Abstract
The success of a phage that infects a bacterial cell possessing a restriction-modification (R-M) system depends on the activities of the host methyltransferase and restriction endonuclease, and the number of susceptible sites in the phage genome. However, there is no model describing this dependency and linking it to observable parameters such as the fraction of surviving cells under excess phage, or probability of plating at low amount of phages. We model the phage infection of a cell with a R-M system as a pure birth process with a killing state. We calculate the transitional probabilities and the stationary distribution for this process. We generalize the model developed for a single cell to the case of multiple identical cells invaded by a Poisson-distributed number of phages. The R-M enzyme activities are assumed to be constant, time-dependent, or random. The obtained results are used to estimate the ratio of the methyltransferase and endonuclease activities from the observed fraction of surviving cells.
Keywords: Enzyme activities ratio, pure birth process with killing, restriction endonuclease, methyltransferase
1. Introduction
The phenomenon of restriction-modification (R-M) was discovered in the 1950s during experiments in which different strains of the same bacterial species were infected with bacterial viruses (bacteriophages or phages for short) (Luria and Human, 1952; Bertani and Weigle, 1953). It was observed that while the efficiency of plating (calculated as the proportion of phage particles capable of productively infecting the host bacterium and ultimately leading to plaques, i.e., observable foci of infection on host bacterium lawns) on permissive, non-restricting strains was close to one, efficiency of plating on non-permissive, restricting strains was about five orders of magnitude lower. However, phage progeny that recovered from rare productive infections of restricting hosts were able to plate with equally high efficiency on both restricting and non-restricting strains. Furthermore, the progeny of “modified” phages lost the ability to productively infect the restricting strain after a single passage on the non-restricting strain. Thus, phages recovered from the restricting-strain infections do not contain a heritable change; they are said to be “modified” by the restricting host.
In experiments that ultimately led to the development of molecular cloning and genetic engineering, the molecular basis of R-M phenomena were uncovered. It was shown that restricting hosts encode two enzymatic activities that are absent in non-restricting bacteria (reviewed in Arber (1978)).
The endonuclease molecules can cut DNA at recognition sites. Consequently, they can destroy both the foreign DNA and the genomic DNA itself.
The cell uses methyltransferase to protect its genome from being killed by its own endonuclease, as a methylated site is not recognized by the endonyclease. Moreover, even a hemimethylated site is not recognized and cut, retaining protection of a newly replicated genomic DNA molecule. These sites are then fully methylated by the methyltransferase, and thus the methylated state is stably maintained in multiple rounds of replication.
On the other hand, if the phage DNA becomes methylated in the bacterial cell, it also cannot be cut by the endonuclease. The progeny phages are methylated as well, and further rounds of the infection proceed without interference from the R-M system. This means that the fate of the cell and the phage largely depends on the competition between the methyltransferase and the endonuclease for the sites in the invading phage genome: if all sites in the phage genome are methylated before endonuclease recognizes any one of them, the phage survives, leading to successful infection.
Over the years, many R-M enzyme pairs (R-M systems) have been isolated from diverse bacteria, the search has been mostly driven by the constant need of restriction endonucleases with novel specificities to be used for molecular cloning (REBASE, http://rebase.neb.com). Cells possessing an R-M system by definition are more resistant to certain phages, obviously an advantageous trait. Analysis of various phages reveals that their genomic DNA contains little or no recognition sequences for restriction endonucleases commonly found in their hosts, or that they use special mechanisms such as heavy methylation of their DNA or specialized antirestriction proteins that bind to and inactivate restriction endonucleases of the host (Tock and Dryden, 2005). Clearly, phages have evolved these mechanisms to avoid the action of the R-M systems of the host.
The protection afforded by the R-M systems against the infecting phage is not absolute, and a cell that is productively infected ends up serving as a source of modified phage progeny that can effectively wipe out the rest of the population. The efficiency of restriction appears to be genetically determined and is both host strain and phage specific. The physiology of the host also appears to play a role. However, the actual mechanisms that lead to and determine the frequency of overcoming the host restriction by phages are unknown. Here, we model the process of phage infection of a bacterial cell harboring an R-M system. The model makes specific predictions about the efficiency of the phage restriction at varying multiplicity of infection for phage containing different numbers of R-M system recognition sites. We specifically take into account the fluctuations in the amount of restriction endonuclease, methyltransferase, and phage infecting a cell. The results set the stage for discriminative experiments that will allow to confirm or refute the mechanism of phage restriction implicitly assumed in the model and thus increase our understanding of the mechanism of restriction of foreign DNA by cells harbouring R-M systems.
2. Model
We model a culture of bacterial cells that harbors an R-M system and is invaded by a phage. The number of restriction sites N in the phage genome is known, the total number of bacteria in the culture is K, and the total number of phages equals V. The bacterial cells are assumed to be identical up to the effective activities (see below) of restriction endonuclease and methyltransferase denoted by ρ and μ, respectively. The effective activity of an enzyme is the product of the number of molecules of the enzyme and its single-molecule activity. The effective activities ρ and μ can be time-dependent, constant, or randomly depending on the number of enzyme molecules per cell. In the next section we provide details on the concept of effective activity. We assume that the phage is restricted (or modified) before the replication commences. Our first goal is to obtain probabilities of survival or death for a single bacterium, and, simultaneously, the probabilities of productive or abortive infection for a single phage. We start by modelling our system for the case of a single bacterium invaded by a single phage assuming time-dependent activities ρ(t) and μ(t). Then we generalize our results to the case of a bacterial culture invaded by multiple identical phages. We assume that the number of phages infecting a single cell is Poisson-distributed. The distribution of the number of R and M molecules per cell is assumed to be Poisson and the single-molecule activities are assumed to be constant. We do not consider conversion to the lysogenic state that is modeled, e.g. in (Avlund et al., 2009). We also do not model the spatial distribution of susceptible and restricting colonies, or colonies possessing different R-M systems (Gregory et al., 2010).
