Abstract
Mitochondrial cytochrome P450 enzymes (CYP or P450, EC 1.14.13.15) play an important role in metabolism of cholesterol. CYP27A1 hydroxylates cholesterol at position 27 and, thus, initiates cholesterol removal from many extrahepatic tissues. CYP11A1 is a steroidogenic P450 that converts cholesterol to pregnenolone, the first step in the biosynthesis of all steroid hormones. We utilized a new approach to study membrane topology of CYPs 27A1 and 11A1. This approach involves heterologous expression of membrane-bound P450 in E. coli, isolation of the P450-containing E. coli membranes, treatment of the membranes with protease, removal of the digested soluble portion and extraction of the membrane-associated peptides, which are then identified by mass spectrometry. By using this approach, we found four membrane-interacting peptides in CYP27A1, and two peptides in CYP11A1. Peptides in CYP27A1 represent a contiguous portion of the polypeptide chain (residues 210-251) corresponding to the putative F-G loop and adjacent portions of the F and G helices. Peptides in CYP11A1 are from the putative F-G loop (residues 218-225) and the C-terminal portion of the G helix (residues 238-250). This data is consistent with those obtained previously by us and others and provide new information about membrane topology of CYPs 27A1 and 11A1.
Keywords: CYP27A1, CYP11A1, membrane topology, mass spectrometry analysis
Introduction
Cytochromes P450 (CYP or P450, EC 1.14.13.15) are obligatory enzymes in metabolism of cholesterol, steroid hormones and vitamin D3, in mitochondria (1). We are interested in delineation of structure/function relationships in P450s that initiate cholesterol transformations to bile acids and steroid hormones. Cholesterol serves as the primary substrate for only four mammalian enzymes (CYPs 7A1, 46A1, 27A1, and 11A1), and two of them (CYPs 27A1 and 11A1) reside in mitochodria (2). CYP27A1 is a ubiquitously expressed multifunctional sterol 27-hydroxylase that catalyzes cholesterol 27-hydroxylation in many extrahepatic tissues. In addition, CYP27A1 also metabolizes bile acid intermediates in the liver and vitamin D3 in the kidney (3). Partial or complete lack of CYP27A1 activity results in a disease cerebrotendinous xanthomatosis manifested by a variety of phenotypes including tendon xanthomas, bilateral cataracts, premature atherosclerosis, osteoporosis, neurological and neuropsychiatric abnormalities (4). Unlike CYP27A1, expression of CYP11A1 is limited to steroidogenic tissues and the brain, where it catalyzes the conversion of cholesterol to pregnenolone, the first step in overall steroid hormone biosynthesis (5). Partial or complete lack of CYP11A1 activity results in a disorder of steroidogenesis in which cholesterol accumulates within the steroidogenic tissues, and the synthesis of all adrenal and gonadal steroids is impaired (2). Several different approaches have been used previously to study membrane topology of mitochondrial P450s (6-10). Ou et al. treated mitochondria from the bovine adrenal cortex with trypsin and digested CYP11A1 present in these mitochondria into two fragments. Mitochondria were then washed with sodium carbonate followed by evaluation of the association of the two fragments with the membrane (6). Usanov et al. studied the interaction of the mitochondrion-associated CYP11A1 with antibodies to the whole enzyme and its fragments (7). We investigated CYPs 27A1 and 11A1 by combining heterologous expression in E. coli and site-directed mutagenesis (8, 11). Residues in the F-G loop, a putative site of the interaction with the membrane, were mutated, the mutant P450s expressed in E. coli and their subcellular distribution and catalytic activities compared with those of the wild type enzyme. Simultaneously with our studies, Headlam et al. investigated the F-G loop as well as several other regions in CYP11A1 by utilizing cysteine mutagenesis followed by fluorescent labeling of the mutated residues and measurements of the changes in the fluorescence upon association of the mutant P450 with phospholipid vesicles. The N- and C-terminal truncated variants of CYP11A1 were also generated (10). Both groups suggested that the F-G loop is the site of interaction of mitochondrial P450s with the membrane. In the present investigation we continued elucidation of the membrane topology of mitochondrial P450s. We tested whether we can identify membrane-interacting areas in mitochondrial P450 other than the F-G loop by combining the methodologies of heterologous expression and proteolytic digestion with advances in mass spectrometry. CYPs 11A1 and 27A1 were expressed in E .coli, the E. coli membranes isolated and treated with trypsin. Peptides remaining after extensive washes of the trypsin-treated membranes with sodium carbonate were extracted and identified by MALDI MS. The data obtained suggest that not only the F-G loop but also a part of the G helix is embedded in the membrane in mitochondrial P450s.
