistry is relatively new, and is still an exception rather than the standard.
Both types of NSA are prevalent in nanobiotechnology and nanomedicine, and can completely obscure the biological purpose of the NP and cause undesirable side effects. To further complicate things, both types of NSA vary with surface chemistry, NP material, size [27,41,42] and shape, and surface chemistry can be highly variable not only between labs but also day to day, making it challenging to ascertain its mechanism. Thus, NSA is typically viewed as an impediment.
Characterizing NSA
If NPs are going to be employed for practical biological applications, it is imperative to characterize and understand their biological interface, so that ultimately it can be controllable and predictable. NSA is challenging to characterize because it is due to formation of non-covalent bonds between the biomolecule and NP surface or ligand. Because these interactions are numerous, non-covalent, and dynamic, they are difficult to directly probe. However, there has been substantial progress in measuring their effect on the biomolecular structure and activity, yielding information on how adsorption occurs and the interactions involved [•43].
RH measurements can infer the effects of NSA on biomolecular structure [21,25,44]. If covalently linked DNA adsorbs to the surface, or if other species adsorb to the NP, RH will change (Figure 3a) [32]. Methods to determine RH such as Ferguson analysis, dynamic light scattering, size exclusion chromatography, surface plasmon resonance (SPR), and ultracentrifugation [35] have been successful in quantifying NSA of proteins and DNA to NPs. Centrifugation assays have also been effective in identifying the adsorbed proteins, a critical issue [••33].
Figure 3.
Characterizing NSA. a) RH is sensitive to self-adsorption and adsorption of other species. b) Measuring the effect of NSA on secondary structure of the linked biomolecule. c) Measuring the effect of NSA on biomolecular function of the linked biomolecule.
Effect of NSA on protein structure can be measured directly for proteins with well-defined structures, elucidating how much the protein is denatured (Figure 3b). Circular Dichroism (CD) spectroscopy [45], NMR, and FRET can [28,30] quantify the degree of denaturation when proteins are interfaced to NPs. These approaches typically measure averages in an ensemble, so single molecule experiments have worked well to complement them [46].
While measurements of protein structure yield information on the interface, they must be coupled with activity measurements (Figure 3c), because if function is compromised, then conjugation is pointless. For DNA, this is simply its ability to bind to complement. For proteins, this may be ligand binding, which can be quantified by spectroscopy [22,47], isothermal titration calorimetry (ITC), and SPR [48]. For enzymatic proteins, activity assays are necessary [49,50], and changes in activity could be due to either NP-induced denaturation or sterics.
Unfortunately, the aforementioned experiments cannot yield molecular information about the NP-biomolecule interface. For example, CD yields only secondary structure. Therefore, molecular dynamics (MD) simulations can naturally complement these techniques, since it can elucidate interactions between the biomolecule, NP, and ligand on a molecular level [51]. Recent experiments combining MD with CD and electrophoresis [29] have been able to elucidate rules for how protein structure is affected by NP labeling.
Exploiting interface effects
Due to the major challenges in characterizing and predicting its behavior, NSA is typically viewed as an impediment for nanobiotechnology [39,52]. However, by shifting to an engineering perspective, one can regard NSA as having unique, exploitable properties. The fact that NP surface chemistry can strongly influence biological response [9] can potentially be a means for manipulating biology in ways not previously possible. By realizing that these interface problems are actually an opportunity, one can potentially engineer the NP-biomolecular interface to achieve new capabilities [53]. Listed below are examples of some new approaches that biologically exploit NP interface effects.