2.1. Mathematical Model
The process of infection of a bacterial cell is modelled by a pure birth process with killing (see, for example, Karlin and Tavaré (1982); van Doorn and Zeifman (2005); Coolen-Schrijner et al. (2006) for some general results on this type of processes). We calculate the stationary distribution for the process for a general situation of time-dependent enzyme activities.
Let R(t) be a continuous time Markov process with N + 1 states i = 0, … , N and a so-called ”killing state” −1. The system is at the state i if exactly i restriction sites of the phage DNA are methylated. Assume that effective activities of the methyltransferase and the restriction endonuclease in a bacterial cell are time-dependent functions μ(t) and ρ(t), respectively.
We suppose that at any state i the methyltransferase and the endonuclease select a site to be processed (methylated or cut) with probability 1 − i/N. Thus, at the state 0 the next site will be methylated/cut with the probability 1. In fact, the enzyme molecules select an unmethylated site with probability 1 − i/N if i sites are already methylated. We assume that the enzyme molecules cannot select the same site simultaneously. We also assume that a methylated site cannot be selected by the methyltransferase again.
If all N sites are methylated, the phage survives and the bacterium dies. In this case the Markov chain hits the absorbing state N. If the restriction endonuclease encounters an unmethylated site, the phage dies and the Markov chain hits the “no-phage state” −1 meaning that the bacterium has survived the phage invasion.
Let μi(t) = (1 − i/N)μ(t), ρi(t) = (1 − i/N)ρ(t). In fact, µi(t) is the transition rate from the state i to the state i + 1 at the time t; ρi(t) is the transition rate to the state −1 from the state i at the time t. Roughly speaking, µi(t)h is the probability of methylating a site in the phage genome during an infinitely small time interval h → 0 if exactly i sites are methylated at the time t, and ρi(t)h is the probability of cutting a site during an infinitely small time interval h → 0 if exactly i sites of the phage are methylated at the time t.
Let Pk(t) = P{R(t) = k} be the probability that k sites are methylated at the time t. Applying the theory of birth-and-death processes (Karlin and McGregor, 1957; Feller, 1968) we obtain the following system of differential equations
| (1) |
with the equations for the absorbing states
where the initial conditions are P0(0) = 1, Pk(0) = 0, k ≠ 0.
2.2. Stationary Distribution
Solving the system of the differential equations (see Appendix), we get the stationary distribution of the process R(t),
where .
3. Estimating the ratio of R-M enzyme activities
Recall that the effective activity is defined as a product of a single enzyme molecule activity and the number of enzyme molecules per cell. Denote by NM and NR the number of molecules of methyltransferase and restriction endonuclease in a cell, respectively. Let aM and aR be single-molecule activities of methyltransferase and restriction endonuclease, correspondingly. Then the corresponding effective activities are given by μ = aMNM and ρ = aRNR. In this section we consider the cases of constant or random effective activities. First, we consider a situation when a single cell with constant effective activities is infected by a single phage. Then we generalize it to the case of multiple cells with constant activities that are infected by a Poisson-distributed number of phages. Finally, we consider the case in which the number of phages is Poisson-random as before, but the numbers of enzyme molecules are not constant but Poisson-random. Our goal is to estimate the ratio of single-molecule activities τ = aR/aM and the ratio of effective activities ρ/μ.
3.1. Constant activities
Consider first an imaginary scenario of a single cell being infected by a single phage. Assume that the R-M enzyme effective activities μ and ρ are constant, µ(t) ≡ μ and ρ(t) ≡ ρ. This means that the number of the enzyme molecules in a cell does not depend on time. We have . The probabilities to hit absorbing states given that R(0) = 0 are
This result has a clear intuitive explanation. The situation with constant effective activities can be modelled by a series of Bernoulli experiments. The outcome of each experiment is either methylating or cutting with probabilities μ/(μ + ρ) and ρ/(μ + ρ), respectively. Thus, the probability of the phage survival is equal to the probability of methylating exactly N sites, (μ/(μ + ρ))N. This implies the first formula. The second formula follows from the first one as its complement with respect to one. The general result for time-dependent activities can be also explained in such a way if we take into account that the probability of exactly one site to be methylated during time t equals .
Imagine that we repeat our experiment of single-cell infection by a single phage n times. Denote by ϕ the probability of phage survival, . Denote by the number of infected bacteria that are killed by a phage with N restriction sites in a series of n experiments. In fact, in the case of an infection of a single bacteria by a single phage this number is exactly the same as the number of surviving phages. From the observed average number of surviving phages (killed cells) we can estimate the ratio of R-M enzyme activities τ = aR/aM. Indeed, by the law of large numbers, for large n, tends to ϕ. Recall the definitions of effective activities, μ = aMNM and ρ = aRNR. Then the probability of phage survival is given by
Using this formula it is easy to obtain an estimator of the ratio of activities τ̂,
Similarly, the ratio of the effective activities ρ/μ is estimated as
Further we will always denote an estimate of a parameter by a hat.
3.2. Random number of phages in a cell
Consider now the situation of K bacterial cells infected by V phages. The numbers of cells and phages are large, K, V → ∞, and V/K → Λ as K, V → ∞, where Λ is the average number of phages per cell. All phages and all cells, respectively, are assumed to be identical in a sense that the phages have the same number of restriction sites N and the enzymes have the same constant effective activities. We assume that the activities of the methyltransferase, μ = aMNM, and the restriction endonuclease, ρ = aRNR, are constant, and the effective activities depend on the numbers of molecules of each enzyme, NM and NR, respectively, in the cells. Denote by ψ the probability of phage death, , obviously, ψ = 1 − ϕ.
