Abstract
The ability to predict the efficacy of molecularly-targeted therapies for non-small cell lung cancer (NSCLC) for an individual patient remains problematic. The purpose of this study was to identify tumor biomarkers, using a refined “coexpression extrapolation (COXEN)” algorithm with a continuous spectrum of drug activity, that predict drug sensitivity and therapeutic efficacy in NSCLC to Vorinostat, a histone deacetylase inhibitor, and Velcade, a proteasome inhibitor. Using our refined COXEN algorithm, biomarker prediction models were discovered and trained for Vorinostat and Velcade based on in vitro drug activity profiles of 9 NSCLC cell lines (NCI-9). Independently, a panel of 40 NSCLC cell lines (UVA-40) was treated with Vorinostat or Velcade to obtain 50% growth inhibition values. Genome-wide expression profiles for both the NCI-9 and UVA-40 cell lines were determined using HG-U133A Affymetrix platform. Modeling generated multi-gene expression signatures for Vorinostat (45-gene, p=0.002) and Velcade (15-gene, p=0.0002), with one overlapping gene (CFLAR). Examination of Vorinostat gene ontogeny revealed a predilection for cellular replication and death, whereas those of Velcade suggested involvement in cellular development and carcinogenesis. Multivariate regression modeling of the refined COXEN scores significantly predicted the activity of combination therapy in NSCLC cells (p=0.007). Through the refinement of the COXEN algorithm, we provide an in silico method to generate biomarkers that predict tumor sensitivity to molecularly-targeted therapies. Use of this refined COXEN methodology has significant implications for the a priori examination of targeted therapies to more effectively streamline subsequent clinical trial design and cost.
Keywords: Lung cancer, histone deacetylase inhibitor, proteasome inhibitor, tumor biomarker, molecularly-targeted agents, chemotherapy
Introduction
The need for new pharmacogenomic approaches to drug discovery and subsequent clinical validation in the treatment of non-small cell lung cancer (NSCLC) is based on two important observations. First, approximately 70% of patients with NSCLC present with stage III/IV disease where standard of care guidelines consists of a platinum-based doublet chemotherapeutic regimen with or without local therapies such as surgery or radiation (1). Unfortunately, the results of these platinum-based doublet therapies to treat advanced stage NSCLC patients are poor (2). Second, several recent phase III clinical trials have demonstrated an 8-10% improvement in 5-year survivals in patients following surgery who receive adjuvant chemotherapy for node-positive NSCLC (3-6). The converse is that 90% of patients who receive that same adjuvant chemotherapy receive no benefit, but incur all the risks of subsequent toxicities and cost of such an all-inclusive approach. Therefore, an unmet need exists for drug discovery platforms to identify new, clinically effective therapeutic agents.
Research is currently focused on identifying novel molecularly-targeted therapies that exploit the underlying mechanisms of tumorigenesis and/or tumor cell signal transduction pathways involved in chemoresistance (7-9). In addition, the inability to predict accurately the efficacy of these molecularly-targeted agents for an individual patient remains problematic. Advances in gene expression profiling have begun to dissect the problems outlined above by using a signature-based therapeutic approach for drug discovery and also for predicting chemosensitivity profiles for individual patients (10-14).
We have investigated the utility of combination histone deacetylase (HDAC) and proteasome inhibitors as a molecularly-targeted treatment strategy in NSCLC (8, 15, 16). Isolated HDAC therapy has little effect on NSCLC cell viability, in large part secondary to activation of the anti-apoptotic transcription factor NF-κB through Akt-mediated enhancement of p300 acetyltransferase activity that promotes acetylation of RelA/p65, the transcriptionally active subunit of NF-κB (16). However, when HDAC inhibitors are combined with a proteasome inhibitor, there was a robust, dose-dependent increase in NSCLC cell apoptosis (8). While this increased NSCLC cell death was encouraging, it was apparent there were varying degrees of combination drug sensitivity in selected NSCLC cell lines, regardless of tumor p53, K-Ras, or p16 mutational profiles. Thus, while in vitro and in vivo studies (8, 15, 16) suggest combined HDAC and proteasome inhibition has promise in the treatment of NSCLC, selection of which patient could benefit from such therapy is uncertain. Furthermore, traditional “bench to market” methodologies (i.e. Phase I-III clinical trials) used to assess drug therapy efficacy are lengthy (17), costly (18, 19), and often fail to yield “positive” results (20); these very real concerns all demand a different approach to the problem of which patient will benefit from which drug(s).
In this report, we use the refined version of the “coexpression extrapolation (COXEN)” algorithm (21, 22) applied to 40 NSCLC cell lines to identify tumor biomarkers that predict drug sensitivity to Vorinostat (Supp. Fig. 1A, Merck, Inc., Whitehouse Station, NJ), a HDAC inhibitor, and Velcade (Supp. Fig. 1B, Millennium Pharmaceuticals, Cambridge MA), a proteasome inhibitor. We demonstrate the high prediction capability of this refined COXEN methodology through which we develop a formula that predicts the probable efficacy of combined Vorinostat and Velcade therapy in NSCLC. Based on this, we provide an in silico method through which in vitro assessment of compounds such as Vorinostat and Velcade, in isolation or in combination, can be used to generate biomarkers that are highly predictive of tumor sensitivity.