Experimental Procedure
Materials
Sequencing grade modified trypsin was from Promega Corp. (Madison, WI, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Membrane fractionation
Recombinant human CYP27A1 and bovine CYP11A1 were expressed in E. coli as described (12, 13). Cells were harvested by centrifugation at 10,000 g for 10 min, resuspended in 10 mM HEPES (pH 7.8) containing 10% sucrose, and treated with 0.2 mg/ml lysozyme on ice for 30 min. Spheroplasts were pelleted at 10,000 g for 10 min, resuspended in 10 mM HEPES (pH 7.8) containing 10% sucrose and sonicated using six 10 seconds pulses at 40% duty cycle (Sonifier 450, Branson Ultrasonics Corp., Dandury, CT). The suspension was then layered over a cushion of 55% sucrose topped with 10% sucrose in 10 mM HEPES (pH 7.8) and subjected to centrifugation in a Ti70 rotor at 35,000 rpm for 60 min. The total membrane fraction was recovered from the 55% sucrose interface, washed twice with 25 mM NH4HCO3 (pH 7.9), and placed in pre-weighted tubes.
Trypsin treatment
Wet membrane pellet (5 mg) was resuspended in 1 ml of 25 mM NH4HCO3 (pH 7.9) followed by addition of 10 μg of sequencing grade modified trypsin. The suspension was sonicated as described above and left for proteolysis for 15 hrs at 37°C. The sample was then subjected to 106,000 g centrifugation for 20 min in Optima TLX ultracentrifuge (Beckman Instruments, Inc., Fullerton, CA, USA). Pellet was collected and washed sequentially with 100 mM Na2CO3 (pH 11.5) containing 0.3 M NaCl (5 times) and with water (1 time). Traces of water from the wet pellet were removed by adding 0.2 ml of methanol/chloroform mixture (4/1, v/v) and drying the sample in Vacufuge (Eppendorf AG, Hamburg, Germany). Dried pellet was then vortexed with 0.2 ml of chloroform, organic phase discarded, and the membranes dried again. Extraction of peptides from the dried membranes was carried out with 100 μl of 50% acetonitrile/0.1% trifluoroacetic acid (TFA). The extracted sample was further purified using ZipTip C18 (Millipore Corporation, Bedford, MA, USA) according to the manufacture protocol.
Mass spectrometry analysis
Peptide samples after ZipTip purification were dried, dissolved in 5 mg/ml α-cyano-4-hydroxycinnamic acid in 50% acetonitrile containing 0.1% TFA, and manually spotted onto the ABI 01-192-6-AB target plate. Mass spectrometry (MS) analyses were performed using an AB4700 Proteomics Analyzer (Applied Biosystems, Framingham, MA, USA). MS-mode acquisitions consisted of 1,000 laser shots averaged from 20 sample positions. In the MS/MS-mode acquisitions, 6,000 laser shots were averaged from 60 sample positions for PSD fragments. Automated acquisition of MS and MS/MS data was controlled by 4000 Series Explorer software 3.0.
Computational analysis
Free energy of transfer from water to bilayer interface (ΔGWW) was computed using Totalizer module of MPEx (http://blanco.biomol.uci.edu/mpex) based on the Wimley-White hydrophobicity scale. A function of “no end groups” as a subsequence of a longer sequence was used for this calculation. Hydrophobic moment (μH) was also computed using the Totalizer module of MPEx.