The distribution of phages between the cells satisfies the Bose-Einstein statistics with the number of possible variants . Let qj be the probability that there are exactly j phages in a bacterium. Then
It is known (Feller, 1968) that for V/K → Λ, K → ∞, V → ∞, this probability converges to the geometric distribution,
Note that for a sufficiently small Λ this distribution can be approximated by the Poisson distribution with the mean Λ.
In practice, not every phage may manage to infect. In this case the real value of Λ will differ from the simple ratio V/K. To estimate the effective number of phages per cell Λe, we can calculate the fraction of survived cells q͂0 for a phage with zero restriction sites. Of course, in this case only uninfected cells will survive. Assuming that q͂0 converges to (Λe + 1)−1 as V, K → ∞ we can estimate the effective number of phages per cell Λe. Inverting the approximate formula for q͂0, we obtain Λe = 1/q͂0 − 1. Hereinafter we assume that the number of phages is geometrically distributed between the cells with mean Λe.
We assume that the restriction events in a cell are independent. Thus, the probability of survival of a single cell infected by j phages is ψj (all j phages must be restricted, i.e., their DNA cut at least once). Then the probability that a single bacterial cell survives equals . Thus, as K, V → ∞, we have
Let ν ≡ νK be the observed fraction of survived bacterial cells over K cells. Using the obtained formula for the probability of single cell survival S, we can estimate the ratio of activities τ. Indeed, since , we can write
By the law of large numbers the fraction of survived bacteria ν ≡ νK converges to SV as K → ∞. Using the above limit for SV we can estimate τ as
Note that τ̂ is negative for ν < (Λe + 1)−1 and is undefined for ν = 1. The probability that ν < (Λe + 1)−1 tends to zero as K → ∞, since the probability q0 that a cell is not infected by any phage converges to (Λe + 1)−1 as V,K → ∞. Thus, the fraction of survived bacteria will be greater than (Λe + 1)−1 with probability tending to 1. The case ν = 1 corresponds to the situation when the restriction endonuclease is much more active that the methyltransferase, so that τ → ∞.
Thus, we can rewrite the estimate as
| (2) |
Here I{a ≤ X ≤ b} denotes the indicator of the set {a ≤ X ≤ b} such that I{a ≤ X ≤ b} = 1, if a ≤ X ≤ b and I{a ≤ X ≤ b} = 0, otherwise. It yields an estimate for the ratio of effective activities,
Let us comment about the performance of τ̂. The estimator τ̂ is asymptotically normal as K → ∞ with the asymptotic mean-square risk
It is not difficult to obtain this result using standard techniques of the estimation theory (see, for example, Borovkov (1998)). Thus, the performance of our estimate depends on N, Λe and τ. In particular, for large τ, τ → ∞,
Obviously, in this case the estimator performs worse for larger values of N. On the other hand, for small values of τ, τ → 0, we have
In this case the accuracy of estimation increases for larger N.
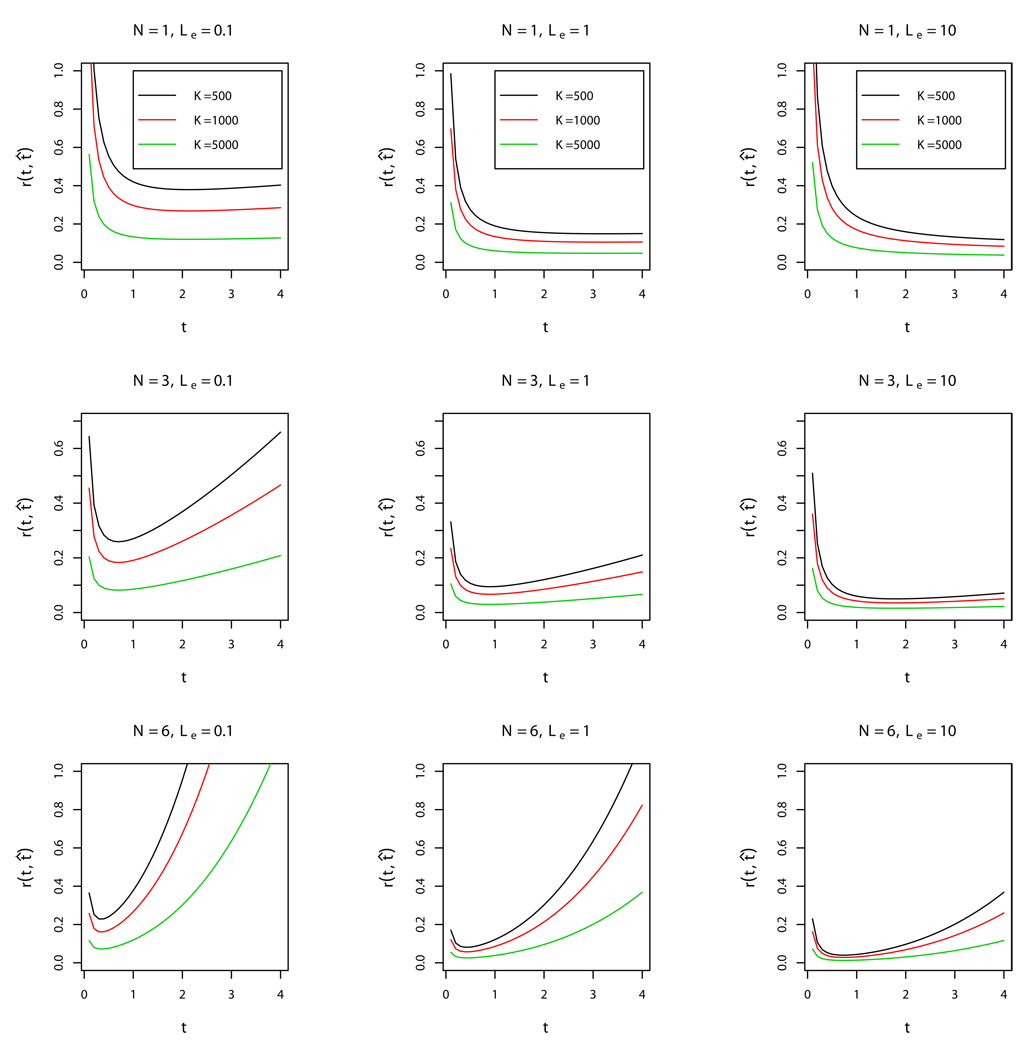
Figure 1 presents the plots of the dependence of relative mean-square error on τ,
for different values of N, Λe, and K. We see that if 0 < τ < 1, τ̂ is more efficient for larger values of N. On the other hand, if τ is large, τ̂ is more efficient for smaller values of N. Efficiency of τ̂ increases as Λe and K increase. We should note that on practice the numbers of phages and infected bacterial cells are of order 105 and larger. Simulations show that the approximation by geometric distribution works fine for K, V ≥ 103 and that the theoretical risk of estimation is very close to the empirical risk. In any case, on practice, we are interested in a large number of bacteria K (of order 105 and above) infected by a relatively large number of phages.
