SUMMARY
The ability of proteins to locate specific sites or structures among a vast excess of nonspecific DNA is a fundamental theme in biology. Yet the basic principles that govern these mechanisms remain poorly understood. For example, mismatch repair proteins must scan millions of base pairs to find rare biosynthetic errors, and they then must probe the surrounding region to identify the strand discrimination signals necessary to distinguish the parental and daughter strands. To determine how these proteins might function we used single-molecule optical microscopy to answer the following question: how does the mismatch repair complex Msh2-Msh6 interrogate undamaged DNA? Here we show that Msh2-Msh6 slides along DNA via one-dimensional diffusion. These findings indicate that interactions between Msh2-Msh6 and DNA are dominated by lateral movement of the protein along the helical axis and have implications for how MutS family members travel along DNA at different stages of the repair reaction.
INTRODUCTION
Postreplicative mismatch repair (MMR) corrects DNA synthesis errors before they lead to genomic instability (Hsieh, 2001; Kunkel and Erie, 2005; Modrich and Lahue, 1996). This repair pathway increases the fidelity of DNA replication by up to 1000-fold, and cells harboring mutations in MMR proteins suffer from an increased frequency of spontaneous mutation (Kunkel and Erie, 2005; Modrich and Lahue, 1996). In E. coli, MutS, MutL, and MutH promote strand-specific repair by taking advantage of the transiently unmethylated state of the newly synthesized strand. MutS binds mismatched DNA and together with MutL activates the MutH endonuclease. This leads to cleavage of the daughter strand at the nearest hemimethy-lated dGATC, which provides entry for other proteins that complete the downstream steps of the pathway (Kunkel and Erie, 2005; Lahue et al., 1989).
Many MMR proteins are conserved throughout evolution (Kunkel and Erie, 2005; Modrich, 2006). In S. cerevisiae and humans, the Msh2-Msh6 heterodimer (MutS homolog) is responsible for the recognition and repair of mispaired bases and small insertion/deletion loops (Kunkel and Erie, 2005; Modrich, 2006). The importance of Msh2-Msh6 is highlighted by the finding that mutations in MMR proteins lead to hereditary nonpolyposis colorectal cancer (HNPCC) and may influence the onset of up to 25% of other sporadic tumors (reviewed in Modrich, 2006).
MutS and Msh2-Msh6 resemble clamps that encircle DNA (Figure 1C; Gradia et al., 1999; Kunkel and Erie, 2005; Lamers et al., 2000; Obmolova et al., 2000; Warren et al., 2007). The bound DNA is kinked by ~45°–60°, and the relative ease with which a mismatch can be distorted may play an important role in target discrimination (Yang, 2006). After mismatch binding, identification of the strand discrimination signals can occur at sites ≥1 kb away from the lesion (Kunkel and Erie, 2005; Modrich, 2006). Several models have been proposed to explain how recognition occurs at the distal location, including: (1) “active translocation,” where the protein uses the free energy released from the hydrolysis of ATP to move along the DNA (Allen et al., 1997; Blackwell et al., 1998); (2) the “molecular switch” model in which ATP binding triggers a conformational change enabling the protein to passively slide along the DNA (Gradia et al., 1999; Mendillo et al., 2005); and (3) “static transactivation” where interactions are governed by a through-space collision between the stationary mismatch-bound protein and the distal site (Ban et al., 1999; Lamers et al., 2000; Obmolova et al., 2000). The mechanism by which this occurs remains controversial; nevertheless, accumulating evidence supports the notion that MutS-related proteins can form “sliding clamps” on DNA after recognition of a lesion and subsequent exchange of ADP for ATP (Gradia et al., 1999; Jiang et al., 2005; Mendillo et al., 2005; Pluciennik and Modrich, 2007).
Figure 1. Experimental Design.
(A) Biotinylated λ-DNA (48,502 bp) was tethered by both ends to solid anchor points on a microfluidic sample chamber surface otherwise coated with a lipid bilayer. Msh2-Msh6 was labeled with QDs and injected into the sample chamber.
(B) Panel shows images of a YOYO1-stained DNA (green) bound by Msh2-Msh6 (magenta). When the DNA is broken, both it and the bound proteins diffuse away from the sample chamber surface (Movie S1). Off-axis signals correspond to proteins that were not bound to the DNA.
(C) Atomic structure illustrating the clamp-like appearance of T. aquaticus MutS in the presence and absence of DNA (Obmolova et al., 2000).
In contrast to the postrecognition events, few studies have addressed how DNA is scanned before a lesion has been located; this inattention is largely due to the lack of experimental methods capable of directly probing detailed dynamic aspects of these mechanisms. As a consequence, many kinetic schemes have inferred that mismatch binding occurs through a random collision mechanism (Blackwell et al., 2001; Yang et al., 2005; Mazur et al., 2006; Jacobs-Palmer and Hingorani, 2007). The problem of lesion recognition is best illustrated by considering the small number of potential targets that Msh2-Msh6 must locate within the genome. The intrinsic error rate of replicative DNA polymerases yields approximately one mispair per 10 million bases, and S. cerevisiae, with a diploid genome of ~2.4 × 107 base pairs, will only incur approximately two mispaired bases with each replication cycle (Fortune et al., 2005; Kunkel, 2004).
Several models have been developed to describe how proteins might search for their targets (Berg et al., 1981; Halford and Marko, 2004; von Hippel and Berg, 1989). These include (1) random collision through three-dimensional (3D) space; (2) hopping, wherein the protein moves along DNA through a series of microscopic dissociation and rebinding events; (3) intersegment transfer, in which a protein containing multiple DNA-binding domains can move from one location to another via a looped intermediate with two simultaneously bound sites; (4) sliding, which posits that the protein can move along the DNA via 1D diffusion; and (5) active translocation, which is a possibility for proteins that have intrinsic nucleotide hydrolysis activity.