Tunable release from NPs
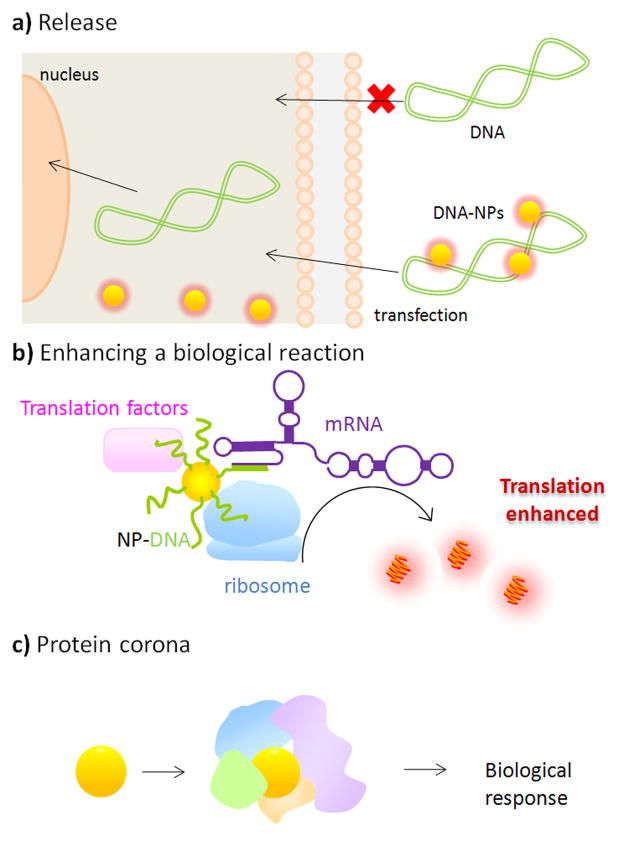
The fact that the non-covalent interactions between NPs and adsorbed molecules change with environment can be used to release a payload from the NPs. Han and Rotello et al. exploited the fact that intracellular concentrations of glutathione are high, and can release NPs bound to DNA [54] to make it available for transcription [55] (Figure 4a). Thus, the NP acts as a smart delivery vehicle.
Figure 4.
Utilizing NSA. a) Tunable intracellular release from NP-DNA “nanoplexes.” Adapted from [55]. b) Enhancing protein translation. From [56]. c) Protein coronas induce a biological response.
Enhancing biological reactions with NP chaperones
Another way that interface effects can be exploited is to use NPs as chaperones for enhancing biological reactions. NPs are approximately the same size as proteins, so adsorbed species are brought within nanometer proximity (Figure 4b). Furthermore, if the reaction involves specific nucleic acids (such as mRNA) the NP can be decorated with DNA that binds specifically to it. Along these lines, NP adsorption has been used to double in vitro protein synthesis selectively by Park and Hamad-Schifferli [56]. The very same aspects that make NSA problematic is actually useful for enhancement – since involves weak binding, it permits species to come on and off for turnover, which would not be possible if binding was strong.
Perturbing protein structure via protein corona
When NPs are introduced to a biological fluid, proteins adsorb to their surface resulting in a dynamic “protein corona” [48]. It is clear that identifying the proteins in the corona and understanding how adsorption occurs and evolves is critical for useful application of NPs. Still, one can imagine ways in which the corona could potentially be used to induce a desired biological function [••33](Figure 4c). Proteins in the corona may be denatured due to interaction with the NP surface, and if this can be controlled, can be used to induce a response. Because the amount of denaturation in the protein can be tuned by changing the surface properties of the NP [28,57], corona properties may be tuned by modifying NP surface. Evidently, this will require “design rules” for how the corona behaves.
Conclusions
The interface of NPs to biomolecules and biological systems presents a formidable challenge for practical application. While it has been challenging understanding NSA, there have been promising advances in its qualitative characterization. Furthermore, there has been a shift in perspective about how to exploit the unique properties of interface effects.