Figure 1.
Dependence of the relative mean-square error r(τ̂, τ) = τ−1Eτ(τ̂, τ) on the ratio of single molecule activities, τ. The graphs are presented for different values of Λe = 0.1, 1, 10 and for the number of sites N = 1, 3, 6. Each subfigure contains three graphs of the relative mean-square error for the number of cells K = 500 (black line), K = 1000 (red line), and K = 5000 (green line).
3.3. Random activities and random number of phages
Let a bacterial culture be infected so that the number of phages V is much smaller than the number of bacteria K in the culture, V ≪ K. Then we may assume that each bacterial cell will be infected by a small number of phages (zero, one or two) such that the probability of infecting a cell with k phages pk is the Poisson distribution with mean Λe,
Here Λe < Λ ≡ V/K.
A cell is not infected with the probability p0 = e−Λe, this is due to the possibility for a phage to get into the intercellular space. Observing the number of killed bacterial cells which is equivalent to the number of colonies formed by surviving phages, we would like to estimate the ratio of R-M enzymes activities τ = aR/aM, where the numbers of molecules of both enzymes are supposed to be random as well as the number of phages in a cell. Denote by the total number of bacterial cells killed by phage with N restriction sites that infected K bacterial cells.
Note that in the case of infection by a phage without restriction sites (N = 0) all phages survive. Thus, in this case the average number of killed bacterial cells (survived phage) is equal to 1 − p0. It means that we can estimate the value of Λe by making an experiment with a phage without restriction sites. We have
and, consequently, we can estimate Λe by .
Let the numbers of molecules of methyltransferase and endonuclease in the i-th cell, and , respectively, be Poisson-distributed (Golding et al., 2005). We assume that all cells are identical in a sense that the number of enzyme molecules per cell has the same Poisson distribution for each cell. Denote for brevity by NM ~ Π(λM) and by NR ~ Π(λR), where λM and λR are the average numbers of molecules of methyltransferase and restriction endonuclease per cell, respectively. Then the probability of a phage survival ϕ(NM, NR) given NM and NR molecules of enzymes in a cell is given by
Note that for NR = 0 we have ϕ = 1, since in this case a cell does not contain molecules of restriction endonuclease and all phages infecting this cell obviously survive. By the same reason we set ϕ = 1 for NM = 0, NR = 0.
Let us now calculate the expected value of the number of killed bacterial cells , where N stands for the number of restriction sites in a phage and K is the total number of infected cells. A cell survives if all phages infected the cell die. It means that a cell dies if at least one phage infected this cell survives. Therefore, if a cell is infected by k phages, the probability of killing the cell is equal to 1 − (1 − ϕ)k. Here the probability of phage survival ϕ depends on the (random Poisson) number of enzyme molecules in this cell. Next, we have to average this probability with respect to the number of enzyme molecules per cell and with respect to the Poisson number of phages per cell and obtain the following average probability of a cell death,
Here EM,R denotes the mean over all possible values of NM and NR and pk is the probability that there are exactly k phage in a cell. Since we have K cells, we have to multiply the above average probability by K to obtain the average number of killed bacterial cells,
The precise formula for in terms of λM and λR is rather complicated (see Appendix). To make our computations easier we assume that a cell can be infected by at most two phages (pk=0 for k > 2). This assumption makes sence if, for example, Λe ≤ 0.1. We have
where
is the average fraction of survived phages given the number of molecules of methyltransferase NM is not equal to zero. Further details on the approximation of Eϕ and are given in Appendix.
Let us introduce the ratio of the average numbers of enzyme molecules per cell,
Obviously, the behavior of will be different for different values of the average activities with the same ratio α.
Consider two important cases. In the first case, λM and λR are large such that λM, λR ≥ 10. In this case we can use the approximate formula for (see Appendix),
| (3) |
The second case is when λM and λR are small, λM, λR → 0 as K → ∞, for example, λM, λR < 1. In this case the behavior of is controlled by the behavior of the term 1 − p0, since
| (4) |
In the case of moderate values of λM, λR as 1 ≤ λM, λR ≤ 5 the approximate formula for will depend not only on the ratio α = λR/λM but also on λM,
Using approximation (3) we can estimate the parameter τ for λR, λM > 5. Indeed, if is the observed number of killed bacterial cells invaded by phages with N restriction sites, then
| (5) |
and the ratio of average effective activities can be estimated as
The condition is obtained by the same reasoning as in Section 3.2. Indeed, for the estimate (5) is undefined (it approaches infinity as ). For the estimate is negative which can happen with a very small probability for sufficiently large K.