Materials and Methods
Cell culture, cell lines, reagents, and RNA isolation
Forty human NSCLC cell lines (UVA-40, Supp. Table 1) were obtained from both American Type Culture Collection (ATCC, Manassas, VA) and the lab of John D. Minna, MD (University of Texas Southwestern Medical Center, Dallas, TX) from October 2008 until April 2009, and were grown as previously described (23). All cell lines were used within 6 months of receipt and have been DNA fingerprinted for provenance using the Promega GenePrint® PowerPlex 1.2 kit (Madison, WI) and confirmed to be the same as the DNA fingerprint library maintained by either ATCC or the Minna/Gazdar lab. Vorinostat was purchased from Sigma-Aldrich (St. Louis, MO). Velcade was provided through a materials transfer agreement with Millennium Pharmaceuticals (Cambridge, MA). Total RNA isolation from these cell lines was performed as previously described (23).
Drug activity assay for cell-line growth inhibition
Human NSCLC cell lines were plated in 96-well culture plates (Costar, Corning, NY) at a density of 1,000 cells per well in 50 μL of complete media as previously described (23) with experimental and control plates, incubated at 37°C. After 6 hours of incubation, the control plates were treated with Alamar Blue (Invitrogen Corporation, Carlsbad, CA) to assess a baseline value for each cell line. After 24 hours of incubation, the experimental plates were treated with Vorinostat, Velcade, or a combination of the two drugs. Each drug dose was plated in eight repeats. The doses for Vorinostat were (in μM): 0, 0.1, 0.5, 1, 5, 10, and 20. The doses for Velcade were (in nM): 0, 5, 10, 25, 50, 100, and 200. The doses for combination therapy were Vorinostat (μM)/Velcade (nM) combination were: 0/0, 0.1/5, 0.5/10, 1/25, 5/50, 10/100, and 20/200. The experimental plates were then incubated for 72 hours (at 37°C) and subsequently treated with Alamar Blue to assess growth inhibition. Drug doses were determined base on previous growth inhibition assays (8).
Microarray expression analysis
The gene profile data of 40 NSCLC cell lines were collected using the Affymetrix HG-U133A platform and are available at Gene Expression Omnibus (GEO) http://www.ncbi.nlm.nih.gov/geo/. Four of these 40 NSCLC cell lines (NCI-H125, NCI-H226, NCI-H292, and NCI-H596) were profiled using Affymetrix HG-U133A GeneChips® (Affymetrix, Santa Clara, CA, USA) at the University of Virginia. Samples of RNA were first assessed for quality using the Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA, USA), performing electrophoretic separations that allowed the inspection for two ribosomal peaks, suggesting no degradation had occurred. Samples were then prepared for analysis using the protocol outlined in the Affymetrix GeneChip® Expression Analysis Technical Manual (http://www.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf).
Statistical analysis
Estimation of GI50 Values
A nonparametric spline regression technique with the constraint that each drug's higher dose concentration provides at least equal or higher drug efficacy (inhibition) than its lower concentration was applied for estimating the drug activities across each drug's experimental range of dose concentration. The smoothness parameter of spline was tuned objectively using the generalized cross validation (GCV) method (24). The software for generalized additive models in mgcv package (25) for R was used for the dose-effect curve estimation, and a combination of golden section search and successive parabolic interpolation for one dimensional optimization (26), implemented with the nlminb routine of R, was utilized to obtain the final GI50 estimates by inverting the dose-effect curves.
Refined COXEN algorithm
The COXEN algorithm (21) is an in silico method to develop molecular-based prediction models by identifying and using the biomarkers that are concordantly expressed between two independent cancer systems or populations (i.e. the NCI-9 and UVA-40 sets in this study). In brief, COXEN is composed of six distinct components or steps: obtaining relevant drug activity data on the training set (step 1), molecular expression data both on the training and test sets (step 2 & 3), initial drug sensitivity biomarker discovery on the training set (step 4), sub-selection of COXEN biomarkers (step 5), and multivariate prediction modeling with these COXEN biomarkers on the training set (step 6). Note that drug activity data on the test set are not needed in these steps and are only prospectively compared with the molecular-based COXEN prediction scores.
In this study, we developed and applied a refined version of COXEN algorithm. Specifically, principal component regression in lieu of linear discriminate analysis was employed for multivariate prediction modeling (step 6). As such, the drug activity data is measured on a continuous scale and no longer divided into either sensitive or resistant groups at the expense of potential information loss. The resultant COXEN scores are predicted GI50 values with a continuous spectrum. Applying the refined COXEN algorithm (Fig 1.), we obtained predictive COXEN biomarkers on a continuous spectrum of drug activity for both individual and combination therapy using Vorinostat and Velcade (Supp. Table 2).
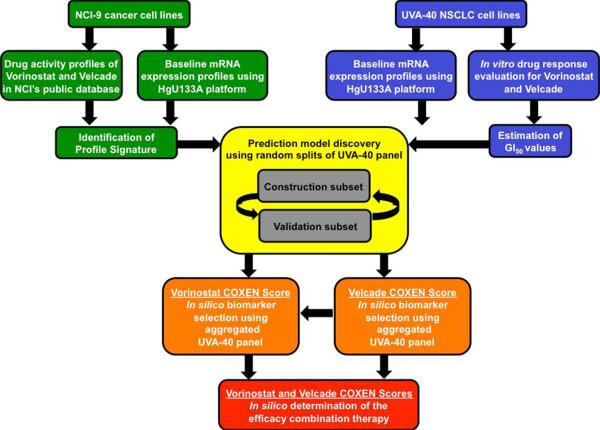
Figure 1. Schematic plot of the COXEN algorithm and associated data sets.