To determine which model(s) might contribute to the mechanisms of MMR, we sought to answer the following question: how does Msh2-Msh6 travel along undamaged duplex DNA? We used total internal reflection fluorescence microscopy (TIRFM) to visualize the behavior of Msh2-Msh6 as it interacted with individual molecules of DNA. These experiments revealed that Msh2-Msh6 could slide along the helical axis of DNA via 1D diffusion. This movement did not require mismatch recognition or ATP hydrolysis, and was driven by thermal energy. Our data were consistent with a model in which Msh2-Msh6 rotates in register with the phosphate backbone while maintaining constant contact with the DNA. We also show that Msh2-Msh6 can reversibly enter a nondiffusing, immobile state. Importantly, the immobile proteins rapidly convert back to a diffusive state in a reaction that is dependent upon nucleotide exchange. We propose a model in which Msh2-Msh6 could slide or be pulled along DNA behind a replication fork while scanning the newly synthesized strands for lesions.
RESULTS
Visualizing Msh2-Msh6 on Single Molecules of DNA
We have developed a TIRFM-based approach for probing the dynamics of protein-DNA interactions at the single-molecule level (Figure 1A; Graneli et al., 2006b). This method relies on λ-DNA (48,502 bp) tethered by both ends to anchor points on a surface otherwise coated with a lipid bilayer. Proteins bound to the DNA are suspended above the bilayer and contained within an inert microenvironment. The DNAs are maintained in an extended configuration and confined within the detection volume defined by the penetration depth of the evanescent field (Figure 1A; Graneli et al., 2006b).
HA-tagged Msh2-Msh6 from S. cerevisiae was labeled with antibody-conjugated quantum dots (QDs; see the Supplemental Data available with this article online). We confirmed that this labeling strategy did not interfere with the biochemical behaviors of Msh2-Msh6 by performing ATP hydrolysis and DNA-binding assays in the presence and absence of the QDs (Figure S1). These assays, and additional evidence presented below, demonstrated that the labeled proteins retained normal biochemical properties.
To probe the behavior of Msh2-Msh6 at the single-molecule level, a tethered DNA was located by staining with YOYO1 (0.5 nM). This low concentration of dye did not perturb the behavior of the protein in any of the bulk assays (data not shown). Msh2-Msh6 was then injected along with ADP and incubated for a brief period, and unbound proteins were rinsed from the flowcell. Using this approach, we detected colocalization of the QD-Msh2-Msh6 complex with the YOYO1-stained DNA (Figure 1B). Controls using QDs with either no Msh2-Msh6 or with Msh2-Msh6 that lacked the HA tag did not yield any DNA-bound complexes (data not shown). This confirmed that the colocalized fluorescent signals were due to specific interactions between Msh2-Msh6 and the DNA. We verified that the protein complexes were bound to DNA by breaking the YOYO1-stained molecules at a high photon flux (Figure 1B). When a break occurred, both the DNA and the proteins drifted out of the evanescent field (Figure 1B and Movie S1), confirming that the proteins were bound to the DNA and were not stuck to the bilayer.
Msh2-Msh6 Slides on DNA via 1D Diffusion
Movies were captured with an electron-multiplying CCD (EMCCD) to monitor the behavior of Msh2-Msh6. As shown in Figure 2, Msh2-Msh6 moved along the individual DNA molecules over distances that spanned several kilo-bases (Movie S2). These experiments were done in the presence of 1 mM ADP (Figure 2), so the observed motion was not dependent upon ATP hydrolysis. This was consistent with previous suggestions that Msh2-Msh6 may be in “search mode” while in the ADP-bound state (Gradia et al., 1999; Jacobs-Palmer and Hingorani, 2007). The same type of movement was observed in experiments performed with either 1 mM ATP or no added nucleotide (see below). Sliding was seen at NaCl concentrations ranging from 50 mM up to 250 mM, but at higher salt concentrations the proteins rapidly dissociated from the DNA (see below). There was no buffer flowing during the data collection, eliminating the possibility that the proteins were being pushed along the DNA. At the outset of the reactions almost all of the proteins (>90%) slide along the DNA, but over longer incubations (several minutes) ~50% of the bound proteins stopped moving (Figure 2 and see below). Immobile proteins occasionally resumed sliding (Figure 2E and Figure S2), suggesting entry into the non-diffusing state was reversible.
Figure 2. Visualizing Movement of Msh2-Msh6 on DNA.
(A)–(F) show kymograms spanning 200 s intervals highlighting the movement of Msh2-Msh6. These examples come from experiments performed in the presence of 1 mM ADP. Kymograms were generated by excising the area that encompassed a single DNA and plotting the resulting images as a function of time. Time (s) is indicated at the top of the panels, and distance (µm) is indicated at the right of each panel. The long axis of the DNA is vertically oriented in each picture, and the starting positions of the protein complexes (magenta) bound to the DNA (green dashed line) are depicted at the left-hand side of each panel. Immobile complexes are also shown to serve as stationary reference points. Time-dependent variations in signal intensity are due to the photophysical characteristics of the QDs (see Figure 3).