Acknowledgments
Support was from the National Institute of Health (R21 EB008156-01) and the National Science Foundation (DMR 0906838).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SE, Liu GL, Kim F, Lee LP. Remote optical switch for localized and selective control of gene interference. Nano Lett. 2009;9:562–570. doi: 10.1021/nl802689k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang X, Neretina S, El-Sayed MA. Gold nanorods: From synthesis and properties to biological and biomedical applications. Adv Mater. 2009;21 :4880–4910. doi: 10.1002/adma.200802789. [DOI] [PubMed] [Google Scholar]
- 3.Niemeyer CM. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew Chem Int Ed. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 5.Bardhan R, Chen W, Perez-Torres C, Bartels M, Huschka RM, Zhao LL, Morosan E, Pautler RG, Joshi A, Halas NJ. Nanoshells with targeted simultaneous enhancement of magnetic and optical imaging and photothermal therapeutic response. Adv Funct Mater. 2009;19:3901–3909. [Google Scholar]
- ••6.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 7.Champion JA, Mitragotri S. Role of target geometry in phagocytosis. Proc Nat Acad Sci. 2006;103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. The effect of particle design on cellular internalization pathways. Proc Nat Acad Sci. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Design considerations for tumour-targeted nanoparticles. Nat Nanotechnol. 2009;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •10.Delehanty JB, Mattoussi H, Medintz IL. Delivering quantum dots into cells: Strategies, progress and remaining issues. Anal BioanalChem. 2009;393:1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- 11.Huff TB, Hansen MN, Zhao Y, Cheng J-X, Wei A. Controlling the cellular uptake of gold nanorods. Langmuir. 2007;23:1596–1599. doi: 10.1021/la062642r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amstad E, Gillich T, Bilecka I, Textor M, Reimhult E. Ultrastable iron oxide nanoparticle colloidal suspensions using dispersants with catechol-derived anchor groups. Nano Lett. 2009;9:4042–4048. doi: 10.1021/nl902212q. [DOI] [PubMed] [Google Scholar]
- 13.Rapino S, Zerbetto F. Dynamics of thiolate chains on a gold nanoparticle. Small. 2007;3:386–388. doi: 10.1002/smll.200600456. [DOI] [PubMed] [Google Scholar]
- 14.Heaven MW, Dass A, White PS, Holt KM, Murray RW. Crystal structure of the gold nanoparticle [N(C8H17)4][Au25(SCH2CH2Ph)18] J Am Chem Soc. 2009;130:3754–3755. doi: 10.1021/ja800561b. [DOI] [PubMed] [Google Scholar]
- 15.Giaume D, Poggi M, Casanova D, Mialon G, Lahlil K, Alexandrou A, Gacoin T, Boilot J-P. Organic functionalization of luminescent oxide nanoparticles toward their application as biological probes. Langmuir. 2008;24:11018–11026. doi: 10.1021/la8015468. [DOI] [PubMed] [Google Scholar]
- 16.Robinson DB, Persson HHJ, Zeng H, Li G, Pourmand N, Sun S, Wang SX. DNA-functionalized MFe2O4 (M = Fe, Co, or Mn) nanoparticles and their hybridization to DNA-functionalized surfaces. Langmuir. 2005;21:3096–3103. doi: 10.1021/la047206o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alper JD, Crespo M, Hamad-Schifferli K. Release mechanism of octadecyl rhodamine B chloride from Au nanorods by ultrafast laser pulses. J Phys Chem C. 2009;115:5967–5973. [Google Scholar]
- 18.Wijaya A, Hamad-Schifferli K. Ligand customization and DNA functionalization of gold nanorods via roundtrip phase transfer ligand exchange. Langmuir. 2008;24:9966–9969. doi: 10.1021/la8019205. [DOI] [PubMed] [Google Scholar]
- •19.Shenhar R, Rotello VM. Nanoparticles: Scaffolds and building blocks. Acc Chem Res. 2003;36:549–561. doi: 10.1021/ar020083j. [DOI] [PubMed] [Google Scholar]
- 20.Gearheart LA, Ploehn HJ, Murphy CJ. Oligonucleotide adsorption to gold nanoparticles: A surface-enhanced Raman spectroscopy study of intrinsically bent DNA. J Phys Chem B. 2001;105:12609–12615. [Google Scholar]
- 21.Zanchet D, Micheel CM, Parak WJ, Gerion D, Alivisatos AP. Electrophoretic isolation of discrete Au nanocrystal/DNA conjugates. Nano Lett. 2001;1:32–35. [Google Scholar]