The COXEN algorithm consists of three main steps: identification of profile signature, construction of prediction model, and validation of prediction model. Identification of the profile signature entails using the 9 NSCLC cell line subset (NCI-9) of the NCI-60 cancer cell line panel and training them for Vorinostat and Velcade using Affymetrix HG-U133A GeneChips® and estimated GI50 values. The UVA-40 panel of NSCLC cell lines was split into Construction and Validation subsets. Generated COXEN scores were used for biomarker selection. The resultant COXEN scores from the single drug prediction modeling for Vorinostat and Velcade served as the input data to create a prediction model for combination Vorinostat and Velcade therapy.
COXEN biomarker selection and prediction modeling for single drug activity
The GI50 values and gene expression profiles of the NCI-9 NSCLC cell lines (NCI-9) among the National Cancer Institute's public database of 60 cancer cell lines (NCI-60) were utilized to rank genes according to their association with the drug activities of each compound. Our application of the refined COXEN algorithm was first rigorously evaluated using a random cross-validation strictly dividing modeling, training, and test subsets. In brief, the UVA-40 panel of NSCLC cell lines was randomly assigned into two independent subsets. The subset used to filter biomarkers and to construct a prediction model was comprised of 19 NSCLC cell lines treated with Vorinostat or Velcade (Construction Subset). The other subset of the UVA-40 panel strictly reserved for validation of the prediction model was comprised of 18 NSCLC cell lines treated with Vorinostat and 19 NSCLC cell lines treated with Velcade (Validation Subset). Note that three NSCLC cell lines treated with Vorinostat and two NSCLC cell lines treated with Velcade were excluded from our prediction modeling due to high experimental variations of drug activities (Supp. Fig. 2). For a robust statistical inference, we performed this random split 100 times.
With each split, to maintain concordant expression patterns between two independent systems, i.e., the NCI-9 and UVA-40 sets, we triaged the top-ranked 200 biomarkers by filtering the ones that showed inconsistent expression patterns between the NCI-9 and the Construction Subset. After the mild filtration, the top-ranked biomarkers served as input variables for constructing multivariate prediction models on the NCI-9. Adding one additional input variable at a time, the candidate prediction models were constructed and the performance of each model was assessed by a rank-based association between its prediction scores and the experimentally measured GI50 values of the Construction Subset along with its 95% bootstrap confidence interval (Supp. Fig. 3). Note that the sequentially enlarged gene space in high dimension was reduced to a few of major orthogonal directions of the input space with most variations. Among the candidate prediction models, the COXEN prediction model was chosen to minimize p-values of the association tests with the narrowest confidence interval, and was independently evaluated by the Validation Subset.
Upon the statistical validation of the refined COXEN algorithm, we obtained the final predictive COXEN biomarkers for Vorinostat or Velcade (Supp. Table 2) on the UVA-40 panel. The performance of the NCI-9 trained COXEN prediction model was then assessed using a rank-based association between its prediction scores and the experimentally measured GI50 values of the UVA-40.
Multvariate prediction modeling for combination drug therapy
The resultant COXEN scores of the single drug prediction models for Vorinostat (Vor) and Velcade (Vel) were utilized for building a prediction model for combination therapy (Comb). A multiple regression form of prediction models with or without interaction terms was considered. Each candidate prediction model for combination therapy was tested using 1000 runs of a “learning-test split,” where for each run half of the COXEN scores, from the single drug prediction modeling, for Vorinostat and Velcade were jointly sampled with corresponding observed GI50 values of combination therapy. This subset analysis was used to fit a prediction model having an identical functional form of the final prediction model. Using the other half of the COXEN scores, the performance of the prediction model was measured by an estimate of the rank-based Spearman's correlation coefficient with its respective p-value.
Results
Biomarker validation for single drug activity in NSCLC cells
Initial application of the COXEN algorithm to the Construction Subsets of the UVA-40 panel resulted in COXEN prediction models that selected a 100 gene model for Vorinostat (p<0.05, Supp. Fig. 3A) and a 45 gene model for Velcade (p<0.05, Supp. Fig. 3B). These models were highly significant, and corresponding 95% bootstrap confidence intervals were simultaneously minimized. COXEN scores were obtained from the constructed prediction model and applied to the Validation Subset of the UVA-40 panel. This subset was strictly reserved for independent prediction where observed GI50 values were plotted against predicted GI50 values for both Vorinostat and Velcade. The resultant plots demonstrated highly significant statistical models with rank-based Spearman's correlations of 0.46 (p=0.0247) for Vorinostat and 0.53 (p=0.0093) for Velcade.
Despite these encouraging observations, it is possible that the performance of the developed COXEN prediction model may depend on the specific random split of the UVA-40 panel. To test the robustness of this schema, 100 random splits were performed for both Vorinostat (Supp. Fig. 4A) and Velcade (Supp. Fig. 4B). The resulting prediction models attained statistical significance (p-value ≤ 0.1) for both Vorinostat (84%) and Velcade (73%). Therefore, the randomization of the UVA-40 panel cell lines to either the Construction or Validation Subset does not play a significant role in the resultant prediction model
With such statistically significant models, we are confident that the COXEN algorithm is capable of developing a prediction model that can be used to predict single drug sensitivity to Vorinostat and Velcade.