Results
Identification of the membrane-interacting peptides in CYP27A1 and CYP11A1
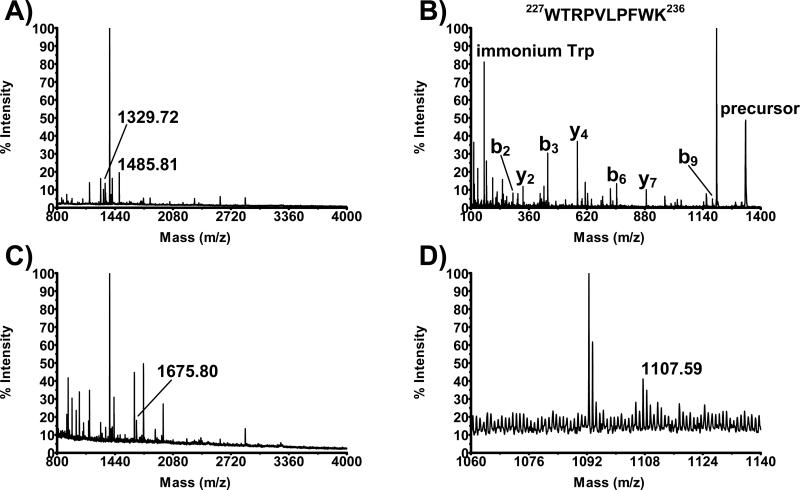
Representative MS spectrum of the extract derived from trypsin-treated CYP27A1-containing E. coli membranes is shown in Fig. 1a. Manual analysis revealed 4 peptides with m/z [M+H]+ values expected for the CYP27A1 hydrolysis with trypsin. Two of these peptides with m/z [M+H]+ values 1329.72 and 1485.81 are labeled in Figure 1a. Signals for two other peptides with m/z [M+H]+ values 1645.78 and 1929.95 were very small and could be seen only after zooming (not shown). In MALDI experiment, signal intensity does not linearly correlate with the concentration of analyte, therefore very small peaks were annotated as well. A summary of all the peptides identified in CYP27A1 is given in Table 1. The expected m/z [M+H]+ values were calculated using MASCOT software and rounded to two decimals. The accuracy of mass measurement in ppm was calculated as a difference between the expected and observed value divided by observed value. Without the internal calibration, the observed m/z [M+H]+ values for these peptides deviate from the expected m/z [M+H]+ values in a range of 40 ppm. Fig. 1b shows the MS/MS spectrum for the peptide ion with m/z [M+H]+ value 1329.72. This spectrum has immonium ion for Trp and various y- and b-type fragment ions that collectively confirm the correct assignment of amino acid sequence. The MS analysis of the CYP11A1 sample revealed two CYP11A1 peptides. One of them with m/z [M+H]+ value 1675.80 is labeled in Fig. 1c. The zoomed spectrum for the second peptide with m/z [M+H]+ value 1107.59 is shown in Fig. 1d. The intensity for this peptide was very low.
Fig. 1.
Membrane-interacting peptides in CYP27A1 and CYP11A1. A) MS spectrum of the extract from the trypsin-treated CYP27A1-containig E. coli membranes. Peptides with m/z [M+H]+ values 1329.72 and 1485.81 correspond to CYP27A1, all the other peptides are of the E. coli origin. B) MS/MS spectrum of the peptide ion with m/z [M+H]+ value 1329.72 that corresponds to the 227WTRPVLPFWK236 peptide in CYP27A1. Multiple fragment ions matching to this sequence are labeled as follows: b2, WT; b3, WTR; b6, WTRPVL; b9, WTRPVLPFW; y2, WK; y4, PFWK; and y7, PVLOFWK. C) and D) Regular and zoomed MS spectrum of the extract from the trypsin-treated CYP11A1-containig E. coli membrane. Peptides with m/z [M+H]+ value 1675.80 and 1107.59 correspond to CYP11A1.
Table 1.
Membrane-interacting peptides in CYPs 27A1 and 11A1. Missed cleavage sites are underlined.
| Peptide | [M+H]+ expected | [M+H]+ observed | Delta (ppm) | ΔGWW (kcal mol-1)a | Hydrophobic moment (μH)a |
|---|---|---|---|---|---|
| CYP27A1 | |||||
| 210SIGLMFQNALYATFLPK226 | 1930.01 | 1929.95 | 31 | -2.36 | 1.98 |
| 227WTRPVLPFWK236 | 1329.75 | 1329.72 | 23 | -2.48 | 5.25 |
| 227WTRPVLPFWKR237 | 1485.85 | 1485.81 | 27 | -1.67 | 5.59 |
| 238YLDGWNAIFSFGKK251 | 1645.84 | 1645.78 | 36 | -1.97 | 6.12 |
| CYP11A1 | |||||
| 218LFRTKTWR225 | 1107.64 | 1107.59 | 45 | -0.65 | 1.39 |
| 238AEKYTEIFYQDLR250 | 1675.83 | 1675.80 | 18 | 4.08 | 6.11 |
Free energy of transfer from water to bilayer interface based on the Wimley-White hydrophobicity scale (ΔGWW) and hydrophobic moment (μH) were computed using Totalizer module of MPEx (http://blanco.biomol.uci.edu/mpex).