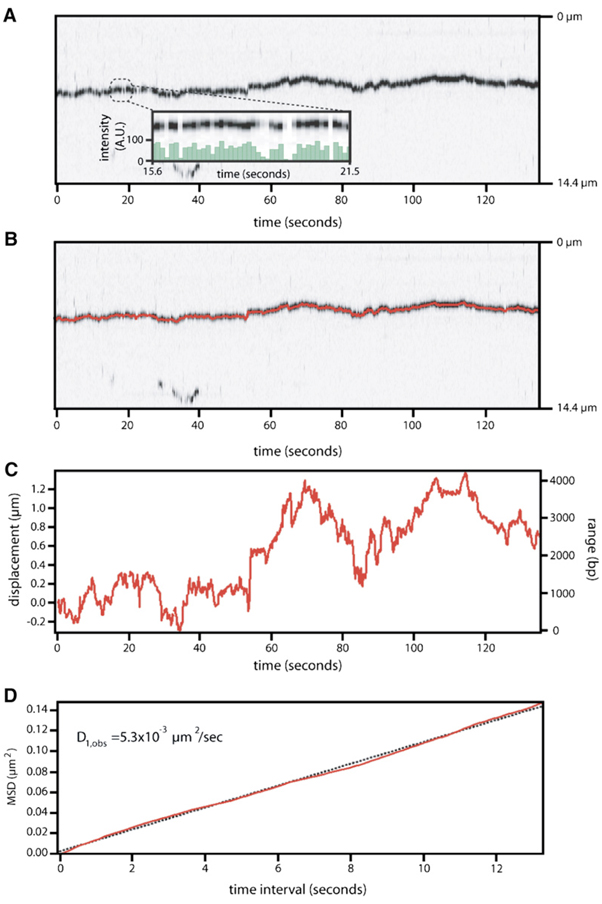
The movement of Msh2-Msh6 was analyzed with a tracking algorithm that located the centroid of each complex (Supplemental Data). Figure 3A shows a kymogram of a diffusing Msh2-Msh6 complex and the inset highlights the rapid blinking behavior of the QD, indicating that the signal is due to a single QD. Figure 3B shows the trace generated from the tracking algorithm superimposed on the kymogram of the mobile protein. The tracking data and a corresponding plot of mean-squared-displacement (MSD) versus time interval are shown in the lower panels (Figures 3C and 3D, respectively). For unbiased 1D diffusion, the MSD varies linearly with time interval, yielding a line whose slope can be used to calculate the diffusion coefficient (Blainey et al., 2006; Graneli et al., 2006b; Qian et al., 1991). Of the 125 complexes that were tracked (~7.5 hr of diffusion data), 58 displayed linear MSD plots. The remaining 67 deviated from linearity over longer time intervals, in an manner characteristic of bounded diffusion (Figure 4A; Movie S3; Supplemental Data; Qian et al., 1991). A histogram of the diffusion coefficients from all 125 of the MSD plots is shown in Figure 4B, revealing a mean of D1,obs = 0.012 ± 0.018 µm2/s, with a distribution of values ranging from 0.0002 µm2/s up to 0.09 µm2/s, and a median of 0.0058 µm2/s.
Figure 3. Msh2-Msh6 Moves on DNA via 1D Diffusion.
(A) Shows a kymogram illustrating the movement of a single QD-tagged Msh2-Msh6 complex over a 140 s duration. Time (s) and relative distance (µm) are indicated at the bottom and right, respectively. The inset shows the rapid blinking of the QD over a 5.9 s period; signal intensity is in arbitrary units (A.U.).
(B) Panel shows the same kymogram with data generated from the tracking algorithm superimposed on the image of the moving complex (also see Movie S2).
(C and D) Panels show details of the tracking and the resulting plot of MSD versus time interval, respectively. The displacement from the origin is indicated in mm (left vertical axis), and the total range spanned is indicated in bp (right vertical axis). The diffusion coefficient for this protein complex was calculated from the slope (dashed line) of the MSD plot.
Figure 4. General Characteristics of Msh2-Msh6 Diffusion.
(A) The graph in (A) shows representative MSD plots for four different Msh2-Msh6 complexes.
(B) A histogram of the diffusion coefficients calculated from the tracked proteins. This panel presents the cumulative information derived from 125 tracked complexes of Msh2-Msh6 in buffer containing 50 mM NaCl and either 1 mM ADP (N = 97; shown in red) or 1 mM ATP (N = 28; shown in blue).
(C) Panel shows a plot of the net displacement of Msh2-Msh6 from the origin after a 120 s period.
(D) Panel shows the range spanned by Msh2-Msh6 as it travels back and forth along the DNA over a 120 s period.
(E) A histogram of the apparent cumulative distance traversed by Msh2-Msh6 in 120 s.
(F) Panel is a histogram showing the average apparent velocities of the proteins calculated from the cumulative distance traveled divided by total time. The means and standard deviations for all plots were determined from Gaussian fits to the binned data. Movie S3 shows all 125 individual diffusion trajectories, the corresponding MSD plots, and the linear fits to each plot.
Based on hydrodynamic considerations, the theoretical maximum for the 1D diffusion coefficient for a complex approximated as a sphere with a 12.9 nm radius (Stokes radius of QD-Msh2-Msh6 complex; Figure S4) that slides on DNA without rotating around the helical axis is D1,calc = 18.3 µm2/s (Figure S5 and Supplemental Data); this was several orders of magnitude greater than the values observed for Msh2-Msh6. As previously reported (Blainey et al., 2006; Schurr, 1979), the theoretical maximum for the diffusion coefficient drops to D1,calc = 0.024 µm2/s if the protein must rotate to maintain constant register with the phosphate backbone (Figure S5). Thus the values measured for Msh2-Msh6 were most consistent with diffusive behavior that included a rotational component. This interpretation was in agreement with the available structures, which show that MutS and Msh2-Msh6 make numerous contacts with the phosphate backbone (Lamers et al., 2000; Obmolova et al., 2000; Warren et al., 2007).