COXEN multi-gene predictors are predictive of tumor sensitivity to Vorinostat and Velcade in NSCLC cells
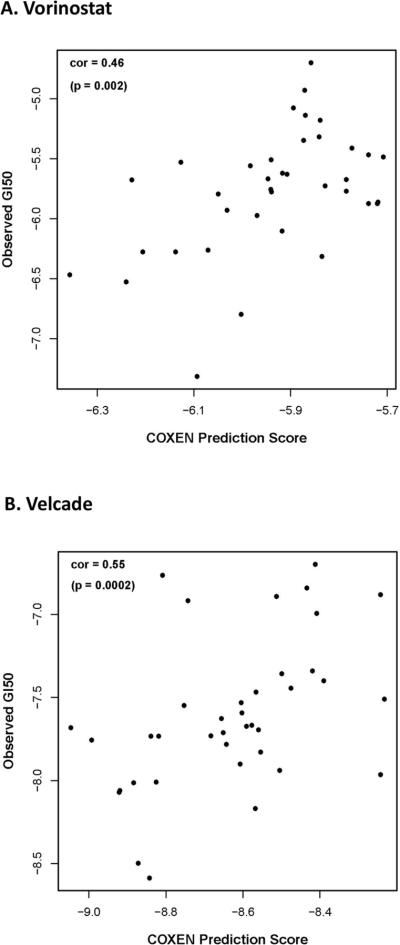
With validation of our COXEN approach to prediction modeling, the multi-gene expression signatures for both Vorinostat and Velcade were developed on the NCI-9 and UVA-40 NSCLC cell lines, resulting in a 45-gene model for Vorinostat (Supp. Fig. 5A) and a 15-gene model for Velcade (Supp. Fig. 5B). Plotting of observed versus evaluated GI50 values for both Vorinostat (Fig. 2A) and Velcade (Fig. 2B) on UVA-40 NSCLC cell lines demonstrated rank-based Spearman's correlations of 0.46 (p=0.002) for Vorinostat and 0.55 (p=0.0002) for Velcade.
Figure 2. Evaluation of COXEN models for UVA-40 NSCLC cell lines.
A) Scatter plot of COXEN prediction scores versus experimentally measured GI50 values for Vorinostat resulted in a rank-based Spearman's correlation of 0.46 (p=0.002). B) Scatter plot of COXEN prediction scores versus experimentally measured GI50 values for Velcade resulted in a rank-based Spearman's correlation of 0.55 (p=0.0002).
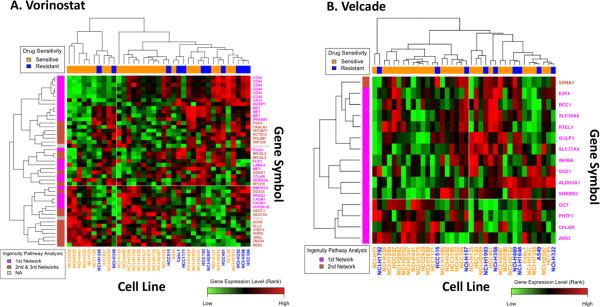
To examine the overall expression patterns of the COXEN biomarkers for Vorinostat (Supp. Table 2A) and Velcade (Supp. Table 2B), we performed a clustering analysis on the UVA-40 (Fig. 3). As shown in Figure 3A for Vorinostat and 3B for Velcade, the UVA-40 cell lines were largely separated based on their drug sensitivity in this unsupervised clustering analysis. In order to demonstrate each gene's relative expression pattern between sensitive and resistant cell lines, the clustering heatmaps were refined from their original output, where there existed a small number of large intensity values which dominated the heatmap landscape, based on their relative ranks of expression intensities. Though simply compared, these clustering heatmaps could show predictive potential of these biomarkers.
Figure 3. Heatmaps of biomarkers highly associated with drug sensitivity to Vorinostat and Velcade.
A) Clustered image map with two-way unsupervised clustering of expression profile data of the 45 most highly ranked genes for Vorinostat using 37 NSCLC cell lines from the UVA-40 panel of NSCLC cell lines. B) Clustered image map with two-way unsupervised clustering of expression profile data of the 15 most highly ranked genes for Velcade using 38 NSCLC cell lines from the UVA-40 panel of NSCLC cell lines.
Red, black, and green indicate high, intermediate, and low expression level, respectively. Orange and blue in the upper bar indicate sensitive and resistant cell types based on mean GI50 values, respectively. Pink and burnt orange in the left bar indicate gene networks. (Top biological functions for each network are summarized in Table 1.)
To determine which, if any, specific biomarkers were essential to the highly predictive nature of the models, each set of biomarkers was tested using random samplings. In testing the robustness of the gene signatures, 1000 random samplings of two-thirds of the genes from each drug compound (Vorinostat – 30 genes and Velcade – 10 genes) demonstrated an overall consistent predictability of the models (p<0.0005, Supp. Fig. 6). This implies that, within the respective gene models, there were no biomarkers that critically affected the performance of the prediction models.
Examination of specific tumor biomarkers for Vorinostat and Velcade
When evaluating the resultant 45 tumor biomarker signature for Vorinostat (Fig. 3A), we find that 4 genes appear multiple times: met proto-oncogene (MET), CD44 molecule (CD44), microtubule associated monoxygenase, calponin and LIM domain containing 2 (MICAL2), and calmodulin 1 (CALM1). The multiple appearances of these genes are a function of how the COXEN algorithm probes for significant biomarkers. In using different probes to scan the genome, genes that span large segments of the genome may be picked up by multiple probes, thereby appearing multiple times. Importantly, the presence of a gene multiple times (i.e. CD44) does not affect the prediction performance of our model (Supp. Fig. 7), since their contributions were mathematically optimized when multiple probes were included in our multivariate model. Thus, our prediction models were quite robust. Additionally, the gene model output renders one biomarker as having only a gene accession number (AK027225), indicating that this is a novel gene yet to be identified. As such, evaluation of the tumor biomarker set for Vorinostat reveals 34 distinct genes that determine the sensitivity of the tumor to single drug therapy with Vorinostat.