- 22.Stoermer RL, Keating CD. Distance-dependent emission from dye-labeled oligonucleotides on striped Au/Ag nanowires: Effect of secondary structure and hybridization efficiency. J Am Chem Soc. 2006;128:13243–13254. doi: 10.1021/ja0637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown KA, Park S, Hamad-Schifferli K. Nucleotide-surface interactions in DNA modified Au-nanoparticle conjugates: Sequence effects on reactivity and hybridization. J Phys Chem C. 2008;112:7517–7521. [Google Scholar]
- 24.Demers LM, Ostblom M, Zhang H, Jang N, Liedberg B, Mirkin CA. Thermal desorption behavior and binding properties of DNA bases and nucleosides on gold. J Am Chem Soc. 2002;124:11248–11249. doi: 10.1021/ja0265355. [DOI] [PubMed] [Google Scholar]
- 25.Park S, Brown KA, Hamad-Schifferli K. Changes in oligonucleotide conformation on nanoparticle surfaces by modification with mercaptohexanol. Nano Lett. 2004;4:1925–1929. [Google Scholar]
- 26.Asuri P, Karajanagi SS, Vertegel AA, Dordick JS, Kane RS. Enhanced stability of enzymes adsorbed onto nanoparticles. J Nanosci Nanotechnol. 2007;7:1675–1678. doi: 10.1166/jnn.2007.453. [DOI] [PubMed] [Google Scholar]
- 27.Vertegel AA, Siegel RW, Dordick JS. Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir. 2004;20:6800–6807. doi: 10.1021/la0497200. [DOI] [PubMed] [Google Scholar]
- 28.Aubin-Tam M-E, Hamad-Schifferli K. Gold nanoparticle-cytochrome c complexes: The effect of nanoparticle ligand charge on protein structure. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 29.Aubin-Tam M-E, Hwang W, Hamad-Schifferli K. Site-directed nanoparticle labeling of cytochrome c. Proc Nat Acad Sci. 2009;106:4095–4100. doi: 10.1073/pnas.0807299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aubin-Tam M-E, Zhou H, Hamad-Schifferli K. Structure of cytochrome c at the interface with magnetic CoFe2O4 nanoparticles. Soft Matter. 2008;4:554–559. doi: 10.1039/b711937b. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch LR, Halas NJ, West JL. Whole-blood immunoassay facilitated by gold nanoshell-conjugate antibodies. Methods in Molecular Biology. 2005;303:101–111. doi: 10.1385/1-59259-901-X:101. [DOI] [PubMed] [Google Scholar]
- 32.Yang W, Zhang L, Wang S, White AD, Jiang S. Functionalizable and ultra stable nanoparticles coated with zwitterionic poly(carboxybetaine) in undiluted blood serum. Biomaterials. 2009;30:5617–5621. doi: 10.1016/j.biomaterials.2009.06.036. [DOI] [PubMed] [Google Scholar]
- ••33.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The nanoparticle–protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv Colloid Interface Sci. 2007;134–135:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 34.Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdörster G, McGrath JL. The influence of protein adsorption on nanoparticle association with cultured endothelial cells. Biomaterials. 2009;30:603–610. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 35.Lees EE, Gunzburg MJ, Nguyen T-L, Howlett GJ, Rothacker J, Nice EC, Clayton AHA, Mulvaney P. Experimental determination of quantum dot size distributions, ligand packing densities, and bioconjugation using analytical ultracentrifugation. Nano Lett. 2008;8:2883–2890. doi: 10.1021/nl801629f. [DOI] [PubMed] [Google Scholar]
- 36.Eck W, Craig G, Sigdel A, Ritter G, Old LJ, Tang L, Brennan MF, Allen PJ, Mason MD. PEGylated Gold Nanoparticles Conjugated to Monoclonal F19 Antibodies as Targeted Labeling Agents for Human Pancreatic Carcinoma Tissue. ACS Nano. 2008;2:2263–2272. doi: 10.1021/nn800429d. [DOI] [PubMed] [Google Scholar]
- 37.Das J, Huh C-H, Kwon K, Park S, Jon S, Kim K, Yang H. Comparison of the nonspecific binding of DNA-conjugated gold nanoparticles between polymeric and monomeric self-assembled monolayers. Langmuir. 2009;25:235–241. doi: 10.1021/la802531d. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka M. Synthesis of phosphorylcholine–oligoethylene glycol–alkane thiols and their suppressive effect on non-specific adsorption of proteins. Tetrahedron Lett. 2009;50:4092–4095. [Google Scholar]