To estimate the thermal barrier to Msh2-Msh6 diffusion, we assumed that the complex underwent a step size of 1 bp (although the calculated values are not dependent upon this assumption; Blainey et al., 2006) and rotated as it moved on the DNA (see Supplemental Data). The random walk stepping rate (klim) for a sphere with a radius of 12.9 nm is ~4.2 × 105 steps/s. From these values we calculated the mean activation barrier 〈ΔG‡〉 required for the protein to move from one position to the next using the observed stepping frequencies (kobs), which were determined from the distribution of diffusion coefficients (Figure S6). These calculations yielded an estimated activation energy of 1.57 ± 1.1 kBT required for Msh2-Msh6 to slide from one position to the next as it traveled along the DNA (Berg, 1993; Blainey et al., 2006). Importantly, because the estimated thermal barrier is not dependent upon the stepping rate, but rather the ratio of the stepping rates, the values are independent of the chosen step size as this parameter affects both klim and kobs equivalently, and is therefore factored out of the calculation (Supplemental Data; Blainey et al., 2006). Similar calculations made with the assumption that the protein did not rotate yielded a barrier of 8.3 ±1.3 kBT. Theoretical predictions suggest that the mean energetic barrier for efficient 1D diffusion cannot exceed ≈2 kBT (Blainey et al., 2006; Slutsky and Mirny, 2004). Therefore the observed diffusion coefficients are consistent with a model in which the protein rotates as it travels along the DNA and maintains constant register with the phosphate backbone. However, because our current experiments do not allow us to visualize the rotation itself we cannot completely rule out other alternative explanations for the observed diffusion coefficients. To verify or refute the rotation-coupled diffusion model, we are designing experiments that will enable us to directly visualize the orientation of the proteins relative to the DNA.
The net displacements Msh2-Msh6 relative to the starting positions were distributed symmetrically around the origin, consistent with a random walk (Figure 4C). Msh2-Msh6 traveled over an average range of 1.20 µm (3.2 kb) during a 120 s period (Figure 4D). The average apparent cumulative distance scanned by Msh2-Msh6 over the 2 min was 34.3 µm (101 kb; Figure 4E). This is the lower bound of the actual number of bases scanned by the protein because there are likely steps occurring below our current resolution limits. The apparent velocity of the diffusing complexes was calculated from the apparent cumulative distance traversed during the observation divided by the total time and yielded an average of 0.28 µm/s (820 bp/s; Figure 4F). These observations confirmed that the movement of Msh2-Msh6 was consistent with the physical predictions for 1D diffusion, and illustrated that thermal energy was sufficient to drive rapid movement of Msh2-Msh6 along the DNA.
Msh2-Msh6 Complexes Appear to Be Topologically Linked to DNA
Our results suggested that Msh2-Msh6 could slide while maintaining constant contact with the DNA and argued against a hopping mechanism. Hopping by definition means that the protein is entirely released from DNA, diffuses a short distance through space, and then rebinds the DNA at a nearby location (Berg et al., 1981; Halford and Marko, 2004). Hopping and sliding can be distinguished from one another by analyzing how the lifetimes and diffusion coefficients of the protein complexes vary at different concentrations of salt (Blainey et al., 2006; Berg et al., 1981). To verify that Msh2-Msh6 maintained continuous contact with the DNA, we determined the diffusion coefficients and lifetimes of the bound complexes at varying concentrations of NaCl. The lifetimes of the bound Msh2-Msh6 decreased at increasing concentrations of salt, but there were no significant changes in the mean diffusion coefficients (Figure S3). These observations were consistent with a model in which Msh2-Msh6 slid along DNA while maintaining constant contact with the phosphate backbone and argued against hopping as a potential mode of travel along the DNA (Blainey et al., 2006).
We also reasoned that if Msh2-Msh6 was capable of hops comparable in magnitude to the dimensions of its DNA-binding domains (i.e., a few nanometers), then two proteins traveling along the same DNA would be able to hop past one another. In contrast, if the proteins remained in contact with the duplex at all times then they would be unable to bypass one another due to steric hindrance. To provide further evidence against hopping, we labeled Msh2-Msh6 with differently colored QDs, λem = 565 nm and λem = 705 nm, represented as green and magenta, respectively (Figure 5A). We then mixed the different colored proteins together and visualized their behavior on a single DNA substrate. An example of these results is presented as a two-color kymogram illustrating the movement of several different proteins traveling along the same DNA molecule over a 10 min period (Figure 5B and Movie S4). Similar results were obtained if the different QDs were pre-mixed and then conjugated to Msh2-Msh6. Even though the proteins moved along the DNA, and often collided with one another, the relative order of the different QD signals did not change over time. These experiments demonstrated that the proteins could not bypass one another and were unlikely to experience large hops as they traveled on the DNA.
Figure 5. Molecular Collisions between Msh2-Msh6 Complexes Bound to the Same Molecule of DNA.
(A) Shows an overview of the experimental design. Here Msh2-Msh6 was labeled with either magenta (λem = 705 nm) or green (λem = 565 nm) QDs, mixed together, and then injected into the same sample chamber.
(B) Shows a kymogram illustrating the magenta and green Msh2-Msh6 complexes diffusing along the same molecule of DNA over a 10 min period (Movie S4). Time (s) is indicated at the top of each panel, and distance is indicated at the right. Apparent collision events are evident as magenta and green complexes approach one another and become white.
To further verify that the protein was not hopping, we performed challenge experiments in which a competitor oligonucleotide (100 nM or 1 µM, either with or without a mismatch; corresponding to ~20,000-fold or ~200,000-fold molar excess [in bp] relative to the amount of λ-DNA tethered to the surface) was injected to the sample chamber either along with Msh2-Msh6 or after prebinding Msh2-Msh6 to the λ-DNA (data not shown). If Msh2-Msh6 was premixed with the competitor oligonucleotide then we detected no binding to the tethered λ-DNA. In contrast, if Msh2-Msh6 was prebound to the λ-DNA then the subsequent addition of the competitor oligonucleotide did not cause it to dissociate from the λ-DNA. Taken together, our data are consistent with a sliding mechanism in which the proteins remain very closely associated with the DNA; however, we cannot completely rule out the possibility that hops occur on an extremely short time and/or length scale undetectable by our assays.