Analysis of the identified peptides
Membrane-interacting protein sequences usually possess a unique combination of hydrophobicity and amphiphilicity. Therefore, the identified peptides were evaluated for two parameters that render peptides “membrane-seeking”: free energy of transfer from water to bilayer interface and hydrophobic moment. Free energy of transfer (ΔGWW) is a measure of solubility in non-polar phase with the positive value of ΔGWW being unfavorable for partitioning the peptide sequence into the membrane interface. Hydrophobic moment (μH) detects periodicities in the hydrophobicity of amino acid sequences and is a measure of the amphiphilicity of a protein fragment (14, 15). The ΔGWW and μH values of the CYP27A1 and CYP11A1 peptides are shown in Table 1. All of the identified peptides except one, 238AEKYTEIFYQDLR250, have negative values of ΔGWW and values of μH from 1.98 to 6.12. This is consistent with the fact that they were extracted from the membrane. The 238A-R250 peptide has a positive ΔGWW value but a high μH value which strongly increases the chances for this sequence to interact with the membrane. Trypsin can not cleave amino acid residues embedded in the membrane. Therefore, the identified peptides contain both membrane-bound and soluble regions. If the soluble region is too long, this may affect the calculated ΔGWW, and could be the case with the 238A-R250 peptide.
In addition to the computational analysis, peptides were examined for the presence of the missed cleavage sites. One may assume that the missed cleavage sites are located either in the membrane or close to the membrane surface, and therefore resistant to proteolysis. Two peptides in CYP27A1, 227W-R237 and 238Y-K251, and one peptide in CYP11A1, 238A-R250, contain one missed cleavage site, and one peptide in CYP11A1, 218L-R225, has two missed cleavage sites. Also, the CYP27A1 peptides with m/z [M+H]+ values 1329.72 and 1485.81 represent the same sequence without and with the trypsin missed cleavage site, respectively.
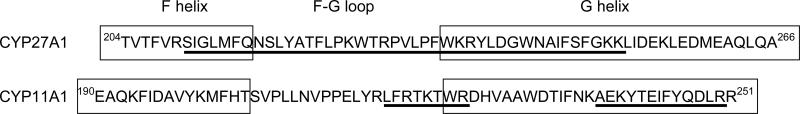
The position of the membrane-interacting peptides in the primary and secondary sequences of CYPs 27A1 and 11A1 is shown in Fig. 2. Peptides in CYP27A1 represent a contiguous portion of the polypeptide chain comprising the putative F-G loop and flanking portions of the F and G helixes. In CYP11A1, the 218L-R225 peptide is located in the F-G loop and the 238A-R250 peptide in the C-terminal portion of the G helix.
Fig. 2.
Location of the membrane-interacting peptides (underlined) in the primary and secondary structures of CYPs 27A1 and 11A1. Boxes indicate putative F and G helices. The secondary structure consensus prediction program (http://npsa-pbil.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_seccons.html) was used to identify these structural elements.