Evaluating the resultant 15 tumor biomarker signature for Velcade (Fig. 3B), we find that no genes appear multiple times and all genes are identifiable. Therefore, the COXEN algorithm indicates 15 distinct genes as being critical in determining the sensitivity to single drug therapy with Velcade.
Examination of the gene ontogeny revealed several gene networks involved in tumor cell replication and death: cellular assembly/organization, cell cycle, and RNA damage/repair gene networks for Vorinostat and cellular development and carcinogenic gene networks for Velcade (Table 1). A review of the selected biomarkers for both drugs revealed no significant overlap with only one shared biomarker, CASP8 and FADD-like apoptosis regulator (CFLAR), for both Vorinostat and Velcade. Though this suggests independent mechanisms of action for the two drugs, CFLAR is a regulator of apoptosis found in both gene signatures. This finding correlates with in vitro studies on NSCLC performed by our group (8, 27, 28) and also by Grant and colleagues on multiple myeloma cells (29, 30), which demonstrated that treatment with combined Vorinostat and Velcade resulted in caspase-mediated apoptosis.
Table 1.
Biologic function of tumor biomarkers selected by modified COXEN algorithm.
| A. Vorinostat | ||
|---|---|---|
| Network ID | Number of Genes | Top Biological Functions |
| 1 | 14 | Tissue Development, Cellular Assembly and Organization, Cellular Function and Maintenance |
| 2 | 12 | Cell Cycle, Cellular Growth and Proliferation, Endocrine System Development and Function |
| 3 | 7 | RNA Damage and Repair, Nucleic Acid Metabolism, DNA Replication, Recombination, and Repair |
| B. Velcade | ||
|---|---|---|
| Network ID | Number of Genes | Top Biological Functions |
| 1 | 12 | Cellular Development, Organ Morphology, Reproductive |
| System Development and Function | ||
| 2 | 1 | Cancer, Neurological Disease, Infection Mechanism |
Multivariate regression modeling of COXEN scores predicts the activity of doublet therapy in NSCLC cell lines
We hypothesized that the efficacy of doublet Vorinostat and Velcade therapy efficacy could be predicted using our refined COXEN algorithm (Fig. 1). Though single drug modeling indicates only one shared tumor biomarker between Vorinostat and Velcade, prior studies on NSCLC (8), multiple myeloma (29, 30), hematologic T-cell leukemia/lymphoma (31), and cutaneous T-cell lymphoma cells (32) confirm that combination therapy results in decreased cell survival compared to single drug therapy, suggesting that some degree of enhanced activity exists when both drugs are used.
In searching for multivariate prediction models for the combination therapy, models with and without a drug interaction term were considered for Vorinostat and Velcade. To examine what type of function (i.e., linear, parabolic, etc.) that would best represent the combined effect of Vorinostat and Velcade therapy, we plotted the GI50 values of combination therapy on the UVA-40 panel against the predicted COXEN scores (GI50 values) for Vorinostat and Velcade (Supp. Fig. 8), which implied a high linear association with single drug effects for Velcade.
Furthermore, with a significant interaction term in the regression modeling, the final fitted multivariate regression model (R2 = 0.9986) included both single drug Vorinostat (Vor) and Velcade (Vel, p=0.06) terms and an interaction term (VorVel, p<0.001). Below is the final regression model for predicting the activity of doublet Vorinostat and Velcade therapy for tumors where gene signatures have been identified:
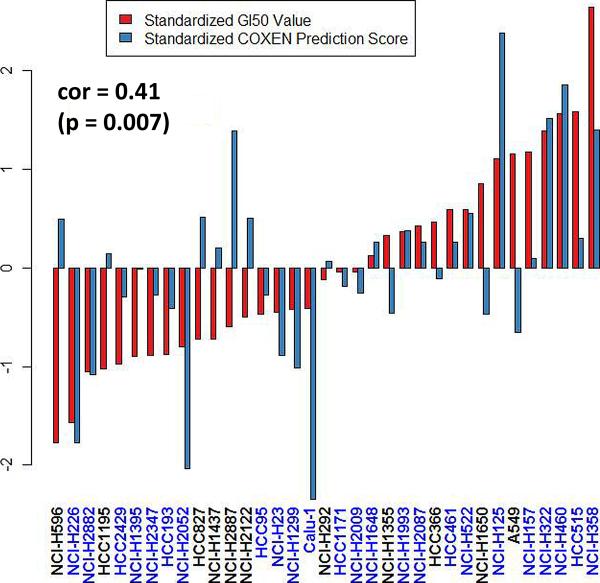
Using the fitted model, observed and predicted GI50 values for combination therapy were significantly correlated (p=0.007, correlation coefficient=0.41, Fig. 4). The underlying assumptions (i.e. constant variance and Gaussian distributional specification) of the fitted model were verified using studentized residual and normal quantile-quantile plots.
Figure 4. Evaluation of a prediction model for combination therapy.
Bar plot of direct comparison between COXEN prediction scores and experimentally measured GI50 values for combination therapy. The cell lines are ordered based on GI50 values. COXEN scores and GI50 values were standardized by subtracting the overall mean and dividing by the standard error across the UVA-40 panel. The statistical significance was assessed by a rank-based Spearman's correlation test (p=0.007, correlation coefficient=0.41). The cell lines were indicated by blue as the signs of COXEN scores and GI50 values were matched.