ADP/ATP Exchange Promotes the Sliding and Release of Msh2-Msh6
We have developed a method for assembling “DNA curtains” at defined positions on the bilayer-coated surface of a sample chamber (Supplemental Experimental Procedures; Graneli et al., 2006b). The DNA molecules that make up the curtain are tethered by a single end, and hydrodynamic force is required to maintain the molecules in an extended configuration confined within the evanescent field. As a consequence, any diffusing proteins are driven in the direction of the flow force and either fall off the ends of the DNA molecules or become trapped behind an immobile protein (Graneli et al., 2006b; see below). Therefore, this assay is uniquely adapted for studying the immobile population of Msh2-Msh6.
To visualize Msh2-Msh6, labeled protein was injected into the sample chamber along with ADP and incubated for several minutes with the λ-DNA curtain, which was sufficient to allow many of the proteins to enter the immobile state. The sample chamber was rinsed to remove unbound Msh2-Msh6, and the remaining proteins were then observed by TIRFM (Figure 6 and Movie S5). Buffer flow was paused (flow ON/OFF control; Figure 6 and Movie S5) to verify that the proteins were bound to the DNA and not simply stuck to the flowcell surface. This procedure caused the DNA molecules to transiently drift out of the evanescent field, but any proteins nonspecifically stuck to the surface remained within view and could be discounted from further analysis. The movies were analyzed to determine the number of proteins on DNA molecules, and 274 complexes were identified in this example (Figure S7A). This represents the minimum number of bound Msh2-Msh6 molecules because the force exerted by the buffer would have pushed any diffusing proteins into the immobile proteins, and therefore each spot can be comprised of multiple Msh2-Msh6 complexes.
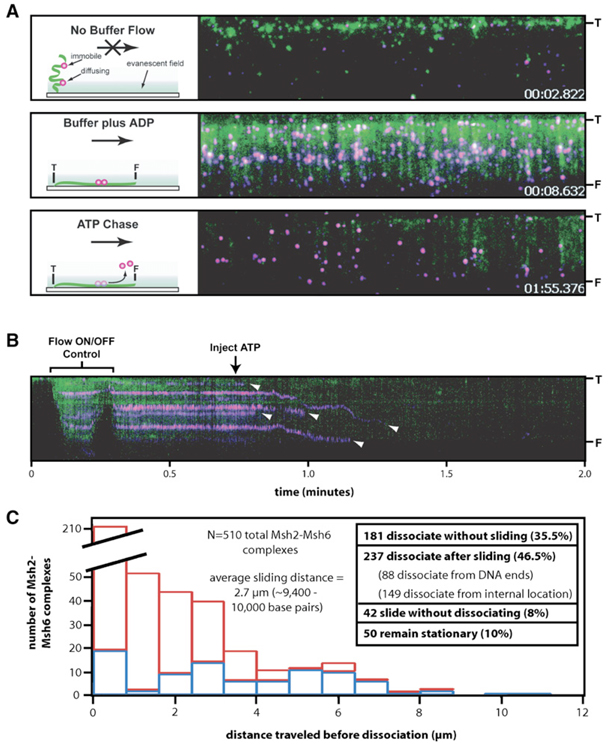
Figure 6. ATP Causes the Immobile Population of Msh2-Msh6 to Begin Diffusing and Then Dissociate from DNA.
(A) Panel shows the assay used to probe the effects of ATP on the immobile population of Msh2-Msh6. The proteins are magenta, and the DNA is green. The DNA is tethered to the surface by only one end (“T”), and the free end (“F”) is only observed when flow is applied. Flow is from top to bottom in each panel, and the distance between T and F is ~13 µm. The panels were extracted from a movie, and the time stamps correspond to the specified frames (Movie S5). The upper panel shows the field after transiently pausing buffer flow (flow ON/OFF control), and the cartoon at the left depicts the behavior of the molecules in the absence of flow. The middle panel shows the same field after resuming flow. Each magenta spot in the image corresponds to at least one Msh2-Msh6 complex, and there are 274 identifiable magenta spots in this experiment and 124 DNA molecules (Movie S5). The lower panel shows the same DNA molecules after the injection of 2 mM ATP.
(B) Panel shows a kymogram showing one DNA molecule and the bound Msh2-Msh6. Arrowheads highlight the dissociation of Msh2-Msh6 (Figure S7).
(C) Panel summarizes the behavior of 510 total Msh2-Msh6 complexes (from three separate experiments) after the injection of ATP into the sample chamber. “Distance traveled before dissociation” corresponds to the distance that Msh2-Msh6 moved prior to falling off the DNA. Dissociation events that occurred from internal positions on the DNA are colored red (N = 149), and those that occurred at the end of the DNA molecules are colored blue (N = 88).
We next tested the effects of replacing ADP with ATP on the immobile Msh2-Msh6 to see whether the change in nucleotide could stimulate release of the proteins. When the ADP was flushed from the sample chamber and rapidly replaced with ATP (or ATPγS; Figure S7B and S7C), most of the proteins (up to ~90%) quickly dissociated from the DNA (t1/2 = 10.2 ± 1.16 s; Figure 6, Figure S7A, and Movie S5). The observed dissociation kinetics were virtually identical to values reported in bulk studies (Mendillo et al., 2005). Visual inspection of the data suggested that the dissociation occurred through two mechanisms: either (1) dissociation after sliding (1D diffusion biased in the direction of buffer flow) along the DNA (46.5%; Figure 6C, Figure S7, and Movie S5) or (2) direct dissociation from the DNA without apparent sliding (35.5%). However, it is possible that proteins that appeared to dissociate directly from the DNA actually slid for a short distance below our resolution limits (~300 bps with this assay). This hypothesis is supported by the fact that the dissociation curves for the entire population of proteins could be fit to a single exponential decay, suggesting that there were not two kinetically distinct populations (Figure S7A). The proteins traveled an average distance of ~10,000 bp prior to dissociating from the DNA molecules (Figure 6C). Of the 237 Msh2-Msh6 complexes that clearly slid along the DNA after ADP/ATP exchange, 88 dissociated from the ends of the DNA whereas 149 dissociated from internal positions prior to reaching the DNA ends. An additional 42 complexes slid along the DNA but did not dissociate, and 50 remained stationary throughout the experiment (Figure 6C). These observations demonstrated that the fluorescently labeled proteins were biologically viable and were not irreversibly trapped on the DNA or stuck to the sample chamber surface.