Discussion
In the current study, we used CYP27A1- and CYP11A1-containing E. coli membranes as a model system to study membrane topology of these mitochondrial enzymes. Previously, we developed E. coli expression for both proteins, showed that they stay associated with the membrane fraction when expressed in E. coli, and are catalytically active in the E. coli membranes (8, 9, 12, 13). The membrane topology of CYPs 27A1 and 11A1 was then studied by selecting a hydrophobic region in their primary sequence, the F-G loop, and evaluating how mutations within this region affect binding to the membrane and activity of the mutant P450s (8, 9). Weakened binding of some the mutants indicated that the F-G loop indeed interacts with the membrane. Mitochondrial P450s are suggested to have a peripheral association with the membrane (16, 17). However, it is not currently clear whether one or several segments of the primary sequence are associated with the membrane. Headlam and colleagues began to address this question by investigating three regions in CYP11A1, the F-G loop, A’ helix, and the 14 amino acid residue-segment at the C-terminus (10). These regions were selected based on hydrophobicity profiling and secondary and tertiary structure predictions. The data obtained indicate the involvement of the F-G loop and possibly A’ helix, but not the C-terminus of CYP11A1, in the interactions with the membrane. Thus, although the computational analysis provided a reasonable prediction, not all of the putative membrane-interacting regions were then validated by other methods. In the present study a principally different approach was undertaken. It includes the protease degradation of the solution-exposed portion of the membrane-associated CYPs 27A1 and 11A1 and subsequent MS analysis of the recovered membrane-bound peptides. Consistent with previous studies, peptide(s) from the F-G loop were extracted from the membrane in CYPs 27A1 and 11A1. Identification of these peptides lends support to the validity of our approach and serves as additional evidence for the role of the F-G loop. We also identified one peptide in CYP27A1 (238Y-K251) and one peptide in CYP11A1 (238A-R250) that are located outside the F-G loop, in the N- and C-terminal parts of the G helix, respectively. The N-terminal part of the G helix represents a continuation of the F-G loop. Therefore identification of the 238Y-K251 peptide in CYP27A1 is not an unexpected finding. In fact, it provides an explanation to our previous observation that CYP27A1 is associated tighter with the E. coli membrane than CYP11A1 (8). A longer stretch of amino acid residues in the F-G loop region seem to interact with the membrane in CYP27A1 than in CYP11A1. Identification of the 238A-R250 peptide in CYP11A1 is a more interesting finding because the C-terminal part of the G helix has never been considered as a potential membrane-binding site in CYP11A1. Further studies are required to confirm this unexpected finding. As any method, the MS-based approach has a limitation. This limitation is hidden in the procedure of analysis. Certain classes of long peptides with high hydrophobicity can escape MS analysis (18), therefore a set of identified peptides may never reach a full completeness. For example, when purified CYPs 27A1 and 11A1 are digested with trypsin in solution, we usually obtain peptides covering about 54% of the primary sequence. If each of the P450s is subjected to SDS PAGE and digested in-gel, the coverage of the primary sequence by the identified peptides is ~46%. When combined, tryptic peptides from solution and in-gel digestion cover ~65% of the primary sequence. Thus, MS studies with purified CYPs 27A1 and 11A1 raise a possibility that not all of the membrane-interacting peptides in CYPs 27A1 and 11A1 were identified in the present study. Nevertheless, our approach provided further insight into membrane topology of CYPs 27A1 and 11A1 and is a valuable addition to the other methods in the field.
Acknowledgements
This research was supported in part by the grants from the National Institutes of Health GM062882 and AG024336 (to I.A.P.).
Certain commercial materials, instruments, and equipment are identified in this manuscript in order to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology (NIST) nor does it imply that the materials, instruments, or equipment identified is necessarily the best available for the purpose.
Abbreviations
- CYP27A1
Cytochrome P450 27A1
- CYP11A1
Cytochrome P450 11A1
- MS
mass spectrometry
- TFA
trifluoroacetic acid
- MALDI MS
matrix assisted laser desorption ionization mass spectrometry
- TOF
time-of-flight
Contributor Information
Irina A. Pikuleva, Department of Pharmacology and Toxicology, University of Texas Medical Branch, Galveston, TX 77555-1031, USA irpikule@utmb.edu
Natalia Mast, Department of Pharmacology and Toxicology, University of Texas Medical Branch, Galveston, TX 77555-1031, USA.
Wei-Li Liao, Center for Advanced Research in Biotechnology, National Institute of Standards and Technology and University of Maryland Biotechnology Institute, Rockville, MD 20850, USA.