If we know λM, we can estimate τ for moderate values 1 < λR, λM ≤ 5 as
| (6) |
Estimate (5) can be compared with the estimate obtained in Section 3.2 for the case of cells with constant activities invaded by a random number of phages. Indeed, we can rewrite the estimate from (2) in terms of the number of killed bacterial cells . We have
Subscript C in τC stands for the case of constant activities. This estimate and the one from (5) are very similar. Indeed, for small Λe we have . Also, for small Λe the average number of killed cells is very small which explains .
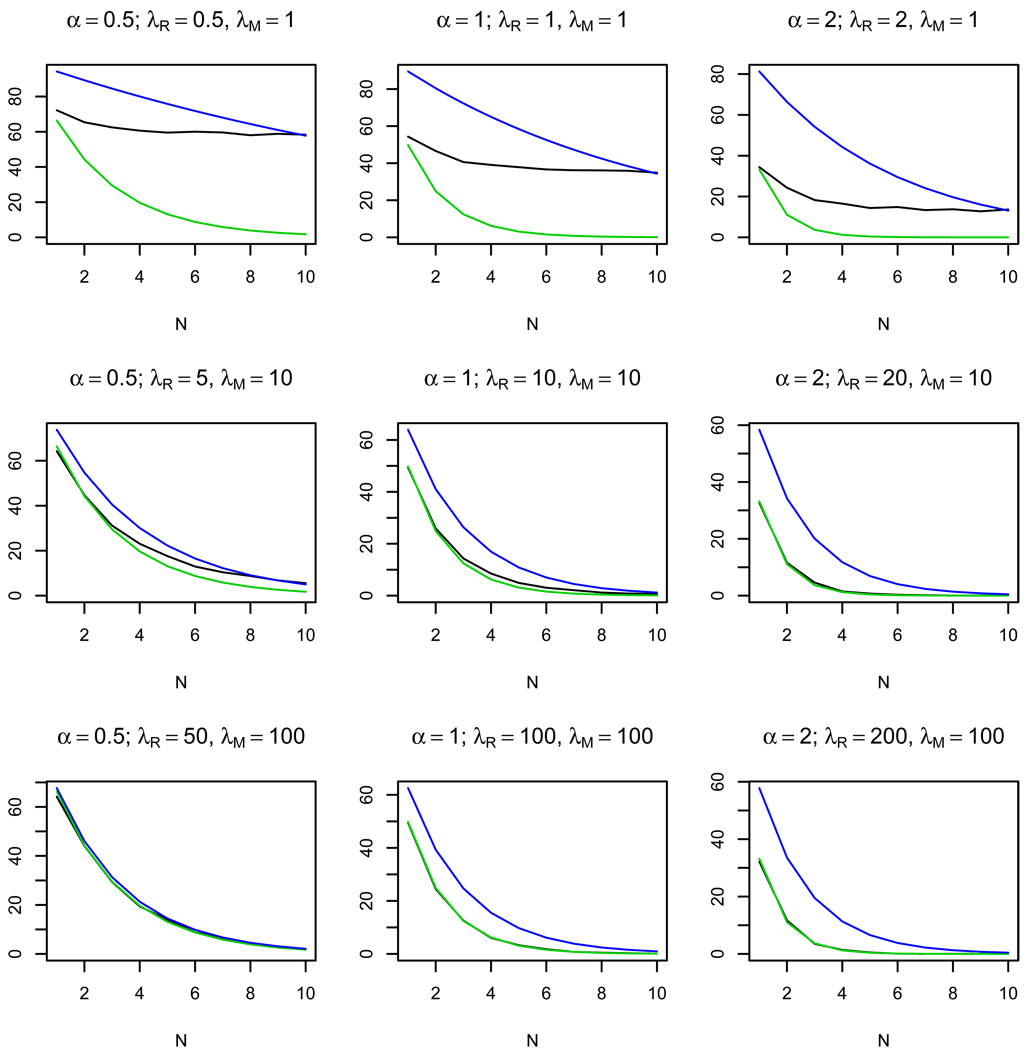
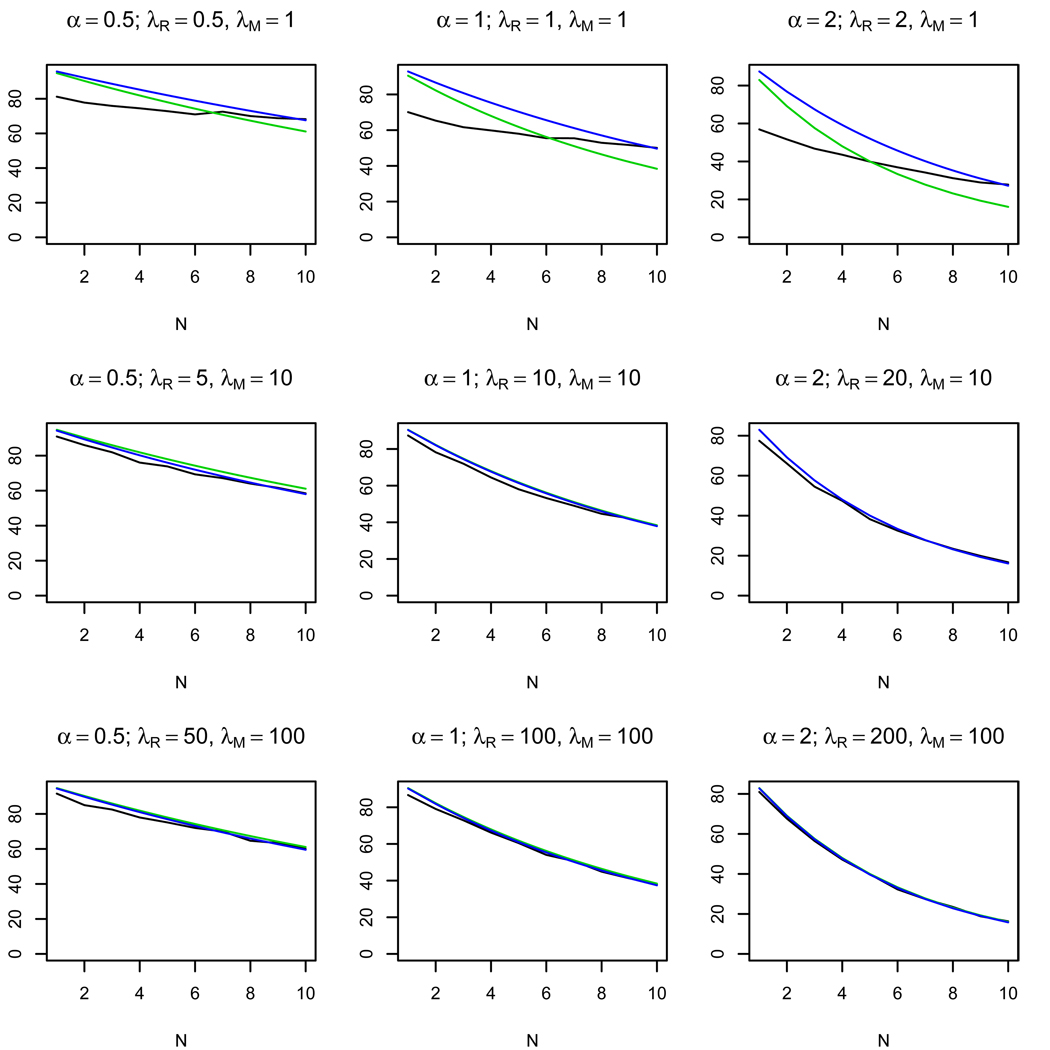
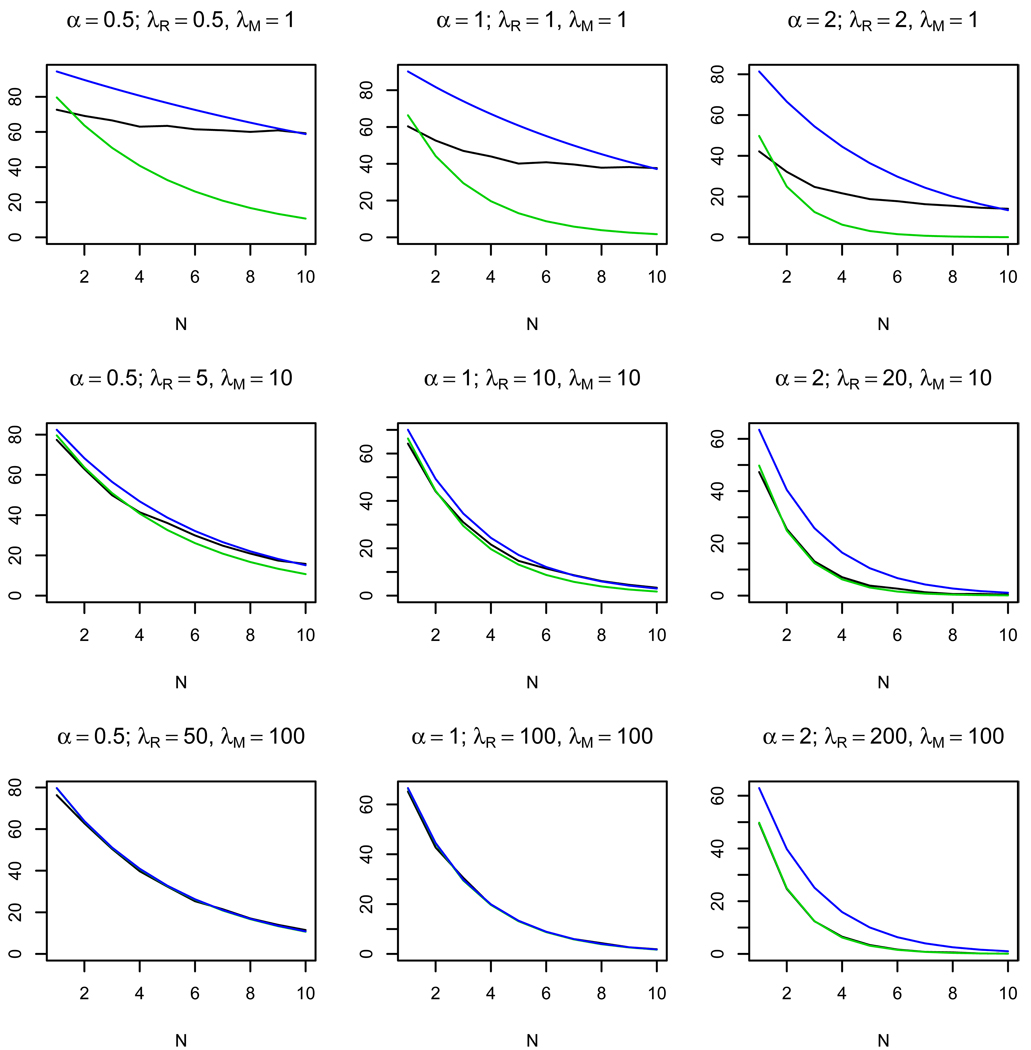
Figures 2–4 show the results of 1000 simulations for K = 103 bacterial cells with the average number of phages per cell Λe = 0.1 and the ratio of activities τ = 1, 0.5, 0.1, respectively. The plots show how the number of restriction sites N influences the observed average number of killed bacteria and of the approximate value of given by formula (3). We also present an estimate of given by the following formula
Figure 2.
Plots of the observed average number of killed cells (black line), the approximate average number of killed cells (green line), and the estimate of the average number of killed cells (blue line) depending on the number of restriction sites N. 103 simulations were made for τ = 1 for K = 103 bacterial cells. The approximate formula (3) works well for large λM and λR and small values of α = λR/λM.
Figure 4.