DISCUSSION
Mechanistic Basis of Msh2-Msh6 Movement along DNA
Our results show that Msh2-Msh6 travels freely along the helical axis of undamaged DNA. Several points argue that Msh2-Msh6 moves via 1D diffusion and suggest that this mechanism is an intrinsic property of the protein. First, the movement did not require ATP nor did it require prior binding to a mispaired base. Second, the movement was consistent with physical predictions for diffusion in that it was unbiased and highly redundant and the MSD plots varied linearly with time interval. Third, our experiments rule out alternative scanning mechanisms: we saw no evidence for active translocation; the only random collisions were those necessary for the initial association; intersegmental transfer was unlikely because the DNA was maintained in a physically extended configuration, which would prevent the juxtaposition of two distal sites; and hopping is inconsistent with the finding that the diffusion coefficients were insensitive to salt concentration, different proteins bound to the same DNA were unable to bypass one another, and they were resistant to challenge with excess competitor oligonucleotide. Finally, we have also observed 1D diffusion for E. coli MutS and S. cerevisiae Msh2-Msh3 (Figure S8), suggesting that lateral motion along DNA is a trait shared among the different members of the MutS family.
We propose a model in which variations in the energy landscape allow Msh2-Msh6 to probe for both lesions and strand discrimination signals (possibly nicks; reviewed in Jiricny, 2006) as it slides along the DNA (Figure 7). A key feature of this model is that initial binding of Msh2-Msh6 yields a stable complex and subsequent events are dominated by lateral movement of the protein along the helix rather than reiterative dissociation and rebinding. This conclusion is consistent with the structures of MutS and Msh2-Msh6, which show that the DNA binding domains completely encircle the bound DNA substrate (Lamers et al., 2000; Obmolova et al., 2000; Warren et al., 2007). Moreover, the DNA binding and clamp domains of MutS (comprising 36% of the protein) are disordered in structures lacking DNA, and only become ordered in the presence of bound substrate, indicating that the initial binding reaction is coupled to extensive local folding (Figure 1C; Spolar and Record, 1994; Lamers et al., 2000; Obmolova et al., 2000). Electron micrographs of human Msh2-Msh6 also revealed that the clamp domains are splayed open in the absence of substrate (Gradia et al., 1999). It is reasonable to conclude that the DNA binding and clamp domains would have to at least partially unfold to allow dissociation, and this requirement for a large-scale structural reorganization may account for the long time periods that Msh2-Msh6 is able to slide on DNA.
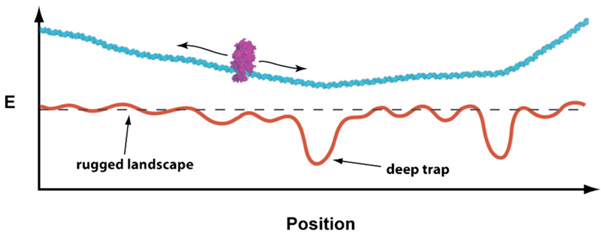
Figure 7. Model for DNA Scanning by Msh2-Msh6.
We present a biophysical model depicting the proposed interactions between Msh2-Msh6 (magenta) and DNA substrates (cyan). Below the DNA is a hypothetical energy landscape describing its bending propensity (Vlahovicek et al., 2003), and we speculate that minima in the landscape correlate with regions that are either intrinsically bent or highly flexible. Positions corresponding to deep depressions in the landscape (e.g., lesions or nicks) will interact favorably with the diffusing protein, and the depth of the energy minima will dictate how long the protein remains at any given site. Proteins that locate deep traps can escape in a reaction driven by the exchange of ADP for ATP.
Importantly, theoretical predictions suggest that diffusion becomes very limited when the roughness of the energy landscape exceeds a threshold of ≈2 kBT (Slutsky and Mirny, 2004), suggesting that the mobile to immobile transitions observed in the diffusion trajectories correspond to local minima or deep traps in the landscape that interact favorably with Msh2-Msh6. Previous models for lesion recognition by MutS proteins have invoked the relative ease with which a mismatch can be kinked as a primary determinant enabling lesion recognition (Wang et al., 2003; Yang, 2006). We envision that intrinsically bent DNA (e.g., poly-dA tracts) and/or more flexible sequences or structures (e.g., mismatches or strand nicks) may serve as deep traps along the energy landscape (Figure 7), and these traps trigger conformational changes in Msh2-Msh6, causing it to reversibly enter an immobile state.
Our results indicate that Msh2-Msh6 can slide freely along DNA while rotating around the helix and that the barrier for each individual step was on the order of 1.57 ± 1.1 kBT. The precise details of the energy barrier calculations are dependent upon our assumption that the majority of the observed diffusing entities were comprised of a single QD bound by one complex of Msh2-Msh6. Several points agree that this is the case. First, Msh2-Msh6 is well behaved in solution, gel filtration studies of our protein stocks reveal a single peak of the appropriate molecular mass, and these stocks are diluted ~1000-fold prior to use. Second, the QDs are well behaved in solution, they show a single peak of the expected size by gel filtration, and prior to use they are purified by gel filtration. Third, the hydrodynamic properties of the complexes are consistent with expectations for approximately one protein complex per QD. If Msh2-Msh6 formed large aggregates, we would expect to see a greater change in the apparent molecular mass. Fourth, Msh2-Msh6 aggregates would contain numerous HA tags and would therefore bind multiple QDs. To verify that this was not the case we show that the QDs blink, indicating only one QD per protein complex. Finally, when labeled with a mixture of different colored QDs the majority of complexes are a single color. If the protein aggregated then these complexes would show two colocalized signals because they would be labeled with both colored QDs. Taken together, our data suggest that the complexes we observed sliding along the DNA are comprised of one QD conjugated to one molecule of Msh2-Msh6. Nevertheless, we cannot entirely rule out the possibility that some of the QDs may be coupled to more than one molecule of Msh2-Msh6. However, the possible presence of more than one protein per QD in no way alters the overall conclusion that Msh2-Msh6 can travel along the DNA via a mechanism involving 1D sliding.