- 39.Chen H, Wang L, Yeh J, Wu X, Cao Z, Wang YA, Zhang M, Yang L, Mao H. Reducing non-specific binding and uptake of nanoparticles and improving cell targeting with an antifouling PEO-b-PγMPS copolymer coating. Biomaterials. 2010:1–11. doi: 10.1016/j.biomaterials.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen PM, Liu W, Chauhan VP, Lee J, Ting AY, Fukumura D, Jain RK, Bawendi MG. InAs(ZnCdS) quantum dots optimized for biological imaging in the near-infrared. J Am Chem Soc. 2010;132:470–471. doi: 10.1021/ja908250r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang W, Nuffer JH, Dordick JS, Siegel RW. Unfolding of ribonuclease A on silica nanoparticle surfaces. Nano Lett. 2007;7:1991 –1995. doi: 10.1021/nl070777r. [DOI] [PubMed] [Google Scholar]
- 42.Goodrich GP, Helfrich MR, Overberg JJ, Keating CD. Effect of macromolecular crowding on DNA: Au nanoparticle bioconjugate assembly. Langmuir. 2004;20:10246–10251. doi: 10.1021/la048434l. [DOI] [PubMed] [Google Scholar]
- •43.Aubin-Tam M-E, Hamad-Schifferli K. Structure and function of nanoparticle-protein conjugates. Biomed Mater. 2008;3:034001. doi: 10.1088/1748-6041/3/3/034001. [DOI] [PubMed] [Google Scholar]
- 44.Park S, Hamad-Schifferli K. Evaluation of hydrodynamic size and zeta-potential of surface-modified Au nanoparticle-DNA conjugates via Ferguson analysis. J Phys Chem C. 2008;112:7611–7616. [Google Scholar]
- 45.Mamedova NN, Kotov NA, Rogach AL, Studer J. Albumin-CdTe nanoparticle bioconjugates: Preparation, structure, and interunit energy transfer with antenna effect. Nano Lett. 2001;1:281–286. [Google Scholar]
- 46.Casanova D, Giaume D, Moreau M, Martin J-L, Gacoin T, Boilot J-P, Alexandrou A. Counting the number of proteins coupled to single nanoparticles. J Am Chem Soc. 2007;129:12592–12593. doi: 10.1021/ja0731975. [DOI] [PubMed] [Google Scholar]
- 47.Chen C, Wang W, Ge J, Zhao XS. Kinetics and thermodynamics of DNA hybridization on gold nanoparticles. Nucl Acids Res. 2009;37:3756–3765. doi: 10.1093/nar/gkp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cedervall T, Lynch I, Lindman S, Thulin TBE, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Nat Acad Sci. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brennan JL, Hatzakis NS, Tshikhudo TR, Dirvianskyte N, Razumas V, Patkar S, Vind J, Svendsen A, Nolte RJM, Rowan AE, et al. Bionanoconjugation via click chemistry: The creation of functional hybrids of lipases and gold nanoparticles. Bioconjugate Chem. 2006;17:1373–1375. doi: 10.1021/bc0601018. [DOI] [PubMed] [Google Scholar]
- 50.Aubin M-E, Morales DG, Hamad-Schifferli K. Labeling ribonuclease S with a 3nm Au nanoparticle by two-step assembly. Nano Lett. 2005;5:519–522. doi: 10.1021/nl0479031. [DOI] [PubMed] [Google Scholar]
- 51.Lee O-S, Schatz GC. Molecular dynamics simulation of DNA-functionalized gold nanoparticles. J Phys Chem C. 2009;113:2316–2321. [Google Scholar]
- 52.Gill R, Willner I, Shweky I, Banin U. Fluorescence resonance energy transfer in CdSe/ZnS DNA conjugates: Probing hybridization and DNA cleavage. J Phys Chem B. 2005;109:23715–23719. doi: 10.1021/jp054874p. [DOI] [PubMed] [Google Scholar]
- 53.You C-C, Chompoosor A, Rotello VM. The biomacromolecule-nanoparticle interface. Nano Today. 2007:2. [Google Scholar]
- 54.Han G, Chari NS, Verma A, Hong R, Martin CT, Rotello VM. Controlled recovery of the transcription of nanoparticle-bound DNA by intracellular concentrations of glutathione. Bioconjugate Chem. 2005;16:1356–1359. doi: 10.1021/bc050173j. [DOI] [PubMed] [Google Scholar]
- 55.Ghosh PS, Kim C-K, Han G, Forbes NS, Rotello VM. Efficient gene delivery vectors by tuning the surface charge density of amino acid-functionalized gold nanoparticles. ACS Nano. 2008;2:2213–2218. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park S. Hamad-Schifferli K. Enhancement of in vitro translation using gold nanoparticle-DNA conjugates. ACS Nano. 2010 doi: 10.1021/nn100362m. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.You C-C, De M, Han G, Rotello VM. Tunable inhibition and denaturation of α-chymotrypsin with amino acid-functionalized gold nanoparticles. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 58.Louie GV, Hutcheon WLB, Brayer GD. Yeast Iso-1-Cytochrome c - A 2.8Å Resolution Three-dimensional Structure Determination. J Molec Biol. 1988;199:205–314. doi: 10.1016/0022-2836(88)90315-4. [DOI] [PubMed] [Google Scholar]