Having created a prediction model for combination therapy, we further statistically validated the performance of the model. Validation of the combination model involved 1000 runs of “learning-test splits,” where 46.1% of the predictions attained statistical significance (p-value ≤ 0.1, Supp. Fig. 9).
Discussion
One of the primary obstacles to the successful treatment of NSCLC has been the dismal performance of platinum-based doublet chemotherapeutic regimens (2, 33). More recently, strategies to overcome this impediment have involved the utilization of genomic signatures to direct use of primary chemotherapy (12). Multiple studies have focused on in vitro modeling using the NCI-60 panel of cancer cell lines coupled with baseline gene expression profiling to develop signatures to predict sensitivity to various chemotherapeutic regimens (22, 34). The methodology commonly used is binary in that cell lines are classified as either either sensitive or resistant to a given agent. Based on that assessment, the respective gene expression data is then used to generate a biomarker profile for drug sensitivity (22, 35). The inherent problem with this approach is that drug sensitivity is really a continuous spectrum, and as such, should not be considered a dichotomous variable. Despite the extensive study of genomic signatures, no other studies prior to this report have developed profiles based on a more natural continuous spectrum of drug activity on novel molecularly-targeted therapies.
In this study, we introduce the application of a refined COXEN algorithm to ascertain the efficacy of two novel molecularly-targeted agents, Vorinostat, a HDAC inhibitor, and Velcade, a proteasome inhibitor, in the treatment of NSCLC. The rationale for using these novel agents stems from preliminary data that has shown in vitro utility of combination histone deacetylase and proteasome inhibitors as a molecularly-targeted treatment strategy in NSCLC (8, 15, 16), multiple myeloma (29, 30), hematologic T-cell leukemia/lymphoma (31), and cutaneous T-cell lymphoma cells (32). Use of Vorinostat is further justified by preclinical and clinical studies that support its use in combination with other cancer therapies for the treatment of NSCLC (36, 37). Finally, it is known that a putative target for HDAC inhibitors lies in the cellular mechanisms for handling misfolded proteins that are degraded by the proteasome (38).
We have previously used the COXEN algorithm to extrapolate in vitro drug sensitivity results to well-established, conventional chemotherapeutic compounds to predict tumor behavior in patients with bladder, breast, and ovarian cancers, with results being validated against independent clinical trial data (21, 39). Previous application of the COXEN algorithm involved developing gene expression profiles based on the inherent sensitivity or resistance of a cell line to a chemical compound (21, 22, 39). Though the COXEN method has been extensively validated, we propose a refined version of the COXEN algorithm (Fig. 1), whereby the gene expression profile is developed based on a continuous spectrum of drug activity. This refined COXEN algorithm eliminates arbitrarily assigned sensitivity/resistance valuations and permits the study of novel molecularly-targeted compounds, where response levels for the drug have not been clinically established. With the aforementioned reports having studied tumor behavior in bladder, breast, and ovarian cancers, we are introducing the first application of COXEN using novel molecularly-targeted agents, Vorinostat and Velcade, in NSCLC. In using the refined COXEN, we were able to identify tumor biomarkers that highly predict single drug sensitivity to Vorinostat and Velcade (Fig. 3, Supp. Table 2).
Another feature of our refined COXEN algorithm is an extension of the methodology to produce a functional form derived from our single drug COXEN output scores for combined Vorinostat and Velcade (Fig. 2), which permits the prediction and internal validation of the probable efficacy of combination therapy for these novel agents on individual tumors. Using this methodology, we developed a model (Fig. 4) that highly predicts the probable efficacy of doublet therapy with Vorinostat and Velcade (Supp. Fig. 8). With the current reported correlation levels of the predicted scores, we can achieve both a >75% positive predictive value and negative predictive value for the top 20% most sensitive and 20% most resistant cell lines. That is, with this degree of correlation, a patient guided by our COXEN prediction will have a >75% certainty either for having a clinical benefit or for avoiding unnecessary toxicity. Given that current results of platinum-based doublet therapies for advanced stage NSCLC yield 5-year survival rates of less than 10% (4-6), the ability to use molecular predictors to guide combined chemotherapeutic therapy, particularly with novel agents, could result in improved survival rates.
While results from previous applications of the COXEN algorithm were validated against independent clinical trial data (22, 39), this methodology will be nearly impossible to replicate as efforts are made to introduce new, more “personalized” drug therapies clinically. With the costs of clinical trials for novel agents steadily rising (40) and the significant number of phase III trials yielding “negative” results after years of patient accrual (41, 42), the current clinical trial methodology is rapidly becoming obsolete. The refined COXEN algorithm provides a framework whereby an in silico method can be applied to screen novel molecularly-targeted therapies in vitro in isolation or in combination to generate biomarkers that predict tumor sensitivity. The resulting biomarkers can be used to prospectively evaluate which tumor types would be responsive to the tested agent. This knowledge would then be used to stratify potential responders into a clinical trial and exclude those non-responders, thereby effectively streamlining clinical trial design and cost (17). Another potential benefit to a priori knowledge of biomarkers that predict tumor sensitivity is the ability for drug salvage and/or repositioning strategy in another tumor system in the case of an ongoing clinical trial that fails to yield a “positive” result. These secondary clinical trials would be particularly efficient as the pharmacology and toxicity of the compound(s) would have been already well documented (13).