Biological Implications of Msh2-Msh6 1D Diffusion
Msh2-Msh6 completely encircles DNA, and therefore its interactions with nonspecific sequences are dominated by lateral movement rather than reiterative binding and dissociation events. This has important consequences for both the regulation of Msh2-Msh6 and the nature of the in vivo search mechanism. For example, in the absence of a 3D component, the time required to complete the search of a DNA of length N can be approximated as TD1 ≈ N2/D1, where D1 is the diffusion coefficient (Gerland et al., 2002). For S. cerevisiae, where N = 2.4 × 107 bp (8160 µm), D1,calc = 0.25 µm2/s (estimated upper limit for Msh2-Msh6 without the QD), and assuming there are on the order of 1000 molecules of Msh2 per cell (i.e., one molecule of Msh2-Msh6 for every 19,512 bp or 6.63 µm of DNA; Ghaemmaghami et al., 2003), the total time required to scan the entire genome by a purely 1D diffusion mechanism would be just 2.9 min.
Sliding alone cannot be the only mechanism involved in scanning DNA because of the difficulty posed by the crowded environment inside of a living cell (Kampmann, 2005). This can be rationalized by considering that sliding in vivo will be physically impeded by the presence of other DNA-bound proteins, such as nucleosomes, and if the diffusing protein is unable to bypass these obstacles it will inevitably become trapped in a futile search between two stationary roadblocks. However, this problem could be overcome if Msh2-Msh6 was pulled along by a motor protein capable of stripping stationary obstacles (e.g., nucleosomes) from DNA. For example, the replication machinery removes nucleosomes from DNA as the genome is being duplicated, and there are ~260 bp of naked DNA behind the progressing fork (Gasser et al., 1996). Interestingly, Msh2-Msh6 binds to PCNA in vitro (reviewed in Kunkel and Erie, 2005; Jiricny, 2006; and Modrich, 2006), and human Msh2-Msh6 colocalizes with DNA replication factories in vivo during S phase (Kleczkowska et al., 2001). Association with the replisome could facilitate Msh2-Msh6 lesion scanning by eliminating any obstacles that might block its path. This also positions Msh2-Msh6 in the ideal location to trail behind the fork and efficiently scan all of the newly replicated DNA as it exits the rear of the polymerase, ensuring that repair could be initiated before loss of the transient signals that allow the protein to distinguish parental and daughter strands (Kunkel and Erie, 2005; Umar et al., 1996; Kleczkowska et al., 2001).
Implications of Nucleotide Exchange-Driven Sliding and Release of Msh2-Msh6
When Msh2-Msh6 was bound to DNA in the presence of either ADP or ATP, the interactions with the DNA were dominated by sliding. However, when the reactions were staged with initial binding in the presence of ADP followed by a switch to ATP (or ATPγS) the proteins dissociated through a mechanism involving sliding along the phosphate backbone followed by either dissociation from an internal location or from the DNA ends. These findings are consistent with previous reports of Msh2-Msh6 release from mismatched DNA (Gradia et al., 1999; Mendillo et al., 2005; Jiang et al., 2005; Blackwell et al., 2001). Although the mechanistic basis for the ADP/ATP exchange-dependent behavior remains unknown, we speculate that the staged delivery of the different nucleotides promotes a rapid and simultaneous conversion of both the Msh2 and Msh6 subunits to the ATP-bound state, enabling the immobile complexes to begin moving along the DNA before dissociation (Mazur et al., 2006). We emphasize that our experiments were done with λ-DNA that did not contain any mispaired bases, and it is possible that the proteins could behave differently when bound to mismatched substrates or when other MMR proteins are present. Nevertheless, the fact that the immobile Msh2-Msh6 rapidly resumed sliding and was eventually released from the DNA after the switch from ADP to ATP demonstrated that these proteins displayed normal biochemical properties and suggested that the immobile population had encountered deep traps in the energy landscape that mimicked damaged DNA. This also indicated that a nucleotide exchange-driven conformational change in the protein allowed it to escape these deep traps and supports the idea that nucleotide exchange destabilizes the “clamp-like” binding mechanism, perhaps by eliciting a further conformational change that eventually releases Msh2-Msh6 from the DNA.
In summary, this work directly demonstrates that Msh2-Msh6 can slide on DNA without the need for ATP hydrolysis or mismatch binding. Our results imply that the overall mechanism of mismatch recognition and initiation of repair occurs via Msh2-Msh6 first sliding along DNA (possibly in association with DNA replication factories, as described above) until a lesion is located. Msh2-Msh6 would then stop sliding and remain tightly bound to the DNA upon encountering the lesion as it exited from the rear of the replication fork (i.e., a deep trap along the energy landscape). We speculate that the dwell time at the lesion may allow Msh2-Msh6 to recruit additional MMR proteins required to complete repair. Finally, Msh2-Msh6 would undergo a conformational change driven by the exchange of ADP for ATP, allowing it to continue diffusing along the DNA while searching for strand discrimination signals, presumably in an altered conformation competent to complete downstream steps in the repair path. Additional TIRFM experiments with damaged DNA substrates and other MMR factors will be required to probe further details of these postrecognition steps.