Plots of the observed average number of killed cells (black line), the approximate average number of killed cells (green line), and the estimate of the average number of killed cells (blue line) depending on the number of restriction sites N. 103 simulations were made for τ = 0.1 for K = 103 bacterial cells. The approximate formula (3) works well for λM, λR ≥ 5.
This formula is obtained by substituting the estimate τ̂ (5) into (3). The plots show that for the same ratio of the average number of enzyme molecules α the approximate formula works well for λM, λR > 5. The quality of the approximation increases as τ decreases (see Fig. 3, 4). On the other hand, for small λR, λM the approximation is bad for all values of τ, which allows us to distinguish between the cases of small and large average numbers of enzyme molecules per cell.
Figure 3.
Plots of the observed average number of killed cells (black line), the approximate average number of killed cells (green line), and the estimate of the average number of killed cells (blue line) depending on the number of restriction sites N. 103 simulations were made for τ = 0.5 for K = 103 bacterial cells. The approximate formula (3) works well for λM, λR ≥ 5 and α = λR/λM ≤ 1.
4. Discussion
In this work, we proposed a mathematical model for the process of infection of a bacterial cell harboring an R-M system with a phage. The model provides an estimate for the ratio of average effective activities τ · (λR/λM) of the methyltransferase and the restriction endonuclease, based on the number of killed bacterial cells observed in experiments.
Numerical simulations (Fig. 2–4) show that the quality of approximation is essentially better for small values of the ratio of single-molecule activities τ and also that the approximation is very bad for small average number of enzyme molecules per cell λM and λR. It allows us to distinguish between the case of the large number of enzymes per cell and the case of the small number of enzymes per cell (see Figures 2–4, the plots for the same ratio α = λR/λM and different values of λR and λM).
To validate the model, a series of experiments should be done with identical phages having different numbers of restriction sites N = 0, 1, 2, … , 10. The experiment with N = 0 allows one to estimate the effective mean number of phages per cell Λe. For N = 1, 2, … one needs to measure the average number of killed bacterial cells in each series and to check whether this number depends exponentially on the number of sites N according to the obtained formula . If the graphs and are close to each other, it means that the approximation works well and the average numbers of enzymes per cell λM, λR are large. In this case we can estimate τ̂ using formula (5) and, correspondingly, obtain an estimate . On the other hand, if the graphs are far from each other, we have the case of small λR and λM. In this case we cannot estimate τ well using estimate (5). To construct a good estimate we have to use formula (6), where λM is unknown. Hence in this case, in the absence of additional data, we cannot say anything except that λM and λR are small.
The efficiency of plating of phage lambda containing two or three recognition EcoRI sites on a restricting host was estimated in Rambach and Tiollais (1974). By design, this experiment was conducted at conditions of large excess of cells over the phage (Λe in our notation). The results indicated that an extra site increased the plating efficiency by an order of magnitude (from 4×10−2 to 5×10−3). This is roughly consistent with the predicted dependence of the probability of successful infection on the number of sites, assuming the plating efficiency close to 1 for a phage with no sites.
One can imagine at least two possible non-overlapping mechanisms of overcoming the protection afforded by an R-M system by an infecting phage (we assume that the phage lacks specific antirestriction mechanisms of the type described in Tock and Dryden (2005)). First, it is possible that a small proportion of cells has no restriction endonuclease (or, conversely, a very large amount of methyl-tranferase) at the time of infection (Mruk and Blumenthal, 2008). The variation in the amount of restriction-modification enzymes in the cell can be the result of a stochastic noise in the levels of R-M gene expression, unequal partitioning of the R-M gene products to daughter cells etc. In this scenario, the proportion of cells that are susceptible to infection should remain constant and should not depend on the multiplicity of infection (i.e., the average number of phage particle infecting each cell).
An alternative scenario may involve fluctuations in the number of phages infecting bacterial cells at a particular multiplicity of infection. One can imagine that restriction endonuclease in cells receiving more than the average number of phage becomes “overwhelmed” allowing productive infection to occur. In this scenario, the proportion of cells that become productively infected should increase together with multiplicity of infection. The second scenario is suggested by the fact that the number of productive infections of restricting cells with a non-modified phage lambda increases when the cells are “preinfected” at high multiplicity with another non-modified phage immediately prior to infection with the first phage (Heip et al., 1974). Since our model explicitly involves different Poisson distributions for the activities of the methyltransferase and endonuclease, and for the number of phage invading a cell, it allows one in principle to distinguish between these scenarios, which may be different for different R-M systems and, possibly, phages.
Our analysis and numerical simulations show that both the best power to discriminate between the continuous model (assuming large amounts of the endonuclease and methyltransferase) and the discrete model (few molecules), and the more robust estimates of the ratio of activities is provided by the systems when the phage has a small number of sites for the R-M system. To compare the models and to estimate the ratio, different types of experiments may be suggested, including changing the number of sites by point mutagenesis, changing the expression rate of the R-M operon (retaining the ratio of activities, but changing the number of molecules), changing the activity of the endonuclease by point mutagenesis, changing the expression rate of either endonuclease or methyltransferase by inroducing additional gene copies in separate operons, etc. Having calculated the number of successful infections, and knowing the number of sites, one can then estimate the degree of fit to either model, and obtain an estimate for the ratio of activities of the endonuclease and the methyltransferase.
Acknowledgements
KVS and MSG conceived the study. FNE and MSG developed the model. FNE performed numerical simulations. FNE, KVS, and MSG wrote the paper. All authors have read and approved the final version.
This study was partially supported by grants from the Russian Foundation for Basic Research (09-04-01098, FNE), the Howard Hughes Medical Institute (55005610, MSG), and the Russian Academy of Sciences (programs “Molecular and Cellular Biology”, MSG, KVS and “Genetics Diversity”, FNE, MSG), and the Russian Science Agency under contract 2.740.11.0101.