Potential limitations of our analysis include the use of an in-vitro analysis of tumorigenic cell lines to determine biomarkers that predict sensitivity to anti-cancer agents. Cancer cell lines derived from human tissue can exhibit heterogeneity (i.e., stemness) (43-45), have differential doubling times, and grow in non-physiologic conditions – all of which that can affect how these cells respond to anti-cancer agents (46). To mitigate these concerns as much as possible, we performed our experiments using a 72-hour assay that has been shown by previous studies (22, 47) to be an effective strategy to minimize these issues. An additional concern is that in vitro gene expression profiling of cancer cell lines may not reflect the genomic status of the primary tumor. A recent study performed by Sos and colleagues, genomically validated 84 NSCLC cell lines, 37 of which make up the UVA-40, where they demonstrated through comparative analysis of orthogonal genomic data sets of these cell lines and primary tumors, that NSCLC cell lines reflect the genetic and transcriptional landscape of primary NSCLC specimens (48). The validation of both the assay methodology and the genetic composition of the UVA-40 NSCLC cells strengthen the experimental foundation of the refined COXEN algorithm as described in our study. Notably, our study focuses on the drug sensitivity prediction of lung cancer cell lines to Vorinostat and Velcade. We believe that this strategy may be applicable to other tumor types and/or novel agents, but we by no means claim that our strategy would work for a wide range of applications in cancer therapeutics.
In conclusion, through the application of a refined COXEN algorithm employing a continuous spectrum of drug activity on 40 NSCLC cell lines, we are the first to identify tumor biomarkers that highly predict drug sensitivity to Vorinostat, a HDAC inhibitor, and Velcade, a proteasome inhibitor. Additionally, by extension of the refined COXEN algorithm to combination therapy, we demonstrate an ability to predict the probable efficacy of this doublet therapy on NSCLC. In silico COXEN models such as these may significantly enhance our ability to predict a priori the efficacy of novel targeted therapeutics such as Vorinostat and Velcade for NSCLC patients and offer important additional data for subsequent clinical trial designs.
Supplementary Material
Acknowledgments
This study was aided by the gracious contribution of NSCLC cell lines from the lab of John D. Minna, MD (University of Texas Southwestern Medical Center, Dallas, TX).
Financial Support: NIH grants R01 CA136705 (to DRJ), R01 HL081690 (to JKL), and the Thoracic Surgery Foundation for Research and Education (TSFRE) Research Fellowship (to ASN).
Footnotes
Conflicts of Interest: David R. Jones is the principal investigator for a Phase I clinical trial involving Vorinostat and Velcade at the University of Virginia (UVA) in patients who have surgically resectable NSCLC. Jae K. Lee is the co-founder of Key Genomics, Inc.
References
- 1.Azzoli CG, Baker S, Jr., Temin S, et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–66. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Tribodet H, Aubert D, et al. Adjuvant cisplatin and vinorelbine for completely resected non-small cell lung cancer: subgroup analysis of the Lung Adjuvant Cisplatin Evaluation. J Thorac Oncol. 5:220–8. doi: 10.1097/JTO.0b013e3181c814e7. [DOI] [PubMed] [Google Scholar]
- 4.Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 2006;7:719–27. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 5.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–91. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 6.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–97. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 7.Bunn PA., Jr. The potential role of proteasome inhibitors in the treatment of lung cancer. Clin Cancer Res. 2004;10:4263s–5s. doi: 10.1158/1078-0432.CCR-040011. [DOI] [PubMed] [Google Scholar]
- 8.Denlinger CE, Keller MD, Mayo MW, Broad RM, Jones DR. Combined proteasome and histone deacetylase inhibition in non-small cell lung cancer. J Thorac Cardiovasc Surg. 2004;127:1078–86. doi: 10.1016/s0022-5223(03)01321-7. [DOI] [PubMed] [Google Scholar]
- 9.Mack PC, Davies AM, Lara PN, Gumerlock PH, Gandara DR. Integration of the proteasome inhibitor PS-341 (Velcade) into the therapeutic approach to lung cancer. Lung Cancer. 2003;41(Suppl 1):S89–96. doi: 10.1016/s0169-5002(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 10.Anguiano A, Nevins JR, Potti A. Toward the individualization of lung cancer therapy. Cancer. 2008;113:1760–7. doi: 10.1002/cncr.23644. [DOI] [PubMed] [Google Scholar]
- 11.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–80. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 12.Potti A, Nevins JR. Utilization of genomic signatures to direct use of primary chemotherapy. Curr Opin Genet Dev. 2008;18:62–7. doi: 10.1016/j.gde.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Smith SC, Baras AS, Lee JK, Theodorescu D. The COXEN Principle: Translating Signatures of In vitro Chemosensitivity into Tools for Clinical Outcome Prediction and Drug Discovery in Cancer. Cancer Res. doi: 10.1158/0008-5472.CAN-09-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9:253–64. doi: 10.1038/nrc2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denlinger CE, Rundall BK, Jones DR. Proteasome inhibition sensitizes non-small cell lung cancer to histone deacetylase inhibitor-induced apoptosis through the generation of reactive oxygen species. J Thorac Cardiovasc Surg. 2004;128:740–8. doi: 10.1016/j.jtcvs.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Denlinger CE, Rundall BK, Smith PW, Jones DR. Suberoylanilide hydroxamic acid induces Akt-mediated phosphorylation of p300, which promotes acetylation and transcriptional activation of RelA/p65. J Biol Chem. 2006;281:31359–68. doi: 10.1074/jbc.M604478200. [DOI] [PubMed] [Google Scholar]