Illarion V. Turko, Center for Advanced Research in Biotechnology, National Institute of Standards and Technology and University of Maryland Biotechnology Institute, Rockville, MD 20850, USA
References
- 1.Omura T. Mitochondrial P450s. Chem Biol Interact. 2006;163:86–93. doi: 10.1016/j.cbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Pikuleva IA. Cholesterol-metabolizing cytochromes P450. Drug Metab Dispos. 2006;34:513–520. doi: 10.1124/dmd.105.008789. [DOI] [PubMed] [Google Scholar]
- 3.Pikuleva IA. Cytochrome P450s and cholesterol homeostasis. Pharmacol Ther. 2006;112:761–773. doi: 10.1016/j.pharmthera.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkhem I, Boberg KM, Leitersdorf E. The metabolic and molecular bases of inherited disease). In: Scriver AL, Beaudet AL, Sly WS, Valle D, Childs B, editors. Inborn errors in bile acid biosynthesis and storage of sterols other than cholesterol. 7th ed. McGraw-Hill; New York: 1995. 1995. pp. 2073–2099. Volume. [Google Scholar]
- 5.Stone D, Hechter O. Studies on ACTH action in perfused bovine adrenals: site of action of ACTH in corticosteroids. Arch Biochem Biophys. 1954;51:457–469. doi: 10.1016/0003-9861(54)90501-9. [DOI] [PubMed] [Google Scholar]
- 6.Ou WJ, Ito A, Morohashi K, Fujii-Kuriyama Y, Omura T. Processing-independent in vitro translocation of cytochrome P-450(SCC) precursor across mitochondrial membranes. J Biochem. 1986;100:1287–1296. doi: 10.1093/oxfordjournals.jbchem.a121835. [DOI] [PubMed] [Google Scholar]
- 7.Usanov SA, Chernogolov AA, Chashchin VL. Is cytochrome P-450scc a transmembrane protein? FEBS Lett. 1990;275:33–35. doi: 10.1016/0014-5793(90)81432-n. [DOI] [PubMed] [Google Scholar]
- 8.Murtazina D, Puchkaev AV, Schein CH, Oezguen N, Braun W, Nanavati A, Pikuleva IA. Membrane-protein interactions contribute to efficient 27-hydroxylation of cholesterol by mitochondrial cytochrome P450 27A1. J Biol Chem. 2002;277:37582–37589. doi: 10.1074/jbc.M204909200. [DOI] [PubMed] [Google Scholar]
- 9.Pikuleva IA. Putative F-G loop is involved in association with the membrane in P450scc (P450 11A1). Mol Cell Endocrinol. 2004;215:161–164. doi: 10.1016/j.mce.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Headlam MJ, Wilce MC, Tuckey RC. The F-G loop region of cytochrome P450scc (CYP11A1) interacts with the phospholipid membrane. Biochim Biophys Acta. 2003;1617:96–108. doi: 10.1016/j.bbamem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Pikuleva IA, Puchkaev A, Bjorkhem I. Putative helix F contributes to regioselectivity of hydroxylation in mitochondrial cytochrome P450 27A1. Biochemistry. 2001;40:7621–7629. doi: 10.1021/bi010193i. [DOI] [PubMed] [Google Scholar]
- 12.Pikuleva IA, Mackman RL, Kagawa N, Waterman MR, Ortiz de Montellano PR. Active-site topology of bovine cholesterol side-chain cleavage cytochrome P450 (P450scc) and evidence for interaction of tyrosine 94 with the side chain of cholesterol. Arch Biochem Biophys. 1995;322:189–197. doi: 10.1006/abbi.1995.1451. [DOI] [PubMed] [Google Scholar]
- 13.Pikuleva IA, Bjorkhem I, Waterman MR. Expression, purification, and enzymatic properties of recombinant human cytochrome P450c27 (CYP27). Arch Biochem Biophys. 1997;343:123–130. doi: 10.1006/abbi.1997.0142. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg D, Weiss RM, Terwilliger TC. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci U S A. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Wachenfeldt C, Johnson EF. Cytochrome P450: Structure, Mechanism, and Biochemistry). In: Ortiz de Montellano PR, editor. Structures of eukaryotic cytochrome P450 enzymes. Plenum Press; New York: 1995. 1995. pp. 183–244. Volume. [Google Scholar]
- 17.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 18.Eichacker LA, Granvogl B, Mirus O, Muller BC, Miess C, Schleiff E. Hiding behind hydrophobicity. Transmembrane segments in mass spectrometry. J Biol Chem. 2004;279:50915–50922. doi: 10.1074/jbc.M405875200. [DOI] [PubMed] [Google Scholar]