EXPERIMENTAL PROCEDURES
Reaction Conditions
For TIRFM experiments, Msh2-Msh6 (100 nM) was mixed with 100– 200 nM anti-HA QDs in reaction buffer containing 40 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 0.2 mg/ml BSA, and reactions were incubated for 15–20 min on ice. Similar results were also obtained at 10:1 QD to protein ratios. The reactions were then diluted to a final volume of 100 µl (2.5–5 nM Msh2-Msh6) immediately prior to injecting the protein into the sample chamber. TIRFM experiments were done using 40 mM Tris-HCl (pH 7.8), 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 0.2 mg/ml BSA with or without 1 mM nucleotide (ADP or ATP), as indicated. In experiments with fluorescent DNA, 0.5 nM YOYO1 was also included, and this low concentration of dye does not affect the length of the DNA as is the case with much higher concentrations of YOYO1. The labeled proteins were injected into the sample chamber under constant flow, and the unbound proteins were quickly flushed from the sample chamber. To minimize laser-induced DNA damage, an oxygen scavenging system comprised of 0.5% β-mercaptoethanol, 0.1X GLOXY, and 1% glucose was included in the reactions. Approximately 75% of the experiments were performed with YOYO1 present and 25% were performed without YOYO1, and there was no detectable difference in the behavior of the proteins with or without the DNA stain. YOYO1 and the oxygen scavenging system were tested in bulk assays and did not affect the DNA binding or ATPase activity of Msh2-Msh6 (data not shown).
DNA Curtain Assay
Assays with the DNA curtains were performed under essentially the same reaction conditions as described above. The only exception is that buffer was continually flowing through the chamber to maintain the DNA molecules aligned along the edges of the diffusion barriers and visible within the evanescent field. For these assays, Msh2-Msh6 was injected over a period of ~5 min in the presence of 1 mM ADP and allowed to bind to the λ-DNA. During this time, some of the Msh2-Msh6 stopped sliding and these immobile proteins functioned as barriers behind which any remaining sliding proteins would accumulate. The flowcells were then rinsed for several additional minutes until all of the unbound proteins were removed from the sample chamber. Data collection was then initiated, and all of these experiments consisted of a series of steps beginning with the flow ON/OFF control and followed by switching to buffer containing 2 mM ATP. Movies were collected continuously throughout this procedure.
All additional details of experimental procedures, calculations, movies, and further discussion are provided as Supplemental Data online.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NSF CAREER Award to E.C.G. and by an NIH grant to E.A. (GM53085). J.A.S. is a research fellow of the National Cancer Institute of Canada supported with funds provided by the Terry Fox Run. We thank Ragan B. Robertson for help with the data analysis software. We thank Dr. Wei Yang and Dr. Christian Biertump-fel (National Institutes of Health) for the generous gift of FLAG-tagged MutS. We also thank Kiyoshi Mizuuchi, Larry Shapiro, Ruben Gonzalez, Jonathan Dworkin, and members of our laboratories for careful reading and insightful comments on the manuscript.
Footnotes
Supplemental Data
Supplemental Data include Supplemental Discussion, Supplemental Experimental Procedures, Supplemental References, eight figures, and five movies and can be found with this article online at http://www.molecule.org/cgi/content/full/28/3/359/DC1/.
REFERENCES
- Allen DJ, Makhov A, Griley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Berg HC. Random Walks in Biology, Expanded Edition. Princeton, NJ: Princeton University Press; 1993. [Google Scholar]
- Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Martik D, Bjornson KP, Bjornson ES, Mod-rich P. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Bjornson KP, Allen DJ, Modrich P. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem. 2001;276:34339–34347. doi: 10.1074/jbc.M104256200. [DOI] [PubMed] [Google Scholar]
- Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PMJ, Kunkel TA. Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- Gasser R, Koller T, Sogo JM. The stability of nucleosomes at the replication fork. J. Mol. Biol. 1996;258:224–239. doi: 10.1006/jmbi.1996.0245. [DOI] [PubMed] [Google Scholar]
- Gerland U, Moroz JD, Hwa T. Physical constraints and functional characteristics of transcription factor-DNA interaction. Proc. Natl. Acad. Sci. USA. 2002;99:12015–12020. doi: 10.1073/pnas.192693599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith JD, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- Graneli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc. Natl. Acad. Sci. USA. 2006b;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P. Molecular mechanisms of DNA mismatch repair. Mutat. Res. 2001;486:71–87. doi: 10.1016/s0921-8777(01)00088-x. [DOI] [PubMed] [Google Scholar]
- Jacobs-Palmer E, Hingorani MM. The effects of nucleotides on MutS-DNA binding kinetics clarify the role of MutS ATPase activity in mismatch repair. J. Mol. Biol. 2005;366:1087–1098. doi: 10.1016/j.jmb.2006.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Bai L, Surtees JA, Zekeriyya G, Wang MD, Alani E. Detection of high-affinity and sliding clamp modes for Msh2-Msh6 by single-molecule unzipping force analysis. Mol. Cell. 2005;20:771–781. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kampmann M. Facilitated diffusion in chromatin lattices: mechanistic diversity and regulatory potential. Mol. Microbiol. 2005;57:889–899. doi: 10.1111/j.1365-2958.2005.04707.x. [DOI] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J.Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lamers M, Perrakis A, Enzlin JH, Winterwerp HHK, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G–T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Mendillo ML, Kolodner RD. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis independent movement along DNA. Mol. Cell. 2006;22:39–49. doi: 10.1016/j.molcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible end-blocking system. J. Biol. Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc. Natl. Acad. Sci. USA. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Sheetz MP, Elson EL. Single particle tracking. Biophys. J. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA. Hydrodynamic considerations. Biophys. Chem. 1979;9:413–414. [PubMed] [Google Scholar]
- Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolar RS, Record MT. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- Vlahovicek K, Kajan L, Ponor S. DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Res. 2003;31:3686–3687. doi: 10.1093/nar/gkg559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, Erie DA. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Bease LS. Structure of the human MutSα DNA lesion recognition complex. Mol. Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair (Amst.) 2006;10:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Yang Y, Sass LE, Du C, Hsieh P, Erie DH. Determination of protein DNA binding constants and specificities from statistical analysis of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005;33:39–49. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.