The authors thank anonymous referees for their constructive comments that helped to improve the paper.
Numerical simulations and figures were made using R.
6. Appendix
6.1. Transitional probabilities
The first equation of system (1) can be easily solved, . Solving recursively the next N equations gives the solutions for k = 0, … , N,
where . The function 1 − G is the distribution of time between two consequent states of R(t). Now, the probability of the phage death can be calculated,
The stationary distribution of the process X(t) is given by
Note that for constant effective activities µ(t) ≡ µ and ρ(t) ≡ ρ we have and the solution to the system for k = 0, … , N is given by
For k = −1 we have
6.2. Average number of killed bacterial cells for random activities
In this section we will estimate the average number of killed bacterial cells
Here are the Poisson probabilities of the number of phages in a cell. Since NM and NR are Poisson distributed Π(λm), Π(λr), respectively, we have the following precise formula
This distribution is non-lattice, which is considerably more difficult to handle than a lattice distribution. Our goal is to obtain an approximate formula for .
We first consider the behavior of the above formula for two boundary cases, τ → 0 and τ → ∞.
In the first case, when τ is small and aR ≪ aM, the restriction endonuclease has much smaller activity, than the methyltransferase. The formula for the average number of killed bacterial cells turns to
We can easily interpret this formula. Here e−λM is the probability that there are no molecules of methyltransferase in a cell and 1−e−λR is the probability that the cell contains molecules of restriction endonuclease. Roughly speaking, in the case of small τ, phages survive if the average number of molecules of methyltransferase λM is large or the average number of molecules of restriction endonuclease λR is small. In fact, a cell survives only if there is no methyltransferase and there is at least one molecule of restriction endonuclease.
In the second case, when τ is very large and aR ≫ aM, the formula for the average number of surviving phages becomes
It means that for the large ratio of activities τ the phages survive only if the amount of restriction endonuclease is small independently of the amount of methyltransferase.
Let us now find the approximate formula for . For simplicity we will consider only the case when the probabilities pk, k ≥ 3 that there are more than two phages in a cell are very small. For example, for Λe = 0.1 the probability that there are 3 phages in a cell is p3 = 0.0001508062. We assume pk = 0, k ≥ 3. Then our formula transforms into
| (7) |
where
Here NR ~ Π(λR), NM ~ Π(λM). We can find the approximation for Eϕ using the method of propagation of error. Define the following random variable
We have
The approximation for is calculated using the method of propagation of error,
Next, we can find the approximation for the mean EX,
Using the same method again we obtain the following approximate formula for Eϕ:
Here we use just the first two terms of the Taylor series expansion around EX to approximate Eϕ. The quality of approximation would be better if we used the terms of second and higher orders, but in this case it would be harder to derive an estimator for τ. The approximation works well for 0 < τ < 1. Since the second order term depends on τ2, the formula will work worse for large τ, however, in the vicinity of τ = 1 it is sufficiently precise.
Finally, we have the following approximate formula
We omit the term 2p2Eϕ2 in formula (7), since its contribution to the total value of is very small compared to the contribution of the first two terms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Farida N. Enikeeva, Email: enikeeva@iitp.ru.
Konstantin V. Severinov, Email: severik@waksman.rutgers.edu.
Mikhail S. Gelfand, Email: gelfand@iitp.ru.
References
- Arber W. Promotion and limitation of genetic exchange. Nobel lecture in Physiology and Medicine. 1978 [Google Scholar]
- Avlund M, Dodd IB, Semsey S, Sneppen K, Krishna S. Why do phage play dice? Journal of Virology. 2009;83:11416–11420. doi: 10.1128/JVI.01057-09. Ref. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. J. Bacteriol. 1953;65:113. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovkov AA. Gordon and Breach. Amsterdam: 1998. Mathematical Statistics. [Google Scholar]
- Coolen-Schrijner P, van Doorn E, Zeifman A. Quasi-stationary distributions for birth-death processes with killing. J. of Appl. Math. and Stoch. Analysis. 2006:1–15. [Google Scholar]
- Feller W. An Introduction to Probability Theory and Its Applications. Vol. 1. New York: Wiley; 1968. [Google Scholar]
- Golding I, Paulsson J, Zawilski S, Cox E. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Gregory R, Saunders VA, Saunders JR. Rule-based simulation of temperate bacteriophage infection: Restriction-modification as a limiter to infection in bacterial populations. 2010 doi: 10.1016/j.biosystems.2010.02.010. In press, doi:10.1016/j.biosystems.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Heip J, Rolfe B, Schell J. Abolition of host restriction by high multiplicity of phage infection. Virology. 1974;59:356–370. doi: 10.1016/0042-6822(74)90450-4. [DOI] [PubMed] [Google Scholar]
- Karlin S, McGregor JL. The differential equations of birth-and-death processes, and the stieltjes moment problem. Trans. Amer. Math. Soc. 1957;85:589–646. [Google Scholar]
- Karlin S, Tavaré S. Linear birth and death processes with killing. J. Appl. Prob. 1982;19:477–487. [Google Scholar]
- Luria SE, Human ML. A nonhereditary, host-induced variation of bacteria viruses. J. Bacteriol. 1952;64:557. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I, Blumenthal M. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucl. Acids Res. 2008:1–13. doi: 10.1093/nar/gkn097. Doi:10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambach A, Tiollais P. Bacteriophage i having ecori endonuclease sites only in nonessential region of the genome. Proc. Natl. Acad. Sci. USA. 1974;71:3927–3930. doi: 10.1073/pnas.71.10.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REBASE. The restriction enzyme database. 2010 URL http://rebase.neb.com. [Google Scholar]
- Tock MR, Dryden DTF. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- van Doorn E, Zeifman A. Birth-death processes with killing. Stat. and Prob. Letters. 2005;72:33–42. [Google Scholar]