- 17.Ohashi W, Mizushima H, Tanaka H. Economic advantage of pharmacogenomics - clinical trials with genetic information. Stud Health Technol Inform. 2008;136:585–90. [PubMed] [Google Scholar]
- 18.Schmidt C. Costly cancer drugs trigger proposals to modify clinical trial design. J Natl Cancer Inst. 2009;101:1662–4. doi: 10.1093/jnci/djp460. [DOI] [PubMed] [Google Scholar]
- 19.Collier R. Rapidly rising clinical trial costs worry researchers. CMAJ. 2009;180:277–8. doi: 10.1503/cmaj.082041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurence J. No more boring science, no more waste in clinical trials. Transl Res. 2009;153:1–3. doi: 10.1016/j.trsl.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Lee JK, Havaleshko DM, Cho H, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104:13086–91. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havaleshko DM, Cho H, Conaway M, et al. Prediction of drug combination chemosensitivity in human bladder cancer. Mol Cancer Ther. 2007;6:578–86. doi: 10.1158/1535-7163.MCT-06-0497. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol. 2006;26:8683–96. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastie T, Tibshirani R. Generalized Additive Models. Chapman & Hall; London: 1990. [DOI] [PubMed] [Google Scholar]
- 25.Wood S. Generalized additive models: an introduction with R. Boca Raton. Chapman & Hall/CRC; FL: 2006. [Google Scholar]
- 26.Brent R. Algorithms for Minimization without Derivatives. Prentice-Hall; Englewood Cliffs, NJ: 1973. [Google Scholar]
- 27.Jones DR, Broad RM, Comeau LD, Parsons SJ, Mayo MW. Inhibition of nuclear factor kappaB chemosensitizes non-small cell lung cancer through cytochrome c release and caspase activation. J Thorac Cardiovasc Surg. 2002;123:310–7. doi: 10.1067/mtc.2002.118684. [DOI] [PubMed] [Google Scholar]
- 28.Jones DR, Broad RM, Madrid LV, Baldwin AS, Jr., Mayo MW. Inhibition of NF-kappaB sensitizes non-small cell lung cancer cells to chemotherapy-induced apoptosis. Ann Thorac Surg. 2000;70:930–6. doi: 10.1016/s0003-4975(00)01635-0. discussion 6-7. [DOI] [PubMed] [Google Scholar]
- 29.Pei XY, Dai Y, Grant S. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor bortezomib and histone deacetylase inhibitors. Clin Cancer Res. 2004;10:3839–52. doi: 10.1158/1078-0432.CCR-03-0561. [DOI] [PubMed] [Google Scholar]
- 30.Yu C, Rahmani M, Conrad D, Subler M, Dent P, Grant S. The proteasome inhibitor bortezomib interacts synergistically with histone deacetylase inhibitors to induce apoptosis in Bcr/Abl+ cells sensitive and resistant to STI571. Blood. 2003;102:3765–74. doi: 10.1182/blood-2003-03-0737. [DOI] [PubMed] [Google Scholar]
- 31.Zhang QL, Wang L, Zhang YW, et al. The proteasome inhibitor bortezomib interacts synergistically with the histone deacetylase inhibitor suberoylanilide hydroxamic acid to induce T-leukemia/lymphoma cells apoptosis. Leukemia. 2009;23:1507–14. doi: 10.1038/leu.2009.41. [DOI] [PubMed] [Google Scholar]
- 32.Heider U, Rademacher J, Lamottke B, et al. Synergistic interaction of the histone deacetylase inhibitor SAHA with the proteasome inhibitor bortezomib in cutaneous T cell lymphoma. Eur J Haematol. 2009;82:440–9. doi: 10.1111/j.1600-0609.2009.01239.x. [DOI] [PubMed] [Google Scholar]
- 33.Ettinger DS. Is there a preferred combination chemotherapy regimen for metastastic non-small cell lung cancer? Oncologist. 2002;7:226–33. doi: 10.1634/theoncologist.7-3-226. [DOI] [PubMed] [Google Scholar]
- 34.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–92. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 36.Ramalingam SS, Maitland ML, Frankel P, et al. Carboplatin and Paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 28:56–62. doi: 10.1200/JCO.2009.24.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traynor AM, Dubey S, Eickhoff JC, et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol. 2009;4:522–6. doi: 10.1097/jto.0b013e3181952478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–68. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 39.Williams PD, Cheon S, Havaleshko DM, et al. Concordant gene expression signatures predict clinical outcomes of cancer patients undergoing systemic therapy. Cancer Res. 2009;69:8302–9. doi: 10.1158/0008-5472.CAN-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frantz S. Why are clinical costs so high? Nat Rev Drug Discov. 2003;2:851–2. [Google Scholar]
- 41.Saijo N, Nishio K, Tamura T. Translational and clinical studies of target-based cancer therapy. Int J Clin Oncol. 2003;8:187–92. doi: 10.1007/s10147-003-0324-x. [DOI] [PubMed] [Google Scholar]
- 42.Saijo N, Tamura T, Nishio K. Problems in the development of target-based drugs. Cancer Chemother Pharmacol. 2000;46(Suppl):S43–5. doi: 10.1007/pl00014049. [DOI] [PubMed] [Google Scholar]
- 43.Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10:336–42. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6:146–55. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 46.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 10:241–53. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 47.McDermott U, Sharma SV, Settleman J. High-throughput lung cancer cell line screening for genotype-correlated sensitivity to an EGFR kinase inhibitor. Methods Enzymol. 2008;438:331–41. doi: 10.1016/S0076-6879(07)38023-3. [DOI] [PubMed] [Google Scholar]
- 48.Sos ML, Michel K, Zander T, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–40. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.